Introduction
Myelodysplastic syndromes (MDS) are clonal stem cell
disorders characterized by dysplastic changes in multiple
hematopoietic lineages and ineffective hematopoiesis, which lead to
acute myeloid leukemia (AML). Multiple factors have been implicated
in the pathogenesis of MDS, including cytogenetic changes and
molecular abnormalities, such as gene mutations and epigenetic
changes, as well as disturbances in cellular immunity and
microenvironment (1,2). Disorder of the immune system serves an
important function in the pathophysiology of MDS, and expansion of
different T cell subpopulations may occur at distinct disease
stages, suggesting that progression of MDS may be facilitated by
immune suppression (3,4).
Natural killer (NK) cells are large granular
lymphocytes that function as a component of the innate immune
defense system. The functions of NK cells depend on the absolute
sum of their simultaneous activation and inhibition signals. For
example, a cluster of differentiation (CD)16-mediated activation
signal may lead to antibody-dependent cellular cytotoxicity (ADCC)
by degranulation- and perforin-dependent target cell lysis, and
this NK-mediated ADCC is a dominant component of effective
antitumor activity (5). Different
levels or mechanisms of NK cells in patients with MDS have been
measured in previous studies using different approaches to analyze
the NK cells, making it challenging to understand the pathogenesis
of NK cytotoxicity (6–8). Therefore, the present study investigated
populations of NK cells and examined their functions by activating
receptors, inhibition signals and cytotoxicity factors in patients
with MDS to determine the function of NK cells as immunological
determinants in MDS.
Patients and methods
Patients and controls
Peripheral blood samples were obtained from 35
patients with MDS, 16 patients with AML and 22 healthy donors
referred to the Department of Hematology at General Hospital of
Tianjin Medical University (Tianjin, China) from June 2012 to
September 2017, following the provision of written informed consent
in accordance with the Declaration of Helsinki. The present study
was approved by the Tianjin Medical University Institutional Review
Board (Tianjin, China). The median age of the patients with MDS was
71 years (range, 40–83 years), and 18 were male and 17 were female.
The median age of the patients with AML was 56 years (range, 30–69
years), and 9 were male and 7 were female. The median age of the
healthy donors was 30 years (range, 23–60 years), and 12 of them
were male and 10 were female. According to World Health
Organization criteria (9), the
patients were classified as refractory anemia, refractory anemia
with ring sideroblasts, refractory cytopenias with multi-lineage
dysplasia or refractory anemia with excess blasts. Based on the
International Prognostic Scoring System (IPSS) the patients were
classified in distinct categories as low, intermediate and high
risk (10). The characteristics of
the patients are presented in Table
I.
 | Table I.Characteristics of the patients with
MDS. |
Table I.
Characteristics of the patients with
MDS.
| Characteristic | n |
|---|
| Sex |
|
|
Male | 18 |
|
Female | 17 |
| WHO subtypes |
|
|
Refractory anemia | 8 |
|
Refractory anemia with ring
sideroblasts | 5 |
|
Refractory cytopenias with
multi-lineage dysplasia | 12 |
|
Refractory anemia with excess
blasts | 10 |
| IPSS |
|
|
Low | 7 |
|
Intermediate 1 | 8 |
|
Intermediate 2 | 13 |
|
High | 7 |
Measurement of NK cells and NK-like T
(NKT) cells from the peripheral blood
NK cells
(CD3−CD56+/CD16+) and NKT cells
(CD56+CD3+) from fresh samples were
identified by single-platform flow cytometric analysis. The NK cell
marker antibodies included in the analysis were phycoerythrin
(PE)-conjugates of anti-CD158a (cat. no. 556063; 1:10), anti-CD158b
(cat. no. 559785; 1:10), anti-NKG2D (cat. no. 561815; 1:10),
anti-NKp44 (cat. no. 558563; 1:10) and anti-CD 226 (cat. no.
559789; 1:10), as well as CD56-allophycocyanin (cat. no. 555518;
1:10), CD16-fluorescein isothiocyanate (cat. no. 555406; 1:10) and
CD3-peridinin chlorophyll protein complex (cat. no. 552851; 1:10),
all of which were obtained from BD Biosciences (Franklin Lakes or
San Jose, USA). In a volume of 100 µl, whole blood was
immunostained with 10 µl aforementioned antibodies at 4°C for 30
min, and red blood cells in the sample were lysed with 1 ml 1X
FACS™ lysing solution (BD Biosciences) for 8 min at room
temperature. Then the red blood cell samples were centrifuged at
500 × g for 5 min at room temperature and decant the supernatant.
The peripheral blood mononuclear cells (PBMNCs) left were washed
twice with PBS prior to examination. A minimum of 50,000 cells were
examined by flow cytometry per sample. Each specimen was
investigated using a FACS-Calibur (BD Biosciences) or a DxFLEX flow
cytometer (Beckman Coulter, Inc., Brea, CA, USA) and was analyzed
using the CellQuest 3.1 software (BD Biosciences) or the CytExpert
1.0 software (Beckman Coulter, Inc.), respectively.
Measurement of the function of NK
cells
NK cells from fresh samples were labeled as
aforementioned. The NK cell receptors included in the analysis were
phycoerythrin (PE)-conjugates of anti-CD158a (cat. no. 556063;
1:10), anti-CD158b (cat. no. 559785; 1:10), anti-NKG2D (cat. no.
561815; 1:10), anti-NKp44 (cat. no. 558563; 1:10) and anti-CD 226
(cat. no. 559789; 1:10), all of which were obtained from BD
Biosciences (San Jose, USA). PE-conjugated anti-CD107a (cat. no.
130095515; 1:10) was obtained from Miltenyi Biotech (Bergisch
Gladbach, Germany). The inhibitory and activating receptors on the
cell membranes were stained with 10 µl appropriate monoclonal
antibodies (mAb) in a 100 µl volume at 4°C for 30 min. Washed twice
with PBS before examination. In order to analyze the cytoplasmic
proteins, perforin and granzyme B, the cells were mixed with 1.0 ml
1X FACS™ permeabilizing solution 2 (BD Biosciences) for 10 min. The
suspensions were washed with PBS containing 0.1% sodium azide prior
to being stained with the perforin or granzyme B mAbs at room
temperature for 30 min in the dark. Both washed twice with PBS
before examination. PE-Perforin (cat. 556437) and PE-Granzyme B
(cat. 561142) were obtained from BD Biosciences (San Jose, USA).
The inhibitory receptors were identified as CD158a and CD158b
(11), and the activating receptors
were identified as NKG2D, DNAM-1 and NKp44 (12,13). The
CD107a expression was measured in CD16+ and
CD16− NK cells. Stained cells were also analyzed using a
flow cytometer and associated software as aforementioned.
Isolation and purification of target
NK lymphocytes
A total of 10 ml fresh human peripheral blood was
obtained from patients with MDS and from normal controls. PBMNCs
were isolated by density gradient centrifugation using Ficoll-Paque
Plus solution (GE Healthcare, Chicago, USA) at 400 × g for 20 min
at room temperature. The purified NK cells were subsequently
obtained using an NK Cell Isolation kit (Miltenyi Biotech, Bergisch
Gladbach, Germany). Isolated PBMNCs were mixed with NK Cell
Biotin-Antibody Cocktail from the kit (cat. no. 130092657) and
incubated for 5 min at 4°C followed by NK Cell MicroBead Cocktail
from the kit (cat. no. 130092657) and subsequent magnetic cell
separation. Following detection using a multi-parametric flow
cytometer (BD Biosciences) and analysis using Cell Quest software
(version 3.1; BD Biosciences), the purity of
CD3−CD56+ NK cells was 90–99%.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA in previously purified NK cells was
extracted using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, USA) and 1 µg RNA was reversed
transcribed using a TIANScript RT kit (Tiangen Biotech Co., Ltd.,
Beijing, China). The thermocycler conditions were; 50 min at 42°C,
followed by 5 min at 95°C. The sequences of primers specific for
perforin, granzyme B, NKG2D, NKP44 and β-actin, which were designed
and synthesized by Sangon Biotech Co., Ltd. (Shanghai, China), are
listed in Table II.
 | Table II.Primers used for reverse
transcription-quantitative polymerase chain reaction. |
Table II.
Primers used for reverse
transcription-quantitative polymerase chain reaction.
| Target gene | Primer
sequences |
|---|
| Perforin |
|
|
Forward |
5′-GAGGAGAAGAAGAAGAAGCACAA-3′ |
|
Reverse |
5′-AGGGGTTCCAGGGTGTAGTC-3′ |
| Granzyme B |
|
|
Forward |
5′-CCAGCAGTTTATCCCTGTGAA-3′ |
|
Reverse |
5′-CACCTCTTGTAGTGTGTGTGAGTG-3′ |
| NKP44 |
|
|
Forward |
5′-TCCAAGGCTCAGGTACTTCAAAG-3′ |
|
Reverse |
5′-GGGCGGGTACTGGCATCT-3′ |
| NKG2D |
|
|
Forward |
5′-TCCATTCTCTCACCCAACCTAC-3′ |
|
Reverse |
5′-CTCTGTTCCTGGCTTTTATTGA-3′ |
| β-actin |
|
|
Forward | 5′-
TTGCCGACAGGATGCAGAA-3′ |
|
Reverse | 5′-
GCCGATCCACACGGAGTACT-3′ |
qPCR was performed on a Bio-Rad iQ5 Real-Time system
(Bio-Rad Laboratories, Inc., USA) using a SYBR Green kit (Tiangen
Biotech Co., Ltd.). The thermocycler conditions were as follows:
Denaturation for 2 min at 94°C, then 40 cycles of 15 sec at 58°C
for elongation followed by the annealing step of 40 sec at 72°C.
β-actin was used as an internal reference gene for standardizing
the expression of targeted mRNA. All reactions were performed in
triplicate and single outliers, with values >5 Ct compared with
the average of the remaining two Ct values discarded. The gene
expression ratio was calculated using the 2−ΔΔCt method
(14).
Statistical analysis
Statistical analysis was performed using GraphPad
Prism software (version 5.0.1; GraphPad Software, Inc., La Jolla,
CA, USA). Quantitative data were presented as mean ± standard
deviation. The differences in the distribution of continuous
variables between two categories were analyzed using Student's
t-test for normally distribution data and Mann-Whitney test for
non-parametric data. For data involving three or more groups, the
Kruskal-Wallis test was used. All reported P-values were two-sided
and P<0.05 was considered to indicate a statistically
significant difference.
Results
NK cell subsets and NKT cells in MDS
cases compared with non-MDS cases
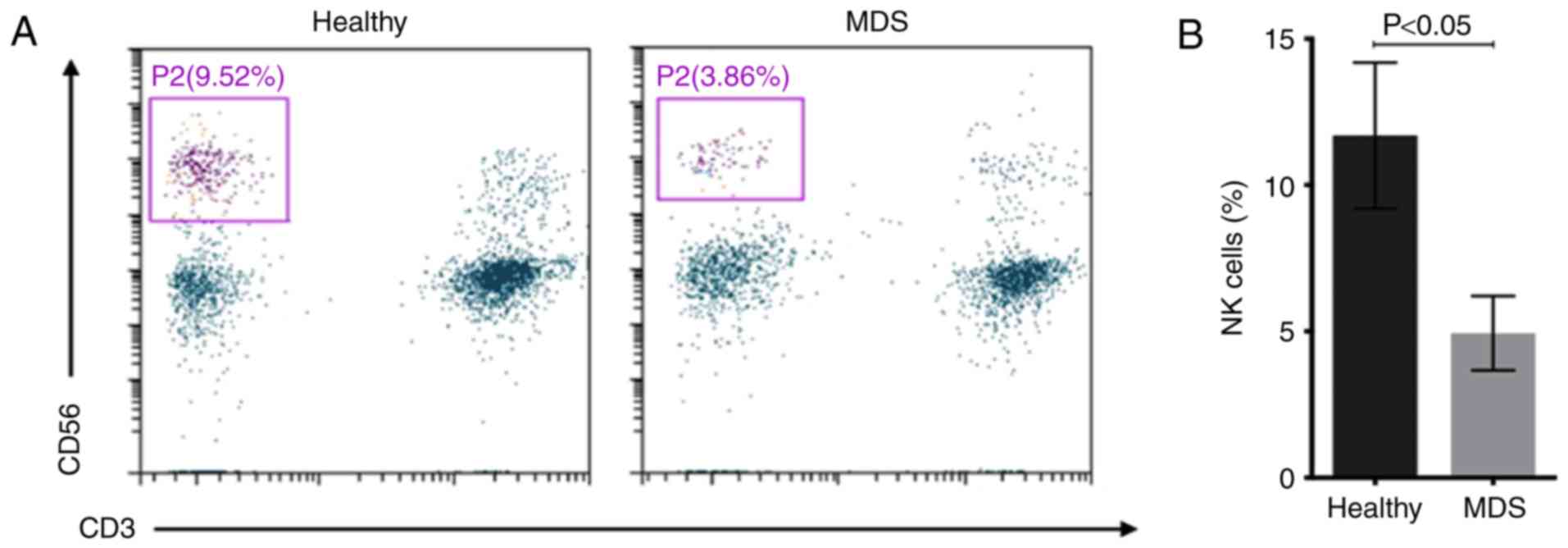
The number of NK cells
(CD3−CD56+) in distinct subgroups of diseases
was measured. The percentage of NK cells in peripheral blood
lymphocytes was 4.94±1.28% in patients with MDS and 11.69±2.49% in
healthy controls (Fig. 1).
Subsequently, the CD16+ NK cell subset and the NKT cells
(CD56+CD3+) were separated into different
disease subgroups, and flow cytometry indicated that the percentage
of peripheral blood CD16+NK cells was significantly
decreased in patients with MDS and AML compared with that in
healthy donors (Fig. 2A). The
percentage of CD16+NK cells in peripheral blood
lymphocytes was 15.20±6.41% in patients with MDS, 9.97±4.26% in
patients with AML and 20.14±3.61% in healthy controls. The
percentage of CD16+ NK cells in patients with MDS was
significantly decreased compared with that in healthy controls
(P<0.001) and the percentage of CD16+NK cells in
patients with AML was significantly decreased compared with that in
patients with MDS (P<0.05). The median percentage of NKT cells
was 2.36% in patients with MDS, 4.31% in patients with AML and
2.14% in healthy controls. There were no significant differences
between the percentage of NKT cells in patients with MDS/AML and
healthy donors (Fig. 2B).
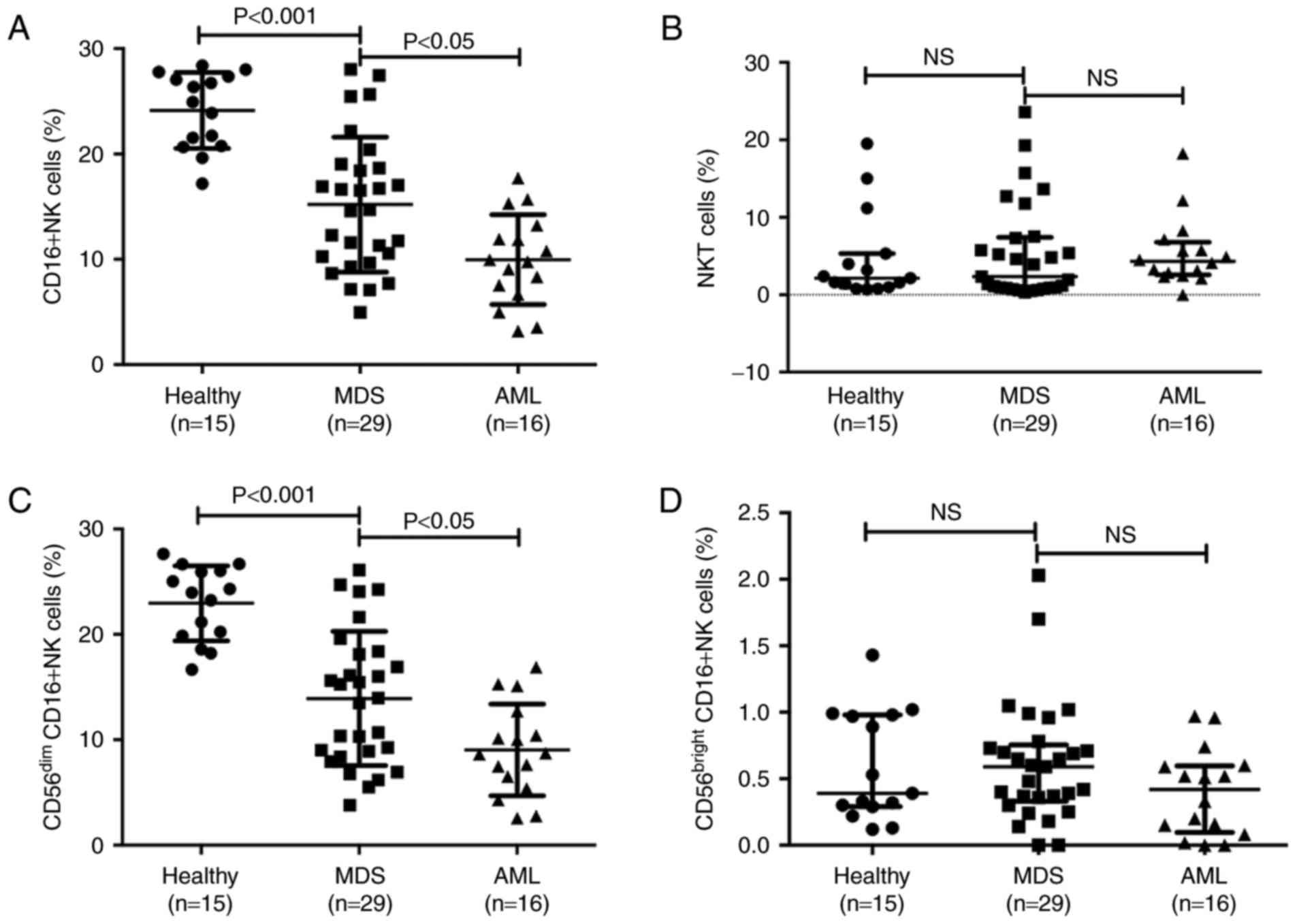 | Figure 2.CD16+NK cells and NKT
cells in different subgroups of diseases. (A) The percentages of
CD16+NK cells
(CD3−CD56+CD16+) in healthy
control, MDS and AML groups. (B) The percentages of NKT cells
(CD56+CD3+) in healthy control, MDS and AML
groups. (C) Difference in CD56dim cells in healthy
control, MDS and AML groups. (D) Level of CD56bright
cells in healthy control, MDS and AML groups. NK cell deficiency is
associated with high-risk subgroups. CD, cluster of
differentiation; NK, natural killer; NKT, NK-like T; MDS,
myelodysplastic syndromes; AML, acute myeloid leukemia; NS, not
significant. |
Subsequently, the distinct subtypes of mature
CD56dim NK cells and less mature CD56bright
cells were investigated. The percentage of CD56dim cells
was 13.92±6.37% in patients with MDS, 9.03±4.35% in patients with
AML and 22.95±3.55% in healthy controls. Consistent with the
results in NK cells, the percentage of CD56dim cells in
patients with MDS was significantly decreased compared with that in
healthy controls (P<0.001), and the percentage of
CD56dim cells in patients with AML was significantly
decreased compared with that in patients with MDS (P<0.05;
Fig. 2C). The median percentage of
CD56bright cells was 0.59% in patients with MDS, 0.42%
in patients with AML and 0.39% in healthy controls; no statistical
difference was observed between patients with MDS/AML and healthy
donors (Fig. 2D).
The association between NK cell deficiency and
specific disease subgroups (data not shown), as defined by the
International Prognostic Scoring System (10), was evaluated. In the present study,
patients in the low risk group were 20%, intermediate-1 group were
23%, intermediate-2 group were 37% and high risk group were 20%.
The percentage of CD16+NK cells was lower in the
high-risk MDS group (12.19±4.56%) than in the low-risk group
(17.20±5.67%; P<0.05).
Activating receptors and adhesion
receptors on NK cells in patients with MDS
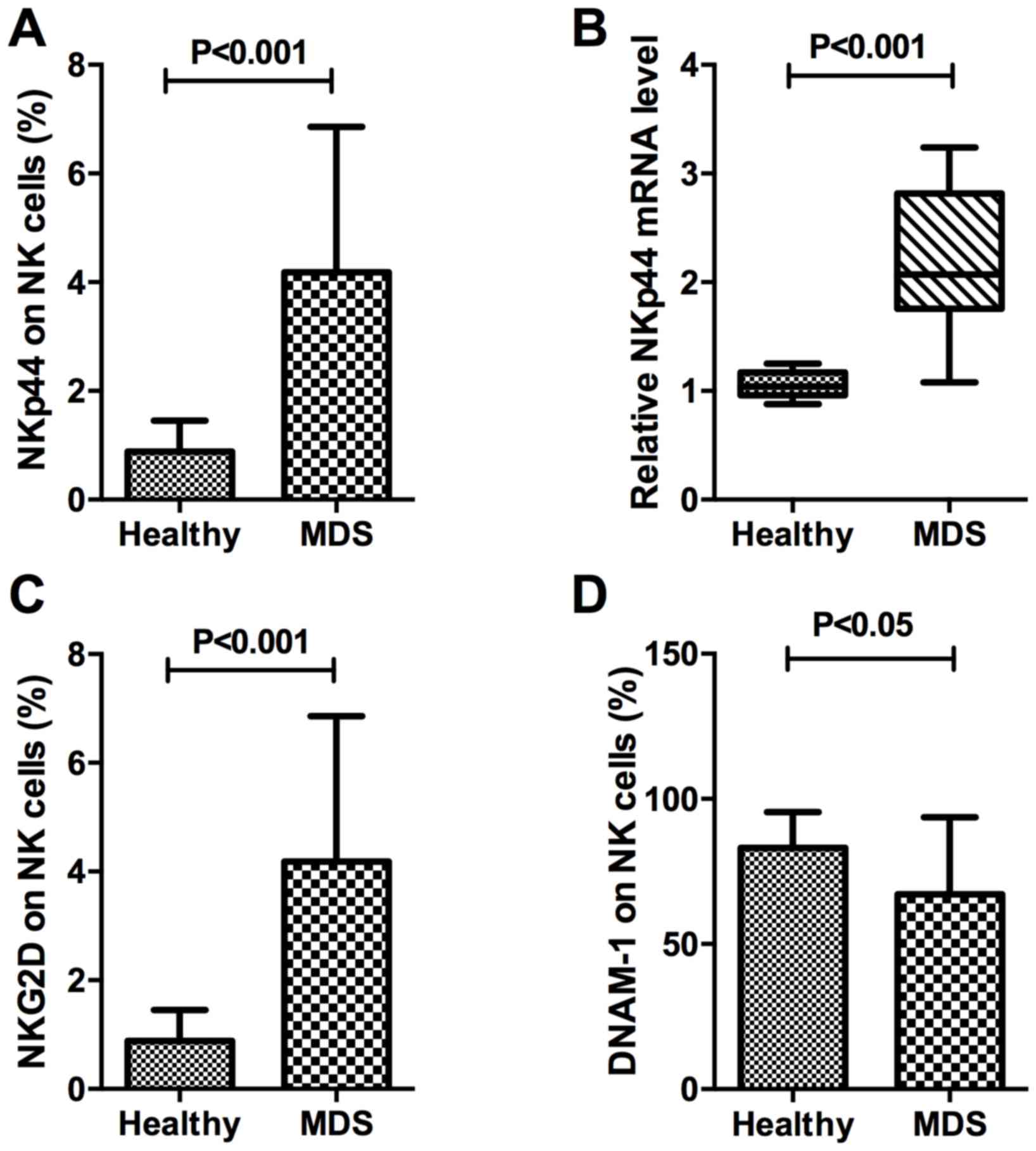
The present study investigated NKp44 protein and
mRNA expression in the natural cytotoxicity receptors (NCR). The
expression percentage of NKp44 on CD16+NK cells in
peripheral blood was 4.18±2.68% in patients with MDS and 0.89±0.56%
in healthy controls, which was significantly different
(P<0.001). In CD16+NK cells, RNA expression of NKp44
in MDS samples was compared with that in controls. The relative
expression of NKp44 in purified target NK lymphocytes was
2.20±0.63% in patients with MDS and 1.05±0.12% in healthy controls,
which was consistent with the results of protein expression
demonstrated a significant difference (P<0.001). Taken together,
these results indicated that NKp44 expression in the NK cells
(CD3−CD56+CD16+) of patients with
MDS was significantly increased compared with that in healthy
donors (Fig. 3A and B).
The present study also measured other stimulatory
signals, including NKG2D and DNAM-1. The percentage of NKG2D in
total NK cells of healthy controls and patients with MDS were
0.89±0.56 and 4.18±2.68%, respectively (P<0.001; Fig. 3C). Glycoprotein DNAM-1 expression on
NK cells in peripheral blood was 67.17±26.55% in patients with MDS
and 83.11±12.31% in healthy controls, a difference that was
statistically significant (P<0.05; Fig. 3D). On the surface of NK cells,
expression of DNAM-1, a protein that mediates cellular adhesion
(15–16), was decreased in MDS, whereas
expression of NKG2D, which mediates lysis of certain tumor cells
(17–18), was increased, compared with the
healthy control.
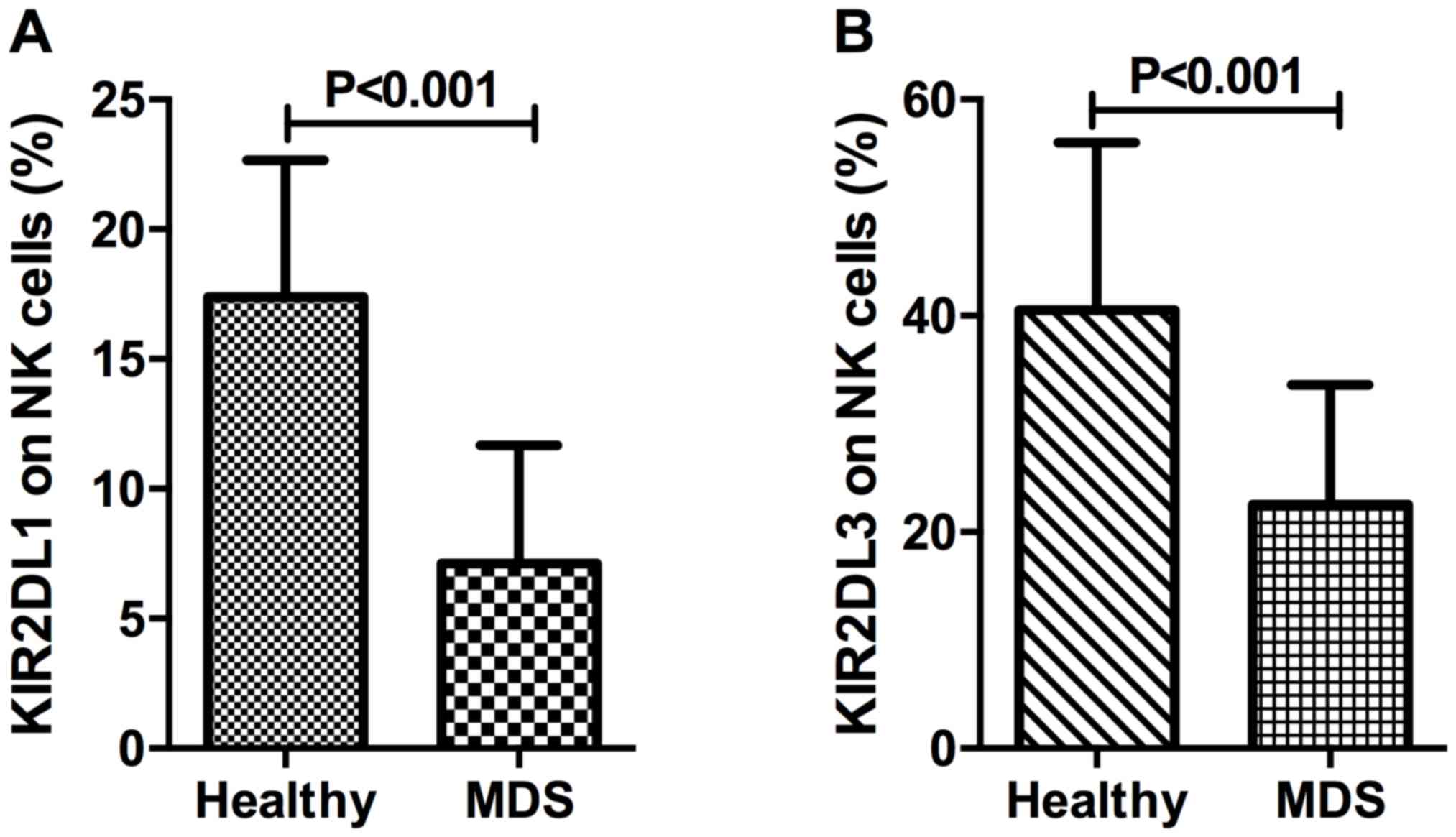
For the identification of inhibitory NK cell
receptors, the present study investigated two killer cell
immunoglobulin-like receptors, CD158a (also known as KIR2DL1) and
CD158b (also known as KIR2DL3). The percentage of KIR2DL1 in NK
cells of patients with MDS and healthy controls was 7.12±4.55 and
17.39±5.27%, respectively (P<0.001; Fig. 4A). The percentage of NK cells
expressing KIR2DL3 was 22.55±11.05% in patients with MDS and
40.51±15.50% in healthy controls, which demonstrated a
statistically significant difference (P<0.001; Fig. 4B).
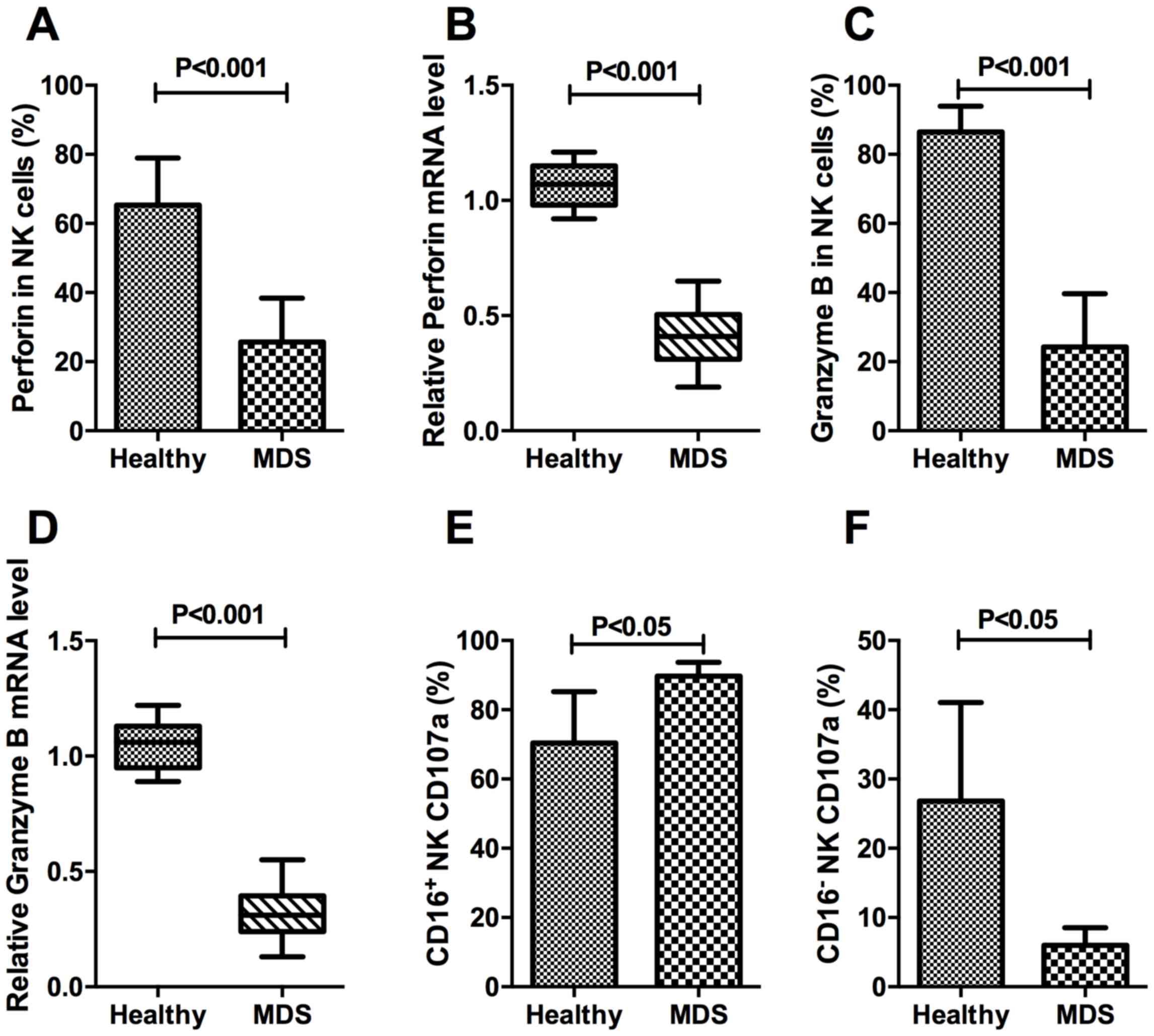
To determine the cytotoxic function of NK cells in
patients with MDS, the expression percentage and relative mRNA
level of perforin and granzyme B on target NK cells was evaluated.
The expression of CD107a was also evaluated in different subsets of
NK cells.
The percentage of perforin-expressing NK cells in
the MDS group was 25.67±12.71%, which was significantly decreased
compared with the healthy control group (65.33±13.65%; P<0.001;
Fig. 5A). In the isolated pure NK
cells, the relative mRNA expression levels of perforin in the MDS
and healthy control samples were 0.42±0.12% and 1.06±0.09%,
respectively, a difference that was consistent with the protein
expression results, as well as demonstrating significance
(P<0.001; Fig. 5B). The percentage
of granzyme B on target NK cells from the MDS group was
24.30±15.36%, which was significantly decreased compared with that
in the healthy control group (86.52±7.46%; P<0.001; Fig. 5C). The relative mRNA expression of
granzyme B in the MDS and healthy control samples was 0.31±0.10 and
1.06±0.11%, respectively, a difference that was statistically
significant (P<0.001; Fig. 5D).
These results indicated that patients with MDS were deficient in
perforin and granzyme B at the protein and the mRNA level. The
differences in CD107a expression on CD16+NK and
CD16−NK subsets demonstrated ADCC-mediated NK activity
in MDS patients. The percentage of CD16+NK expressing
CD107a was increased in patients with MDS compared with that in
controls (P<0.05; Fig. 5E).
Conversely, the expression of CD107a was decreased in
CD16−NK subsets of MDS compared with the normal control
(P<0.05; Fig. 5F).
Discussion
NK cells are lymphocytes that are able to kill any
target that lacks self-major histocompatibility complex (MHC) class
I molecules, also known as ‘missing self’, by mediating lysis of
certain tumor cells and virus-infected cells without previous
activation (5). Based on flow
cytometric analysis, the traditional phenotype used to define human
NK cells was the expression of CD56 and the absence of CD3,
excluding T cells within the lymphocyte gate (5,6,11). In the present study, the
CD3−CD56+NK cell population in peripheral
blood lymphocytes was decreased in patients with MDS compared with
that in healthy controls. Furthermore, CD16 was also used to
identify NK cells that may lead to ADCC, which is a dominant
component of effective antitumor activity (19). Additionally, the proportion of
CD3−CD56+CD16+-expressing NK cells
in the peripheral blood was decreased in patients with MDS or AML
compared with that in healthy donors, and the percentage of
CD16+ NK cells in patients with AML was decreased
compared with that in patients with MDS. As CD16+ NK
cells are important in immunological tumor surveillance (18,20), a
lack of CD16+ NK cells in patients with MDS or AML may
explain their increased tumor burden. The target recognition and
host defense functions of NK and cytolytic T cells have suggested
that they may evolve from a common ancestral cytolytic effector
cell; however, the proportion of
CD3+CD56+-expressing NKT cells did not differ
significantly between patients with MDS or AML and healthy donors,
in the present study. T-cells with expression of CD56 and/or
CD16/57 were defined as NKT cells in previous study, but only the
CD3+CD56+-expressing cells were included in
our study. The specific gating strategy utilized in the present
study may explain why the frequency of NKT observed in patients
with MDS was decreased competed with that in previously published
data (21). Therefore, NK cells
displaying cytolytic capacity against target cells were further
investigated. Human NK cells may be distinguished by CD56 surface
expression density; high expression of CD56 subsets may produce
significantly increased levels of cytokines in response to
activation, however may exhibit weaker target function (12,22). The
majority of circulating NK cells are CD56dim and express
high levels of CD16, which exhibit a decreased ability to produce
cytokines compared with the CD56bright NK cells
(22), and improved ability to
effectively mediate ADCC and natural cytotoxicity (5,19). The
results of the present study demonstrated that an increased number
of CD16+ NK cells were CD56dim and that the
percentages of CD56dim CD16+ NK cells in the
peripheral blood were decreased in patients with MDS, compared with
healthy control. Therefore, we hypothesized that the number of
CD56dim CD16+ NK cells was too limited for
them to be able to recognize the cancerous cells, thereby resulting
in the ineffective hematopoiesis observed in MDS.
NK cells exhibit a dynamic balance of several
distinct families of activating and inhibitory signals that engage
potential target cell recognition (5,23–25). In humans, the major receptors
responsible for tumor cell recognition are NKp46, NKp30, NKp44,
DNAM1 and NKG2D (25). The NCR family
consists of NKp46, NKp30 and NKp44. NKp46 is a popular maker for NK
cells, and resting and activated NK cells express NKp30 and NKp46,
whereas NKp44 is limited to IL-2-activated NK cells (5,12). The
present study observed an increased expression of NKp44 in
CD16+NK cells of patients with MDS patients at the
protein and RNA level compared with the healthy donors, indicating
that an increased number of NK cells favored the activated
condition in MDS. The present study also measured other stimulatory
signals, including NKG2D and DNAM-1. NKG2D, also known as CD314, is
characterized by the presence of a calcium-dependent lectin domain
that is preferentially expressed in NK cells, and binds to ligands,
including MHC class I chain-related A and B proteins, which may
result in the activation of NK cells (17,23).
DNAM-1 (also known as CD226) is a member of the immunoglobulin
(Ig)-superfamily containing two Ig-like V-set domains and is
categorized as a cellular adhesion receptor of NK cells (16,25). In
the present study, the percentage of NKG2D expression in total NK
cells of the MDS group was significantly increased compared with
that of the healthy control group, and glycoprotein DNAM-1
expression on NK cells in the peripheral blood was decreased in
patients with MDS compared with that in healthy controls. Killer
cell Ig-like receptors (KIRs) are glycoproteins classified by the
number of extracellular Ig domains and by whether they possess a
long or short cytoplasmic domain; KIRs with a long cytoplasmic
domain containing an immune tyrosine-based inhibitory motif (ITIM)
interact with MHC class I as inhibitory signals upon ligand binding
(23). With regards to inhibitory
receptors expressed on human peripheral blood NK cells, the
percentage of KIR2DL1 and KIR2DL3 in NK cells of the MDS group was
decreased compared with that of the healthy control group. In
conclusion, the coordinated expression of NCRs, NKG2D and
inhibitory signals, including KIRs, may trigger the process of
tumor cell lysis in MDS while cellular adhesion may be the
underlying biological mechanism.
In NK cell effector signaling pathways, activating
receptors initiate protein tyrosine kinase-dependent and DAP10
signaling pathways. Inhibitory receptors characterized by ITIMs
antagonize activating signaling pathways through protein tyrosine
phosphatases. Therefore, expression of certain receptors may
trigger cytolytic programs and cytokine or chemokine secretion
through these pathways (24). In the
present study, as a result of effector signaling pathways of NK
cells, the expression percentage and relative mRNA level of
perforin and granzyme B on target NK cells were downregulated in
patients with MDS. The increased expression of CD107a on
CD16+NK cells demonstrated ADCC-mediated NK activity in
patients with MDS, whereas the decreased CD107a expression on
CD16−NK cells revealed the inability of degranulation in
patients with MDS, compared with the healthy control. As a result
of the lack of key effector molecule for cytolysis, the cancerous
cells in patients with MDS may fail to undergo lysis, resulting in
progression to AML.
In conclusion, CD16+-expressing NK cells
and subset CD56dim NK cells were decreased in the
peripheral blood of patients with MDS, compared with healthy
controls. Altered expression of activating receptors and inhibitory
signals on NK cell effector signaling pathways may trigger the
process of tumor cell lysis in patients with MDS; however, the weak
cellular adhesion and decreased cytotoxicity of NK cells may lead
to ineffective antitumor activity in MDS. The conclusion of the
present study that NK cell function is decreased in patients with
MDS is limited by the lack of functional assays. Therefore, it is
important that future studies explore functional assays, including
cytotoxicity against K562 in NK cells from patients with MDS.
Future analysis comprising larger MDS cohorts and different stages
of MDS is required. The observations of the present study indicated
the function of NK cells as immunological determinants in MDS and
may allow the development of NK cell-based immunotherapy or
newly-validated immunomodulatory agents for the treatment of
patients with MDS (19,26).
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (grant nos. 81570111
and 81400085) and the Foundation of Tianjin Medical University
(grant no. 2012KYQ15).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
All authors have read and approved the final
manuscript. HM, JS, SD, LL and HL performed the experiments. WZ, RF
and ZS designed the study. XX performed the FCM experiments of
CD107a, analyzed and interpreted the data. HW contributed to the
conception of the work, interpretation of the data and revision for
important intellectual content. XX and HW wrote the paper.
Ethics approval and consent to
participate
The present study was approved by the Tianjin
Medical University Institutional Review Board (Tianjin, China).
Patients consented to participate in this study.
Consent for publication
All patients provided written informed consent in
accordance with the Declaration of Helsinki.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Adès L, Itzykson R and Fenaux P:
Myelodysplastic syndromes. Lancet. 383:2239–2252. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tefferi A and Vardiman JW: Myelodysplastic
syndromes. N Engl J Med. 361:1872–1885. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Alfinito F, Sica M, Luciano L, Della Pepa
R, Palladino C, Ferrara I, Giani U, Ruggiero G and Terrazzano G:
Immune dysregulation and dyserythropoiesis in the myelodysplastic
syndromes. Br J Haematol. 148:90–98. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kordasti SY, Ingram W, Hayden J, Darling
D, Barber L, Afzali B, Lombardi G, Wlodarski MW, Maciejewski JP,
Farzaneh F and Mufti GJ: CD4+CD25high Foxp3+ regulatory T cells in
myelodysplastic syndrome (MDS). Blood. 110:847–850. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Caligiuri MA: Human natural killer cells.
Blood. 112:461–469. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hejazi M, Manser AR, Frobel J, Kündgen A,
Zhao X, Schönberg K, Germing U, Haas R, Gattermann N and Uhrberg M:
Impaired cytotoxicity associated with defective natural killer cell
differentiation in myelodysplastic syndromes. Haematologica.
100:643–652. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chamuleau ME, Westers TM, van Dreunen L,
Groenland J, Zevenbergen A, Eeltink CM, Ossenkoppele GJ and van de
Loosdrecht AA: Immune mediated autologous cytotoxicity against
hematopoietic precursor cells in patients with myelodysplastic
syndrome. Haematologica. 94:496–506. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Epling-Burnette PK, Bai F, Painter JS,
Rollison DE, Salih HR, Krusch M, Zou J, Ku E, Zhong B, Boulware D,
et al: Reduced natural killer (NK) function associated with
high-risk myelodysplastic syndrome (MDS) and reduced expression of
activating NK receptors. Blood. 109:4816–4824. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vardiman JW, Thiele J, Arber DA, Brunning
RD, Borowitz MJ, Porwit A, Harris NL, Le Beau MM,
Hellström-Lindberg E, Tefferi A and Bloomfield CD: The 2008
revision of the World Health Organization (WHO) classification of
myeloid neoplasms and acute leukemia: Rationale and important
changes. Blood. 114:937–951. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Greenberg P, Cox C, LeBeau MM, Fenaux P,
Morel P, Sanz G, Sanz M, Vallespi T, Hamblin T, Oscier D, et al:
International scoring system for evaluating prognosis in
myelodysplastic syndromes. Blood. 89:2079–2088. 1997.PubMed/NCBI
|
|
11
|
Kogure T, Fujinaga H, Niizawa A, Hai LX,
Shimada Y, Ochiai H and Terasawa K: Killer-cell inhibitory
receptors, CD158a/b, are upregulated by interleukin-2, but not
interferon-gamma or interleukin-4. Mediators Inflamm. 8:313–318.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
El-Sherbiny YM, Meade JL, Holmes TD,
McGonagle D, Mackie SL, Morgan AW, Cook G, Feyler S, Richards SJ,
Davies FE, et al: The requirement for DNAM-1, NKG2D and NKp46 in
the natural killer cell-mediated killing of myeloma cells. Cancer
Res. 67:8444–8449. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Baychelier F, Sennepin A, Ermonval M,
Dorgham K, Debré P and Vieillard V: Identification of a cellular
ligand for the natural cytotoxicity receptor NKp44. Blood.
122:2935–2942. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shibuya A, Campbell D, Hannum C, Yssel H,
Franz-Bacon K, McClanahan T, Kitamura T, Nicholl J, Sutherland GR,
Lanier LL and Phillips JH: DNAM-1, a novel adhesion molecule
involved in the cytolytic function of t lymphocytes. Immunity.
4:573–581. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Carlsten M, Baumann BC, Simonsson M,
Jädersten M, Forsblom AM, Hammarstedt C, Bryceson YT, Ljunggren HG,
Hellström-Lindberg E and Malmberg KJ: Reduced DNAM-1 expression on
bone marrow NK cells associated with impaired killing of CD34+
blasts in myelodysplastic syndrome. Leukemia. 24:1607–1616. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Schlegel P, Ditthard K, Lang P, Mezger M,
Michaelis S, Handgretinger R and Pfeiffer M: NKG2D signaling leads
to NK cell mediated lysis of childhood AML. J Immunol Res.
2015:4731752015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Konjević G, Mirjacić Martinović K, Jurisić
V, Babović N and Spuzić I: Biomarkers of suppressed natural killer
(NK) cell function in metastatic melanoma: decreased NKG2D and
increased CD158a receptors on CD3-CD16+ NK cells. Biomarkers.
14:258–270. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Battella S, Cox MC, Santoni A and Palmieri
G: Natural killer (NK) cells and anti-tumor therapeutic mAb:
Unexplored interactions. J Leukoc Biol. 99:87–96. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ebert LM, Meuter S and Moser B: Homing and
function of human skin gammadelta T cells and NK cells: Relevance
for tumor surveillance. J Immunol. 176:4331–4336. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Aggarwal N, Swerdlow SH, TenEyck SP,
Boyiadzis M and Felgar RE: Natural killer cell (NK) subsets and
NK-like T-cell populations in acute myeloid leukemias and
myelodysplastic syndromes. Cytometry B Clin Cytom. 90:349–357.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cooper MA, Fehniger TA, Turner SC, Chen
KS, Ghaheri BA, Ghayur T, Carson WE and Caligiuri MA: Human natural
killer cells: A unique innate immunoregulatory role for the CD56
(bright) subset. Blood. 97:3146–3151. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bryceson YT, March ME, Ljunggren HG and
Long EO: Activation, coactivation, and costimulation of resting
human natural killer cells. Immunol Rev. 214:73–91. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Vivier E, Nunès JA and Vély F: Natural
killer cell signaling pathways. Science. 306:1517–1519. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Vivier E, Raulet DH, Moretta A, Caligiuri
MA, Zitvogel L, Lanier LL, Yokoyama WM and Ugolini S: Innate or
adaptive immunity? The example of natural killer cells. Science.
331:44–49. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bachanova V and Miller JS: NK cells in
therapy of cancer. Crit Rev Oncog. 19:133–141. 2014. View Article : Google Scholar : PubMed/NCBI
|