Introduction
Glioma is a common intracranial malignant tumor,
accounting for more than 50% of intracranial tumors (1). Gliomas are classified into four grades
(I, II, III and IV) based on the 2016 World Health Organization
(WHO) classification system. Grade I and grade II tumors are termed
low-grade gliomas, and grade III and grade IV tumors are termed
high-grade gliomas (2). Glioma has
high disability and mortality rates associated with high incidence
of metastasis and relapse. It is one of the major causes of
disability and death in patients. Currently, glioma is treated
mainly by surgery. However, the cancerous tissue is hard to be
removed completely by surgery due to the infiltrative growth nature
of glioma. A relapse often occurs after surgery (3). In addition, a number of challenges stand
in the way of successful treatment for glioma, including sensitive
tumor location, the blood-brain barrier that restrains anticancer
drugs from entering into brain and tumor tissue, and drug
resistance of cancer cells in glioma (4). There is evidence suggesting miRNA might
render drug-resistant cancer cells in glioma sensitive to
chemotherapy through modulation of target gene expression, thus
enhancing chemotherapeutic efficacy (5). miR-200a is a member of the miR-200
family. Its loss is closely associated with onset of multiple
cancers, and upregulated miR-200a expression is beneficial in
inhibition of tumor growth (6). In
the present study, the expression levels of miR-200a in cancer
tissue, paracancerous tissue and serum were determined using
real-time fluorescence-based quantitative polymerase chain reaction
(qRT-PCR). Based on the acquired data, the correlation between
miR-200a expression and chemotherapeutic treatment efficacy of
glioma was explored.
Materials and methods
Subjects
Forty-five patients with glioma treated in the Sixth
People's Hospital of Jinan from March 2015 to June 2017 were
enrolled in this study as observation group. There were 19 males
and 26 females, aged 26–68 years, with an average age of 52.3±9.6
years. All patients with glioma underwent surgical resection. The
cancer tissue and paracancerous tissue were harvested and stored in
liquid nitrogen immediately. Patient's serum samples were also
collected before chemotherapy treatment. In the meantime, serum
samples were collected from 23 healthy subjects of similar age as
control group who took routine physical examination in the Sixth
People's Hospital of Jinan during the same time period.
Inclusion and exclusion criteria of
patients
Inclusion criteria: Patients had a high-grade glioma
identified by histopathological assay; expected survival was >1
year; the Karnofsky Performance Score (KPS) was >60 points.
Exclusion criteria: Patients recently received immunotherapy;
patients had a history of another kind of malignant cancer;
patients had serious heart, liver, spleen, lung, or kidney
diseases. The present study was approved by the Ethics Committee of
the Sixth People's Hospital of Jinan (Jinan, China). Signed written
informed consents were obtained from the patients and/or
guardians.
Chemotherapy and efficacy
evaluation
Forty-five patients with glioma in the observation
group received temozolomide-based chemotherapy (SFDA Approval no.
H20143117; Jiangsu Sheng Di Pharmaceutical Co., Ltd.). Temozolomide
was administered in normal saline via IV injection at a dose of 75
mg/m2 once a day. Four weeks constitute a course of
treatment. The chemotherapeutic efficacy was evaluated after 5
months of treatment according to the WHO response evaluation
criteria. Complete response (CR) was defined as complete
disappearance of the tumor with no new lesions. The response should
last for at least 4 weeks. Partial response (PR) was defined as at
least 50% reduction in the tumor size with no new lesions. The
response should last for at least 4 weeks. Stable disease (SD)
represents <50% reduction to <25% increase in the tumor size.
Progressive disease (PD) represents at least 25% increase in the
tumor size or appearance of new lesions.
Total RNA extraction and cDNA
synthesis
Total RNA was extracted from tissues and serum using
the TRIzol kit (Takara Bio, Dalian, China) and the mirVana PARIS
kit (Shanghai Ponsure Biotechnology, Inc., Shanghai, China) in
accordance with the manufacturers instructions. RNA purity and
concentration were determined by spectrophotometry at 260 and 280
nm. cDNA synthesis was performed by reverse transcription of 1 µg
RNA using the PrimeScript™ RT Master Mix kit (Takara Bio) in
accordance with the manufacturers instruction.
Determination of miR-200a expression
levels by qRT-PCR
The qRT-PCR assay was performed using an ABI 7100
real-time fluorescence-based quantitative PCR instrument. The PCR
reaction system contained 1.0 µl of cDNA, 0.8 µl of primer, 10 µl
of 2X miRNA qPCR Mix, and 8.2 µl of ddH2O. The PCR
reaction was run as follows: pre-denaturation at 95°C for 10 min,
followed by 45 cycles of 95°C for 10 sec, 60°C for 20 sec and 72°C
for 12 sec. Using U6 as the reference gene, the expression level of
miR-200a was calculated by the method of relative quantification
2−ΔΔCq. Specific primer sequences are as follows: for
miR-200a the forward sequence is TAACACTGTCTGGTAACGATGT and the
reverse sequence is ATCGTTACCAGACAGTGTTATT; for U6 the forward
sequence is CTCGCTTCGGCAGCACATTTT and the reverse sequence is
AACGCTTCACGAATTTGCGT. Primers were synthesized by Shanghai Sangon
Biotech Co., Ltd. (Shanghai, China).
Statistical analysis
Data were statistically analyzed by SPSS 17.0
software (SPSS, Inc., Chicago, IL, USA). The t-test was used for
comparison of differences between two groups. One-way ANOVA was
used for comparison of differences between the above groups.
Differences were considered statistically significant at
P<0.05.
Results
General characteristics
The t-test was used to analyze the differences of
age and sex between the observation group and the control group.
The results in Table I show that the
differences of age and sex between the two groups were not
statistically significant (P>0.05).
 | Table I.Comparison of the general
characteristics between the observation group and the control
group. |
Table I.
Comparison of the general
characteristics between the observation group and the control
group.
| Items | Observation
group | Control group | P-value | t-value |
|---|
| Sex |
|
|
|
|
| Male | 19 | 10 | 0.190 | 1.674 |
|
Female | 26 | 13 |
|
|
| Age (years) |
|
|
|
|
| ≤50 | 29 | 14 | 0.127 | 1.765 |
| >50 | 16 | 9 |
|
|
miR-200a expression levels in cancer
tissue and paracancerous tissue
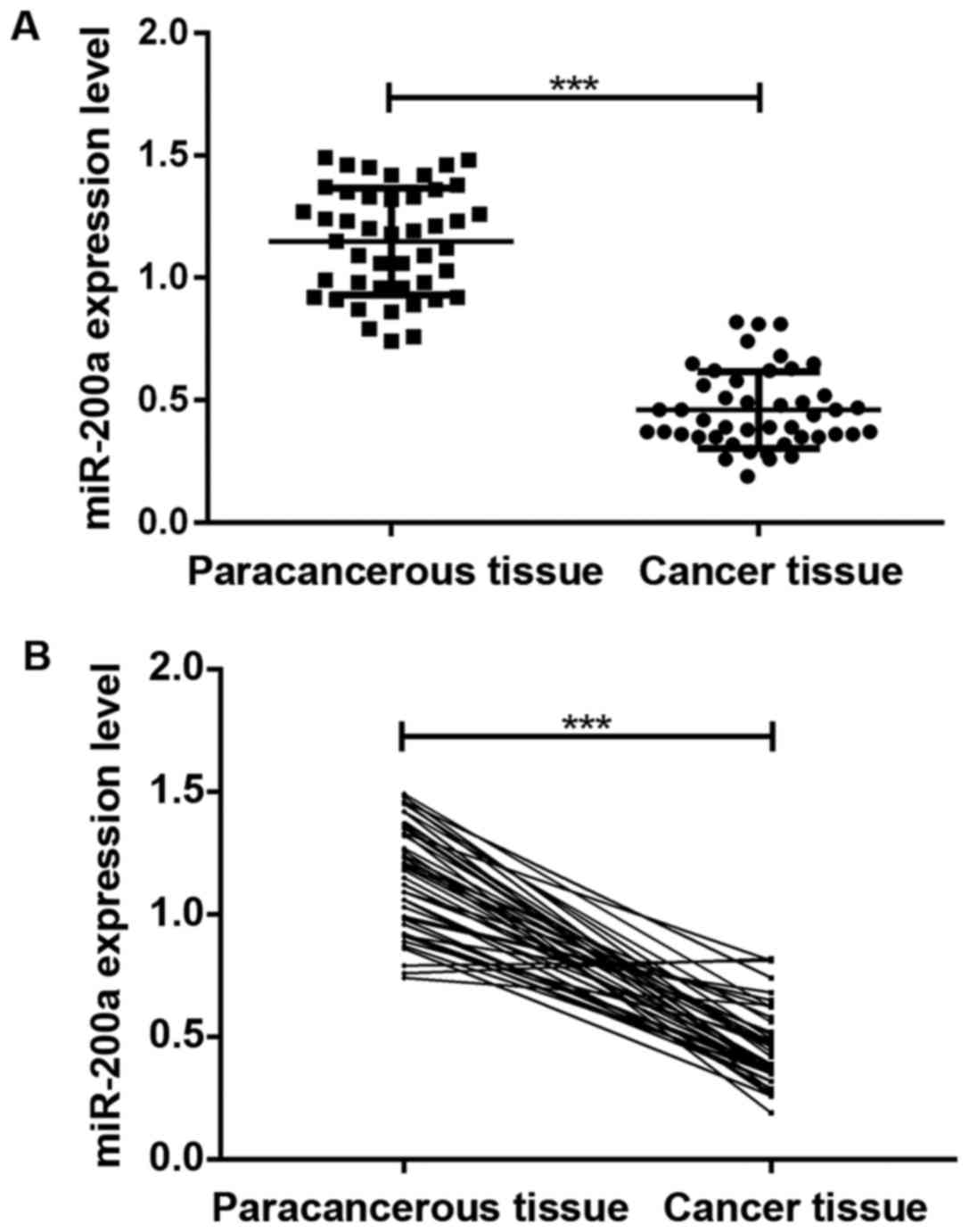
The results of qRT-PCR assay in Fig. 1 show that the expression level of
miR-200a in cancer tissue of patients with glioma was significantly
lower than that in paracancerous tissue (P<0.05), and the
difference was statistically significant.
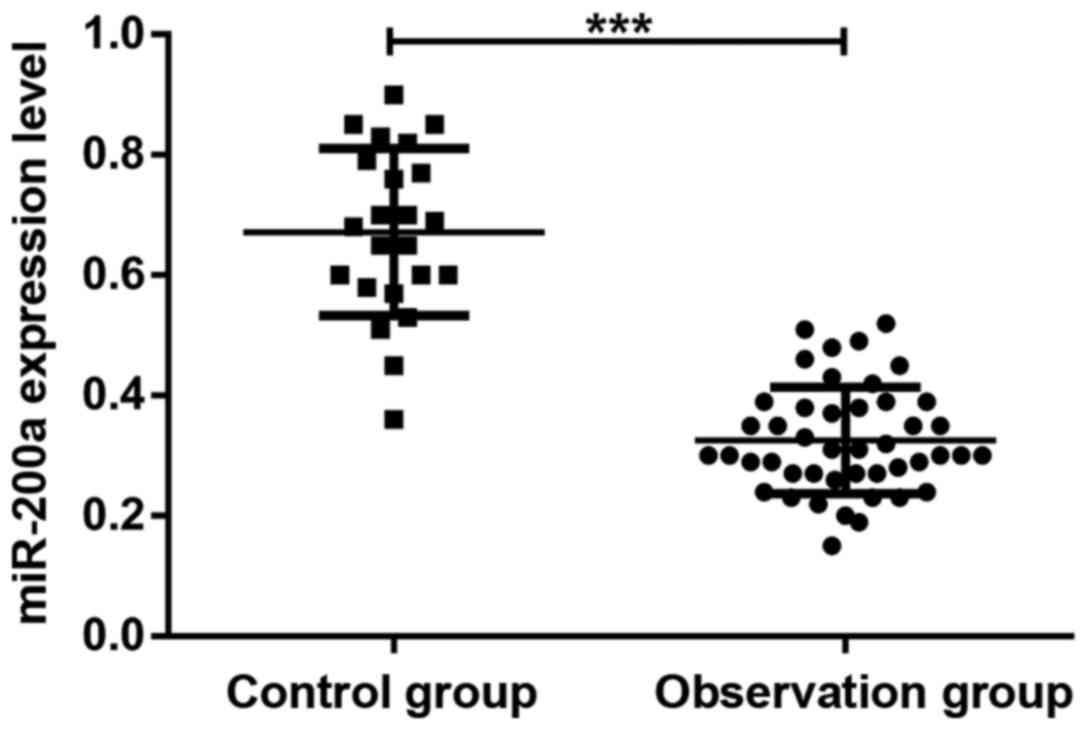
Serum miR-200a expression levels in patients with
glioma before chemotherapy and healthy subjects in the control
group. As shown in Fig. 2, the serum
level of miR-200a in patients with glioma before chemotherapy was
significantly lower than that in healthy subjects (P<0.05) and
the difference was statistically significant.
Correlation between miR-200a
expression and clinical features of glioma patients
The expression levels of miR-200a in cancer tissue
and serum were not correlated with patient age, sex and tumor
location (P>0.05), but were correlated with pathological grade
and tumor size (P<0.05). The detailed results were shown in
Table II.
 | Table II.Correlation between miR-200a
expression level and clinical features of glioma patients. |
Table II.
Correlation between miR-200a
expression level and clinical features of glioma patients.
|
|
| Cancer tissue | Serum |
|---|
|
|
|
|
|
|---|
| Clinical feature | n | Relative expression
level of miR-200a | P-value | t-value | Relative expression
level of miR-200a | P-value | t-value |
|---|
| Age (years) |
|
|
|
|
|
|
|
| ≤50 | 29 | 0.41±0.08 | >0.05 | 0.1218 | 0.35±0.04 | >0.05 | 0.9015 |
|
>50 | 16 | 0.45±0.03 |
|
| 0.36±0.07 |
|
|
| Sex |
|
|
|
|
|
|
|
| Male | 19 | 0.40±0.11 | >0.05 | 0.9145 | 0.32±0.06 | >0.05 | 0.3419 |
|
Female | 26 | 0.47±0.06 |
|
| 0.36±0.02 |
|
|
| Pathological
grade |
|
|
|
|
|
|
|
| III | 12 | 0.53±0.05 | <0.05 | 0.0126 | 0.39±0.05 | <0.05 | 0.0063 |
| IV | 33 | 0.33±0.04 |
|
| 0.28±0.03 |
|
|
| Tumor diameter
(cm) |
|
|
|
|
|
|
|
| ≤5 | 30 | 0.51±0.07 | <0.05 | 0.0083 | 0.34±0.04 | <0.05 | 0.0168 |
|
>5 | 15 | 0.42±0.09 |
|
| 0.26±0.04 |
|
|
| Tumor location |
|
|
|
|
|
|
|
| Frontal
lobe | 16 | 0.41±0.06 | >0.05 | 1.2462 | 0.31±0.06 | >0.05 | 0.8761 |
| Temporal
lobe | 20 | 0.39±0.04 |
|
| 0.29±0.03 |
|
|
| Occipital
lobe | 9 | 0.43±0.04 |
|
| 0.33±0.01 |
|
|
Expression levels of miR-200a before
chemotherapy in cancer tissue and serum for different efficacy
outcomes after chemotherapy
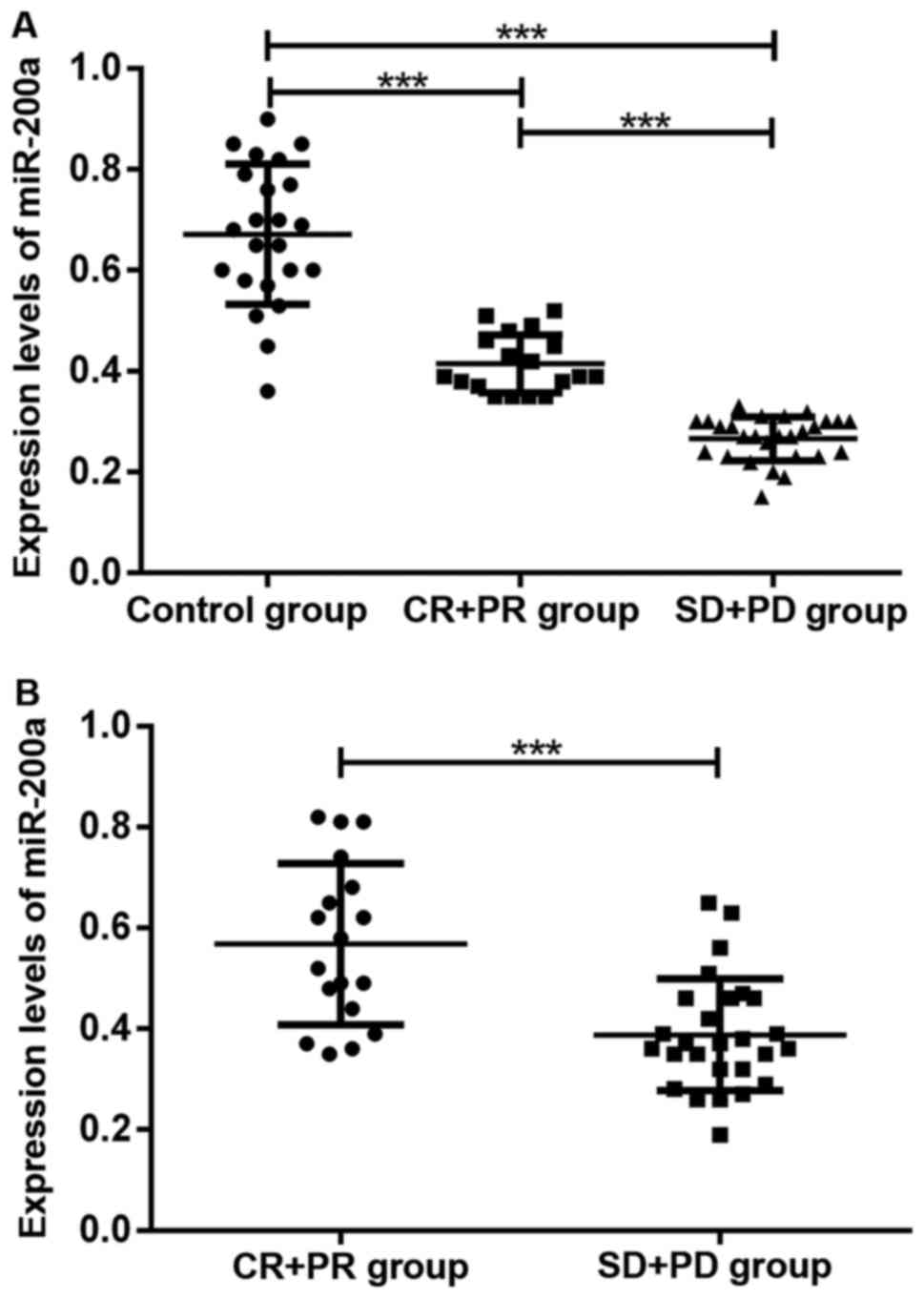
Among 45 patients with high-grade gliomas, there
were 1, 17, 15 and 12 cases that experienced CR, PR, SD and PD,
respectively, after 5 months of chemotherapy treatment. The
expression levels of miR-200a in serum and cancer tissue in
chemotherapy-non-responsive patients were lower than those in
chemotherapy-responsive patients (P<0.05). The serum levels of
miR-200a in chemotherapy-responsive patients were lower than those
in healthy subjects in the control group (P<0.05). As shown in
Fig. 3, the expression level of
miR-200a was correlated with chemotherapeutic efficacy for
treatment of glioma.
Discussion
With continued advances of human genome studies, it
was found that there are only approximately 20,000 gene sequences
with coding function, accounting for less than 2% of the whole
genome sequences. Most of the gene sequences are non-coding RNAs,
including miRNAs (7). miRNAs are
short RNA molecules with 17–25 nucleotides in length and do not
have a coding function. miRNAs can inhibit target gene expression
by binding to the 3-untranslated region of the target gene, thereby
regulating biological processes including cell proliferation,
differentiation, apoptosis and innate immunity (8). A large number of studies suggested that
miRNAs may be involved in onset and progression of many cancers,
including gliomas, as either oncogenes or tumor suppressor genes
(9).
The major members of the miR-200 family that have
been discovered so far are miR-200a, miR-200b, miR-200c, miR-141
and miR-429, of which miR-200a, miR-200b and miR-429 are located on
chromosome 1, while miR-200c and miR-141 are located on chromosome
12 (10). Among these members,
miR-200a played a role as tumor suppressor in a variety of cancers,
including gliomas. In nasopharyngeal carcinoma, miR-200a expression
was downregulated, and the expression level of miR-200a in
undifferentiated nasopharyngeal carcinoma cells was significantly
lower than that in highly differentiated nasopharyngeal carcinoma
cells. As a tumor suppressor, miR-200a can inhibit nasopharyngeal
carcinoma cell proliferation, invasion and metastasis by
attenuating ZEB2 and CTNNB1 expression (11). In liver cancer, miR-200a was
downregulated also in cancer tissue. As a tumor suppressor,
miR-220a can inhibit cancer cell growth and metastasis by blunting
MACC1 gene expression. In addition, the 1-, the 3- and the 5-year
survival rates were lower for patients with low miR-200a expression
than those with high miR-200a expression (12). In breast cancer, miR-200a was also
found to be downregulated in cancer tissue, and as a tumor
suppressor, miR-200a can exert an inhibitory effect on growth of
breast cancer cells through downregulating TFAM expression
(13). miR-200a also played a role in
inhibiting gastric adenocarcinoma growth by reducing ZEB1 and ZEB2
expression, inactivating the wnt/β-catenin signaling pathway, and
attenuating EMT expression (14). For
gliomas, Berthois et al (15)
reported that the miR-200a expression level in grade IV glioma
tissue was lower than those in grade II and III gliomas. Su et
al (16) reported that miR-200a
was downregulated in glioma tissue, and overexpression of miR-200a
in T98G cells reduced glioma cell proliferation, invasion and
metastasis. Inhibition of miR-200a expression led to upregulation
of SIM-2 expression, thereby promoting tumor growth. Ma et
al (17) reported that lncRNA-ATB
promoted glioma cell proliferation, invasion and metastasis by
inhibiting miR-200a expression. If lncRNA-ATB was inactivated, then
miR-200a was re-activated, which lead to reduction of glioma cell
proliferation, invasion and metastasis.
Drug resistance of cancer cells in chemotherapy is
an important factor attenuating chemotherapeutic efficacy, thereby
rendering lower survival rate for cancer patients. Residual cancer
cells surviving chemotherapy gradually developed resistance to the
therapeutic drug, resulting in failed chemotherapy leading to
cancer recurrence (18). Temozolomide
is a newly discovered broad spectrum anticancer drug, and it is the
safest first-line chemotherapy drug currently used in the treatment
of glioma world-wide. Temozolomide is rapidly converted at
physiologic pH to the active form
3-methyl-(triazen-1-yl)imidazole-4-carboxamide (MTIC). Its
cytotoxicity is mediated by alkylation of DNA in cancer cells,
resulting in apoptosis or necrosis (19). In the mid-to-late phase of
chemotherapy for gliomas, varying degrees of drug resistance to
temozolomide was developed in some patients, leading to the failure
of chemotherapy. Induction of chemotherapy drug resistance is a
major factor that compromises chemotherapeutic efficacy (20). With continued advances of gene chip
and sequencing technologies, it was found that abnormal miRNA
expression was an important factor affecting sensitivity of cancer
cells to chemotherapeutic drugs. miRNAs may be involved in onset
and progression of cancer cell resistance to chemotherapeutic drugs
as either oncogenes or tumor suppressor genes (21). In glioma cell line U87MG, miRNA-21
induced upregulation of Bcl-2 and downregulation of Bax. While in
chemotherapy treatment, temozolomide induced cancer cell apoptosis
by inhibiting Bcl-2 expression and promoting Bax expression.
Therefore, overexpression of miRNA-21 attenuated
temozolomide-induced apoptosis of cancer cells, rendering
resistance of cancer cells to temozolomide (22). However, overexpression of miR-200a in
grade IV glioma cells increased cell sensitivity to temozolomide.
In addition, the miR-200a expression level in sensitive glioma
cells to temozolomide was significantly higher than that in less
sensitive glioma cells to temozolomide (15).
In the present study, the expression levels of
miR-200a in cancer tissue, paracancerous tissue and serum of
patients with glioma, as well as in serum of healthy subjects were
determined using qRT-PCR assay. It was found that the expression
level of miR-200a in cancer tissue was significantly lower than
that in paracancerous tissue (P<0.05). In addition, it was also
found that the serum level of miR-200a in the observation group was
significantly lower than those in the control group (P<0.05).
Correlation between miR-200a expression and clinical features of
patients with glioma was explored. It was found that the expression
level of miR-200a in serum was correlated with pathological grade
and tumor size, but not correlated with the patient's age, sex and
tumor location. Thus, it was presumed that miR-200a was involved in
onset and progression of glioma. In this study, the efficacy of
temozolomide in combination with radiotherapy in treatment of
high-grade gliomas was also evaluated. It was found that the
expression levels of miR-200a in serum and cancer tissue in
chemotherapy-non-responsive patients (SD and PD) were lower than
those in chemotherapy-responsive patients (CR and PR, P<0.05).
The serum level of miR-200a in chemotherapy-responsive patients (CR
and PR) was lower before chemotherapy than that in healthy subjects
in the control group (P<0.05). Thus, it was presumed that
changes in miR-200a expression were correlated with
chemotherapeutic efficacy of glioma treatment.
In conclusion, the expression levels of miR-200a in
cancer tissue and serum of patients with glioma were reduced.
miR-200a expression was correlated with glioma pathological grade,
tumor size, as well as chemotherapeutic efficacy, but was not
correlated with patient's age, sex and tumor location. Low
expression of miR-200a represented involvement in onset and
progression of glioma, and was related to cancer cell drug
resistance. The results in this study suggest that miR-200a may
become a new target for the treatment of gliomas.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
XW and YL were responsible for the conception and
design of the study. CW and LK were responsible for collecting the
data. YL and XZ analyzed and interpreted the data. CW drafted this
manuscript. XW revised it critically for important intellectual
content. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
the Sixth People's Hospital of Jinan (Jinan, China). Signed written
informed consents were obtained from the patients and/or
guardians.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bush NA, Chang SM and Berger MS: Current
and future strategies for treatment of glioma. Neurosurg Rev.
40:1–14. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hempel JM, Schittenhelm J, Brendle C,
Bender B, Bier G, Skardelly M, Tabatabai G, Castaneda VS, Ernemann
U and Klose U: Effect of perfusion on diffusion kurtosis imaging
estimates for in vivo assessment of integrated 2016 WHO glioma
grades: A cross-sectional observational study. Clin Neuroradiol.
1-11:2017.https://doi.org/10.1007/s00062-017-0606-8
|
|
3
|
Feng X, Yu Y, He S, Cheng J, Gong Y, Zhang
Z, Yang X, Xu B, Liu X, Li CY, et al: Dying glioma cells establish
a proangiogenic microenvironment through a caspase 3 dependent
mechanism. Cancer Lett. 385:12–20. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Madsen SJ, Christie C, Hong SJ, Trinidad
A, Peng Q, Uzal FA and Hirschberg H: Nanoparticle-loaded
macrophage-mediated photothermal therapy: Potential for glioma
treatment. Lasers Med Sci. 30:1357–1365. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Munoz JL, Rodriguez-Cruz V, Ramkissoon SH,
Ligon KL, Greco SJ and Rameshwar P: Temozolomide resistance in
glioblastoma occurs by miRNA-9-targeted PTCH1, independent of sonic
hedgehog level. Oncotarget. 6:1190–1201. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Su Y, He Q, Deng L, Wang J, Liu Q, Wang D,
Huang Q and Li G: MiR-200a impairs glioma cell growth, migration,
and invasion by targeting SIM2-s. Neuroreport. 25:12–17.
2014.PubMed/NCBI
|
|
7
|
Ganju A, Khan S, Hafeez BB, Behrman SW,
Yallapu MM, Chauhan SC and Jaggi M: miRNA nanotherapeutics for
cancer. Drug Discov Today. 22:424–432. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cheng CJ, Bahal R, Babar IA, Pincus Z,
Barrera F, Liu C, Svoronos A, Braddock DT, Glazer PM, Engelman DM,
et al: MicroRNA silencing for cancer therapy targeted to the tumour
microenvironment. Nature. 518:107–110. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jonas S and Izaurralde E: Towards a
molecular understanding of microRNA-mediated gene silencing. Nat
Rev Genet. 16:421–433. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ning X, Shi Z, Liu X, Zhang A, Han L,
Jiang K, Kang C and Zhang Q: DNMT1 and EZH2 mediated methylation
silences the microRNA-200b/a/429 gene and promotes tumor
progression. Cancer Lett. 359:198–205. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xia H, Ng SS, Jiang S, Cheung WKC, Sze J,
Bian XW, Kung HF and Lin MC: miR-200a-mediated downregulation of
ZEB2 and CTNNB1 differentially inhibits nasopharyngeal carcinoma
cell growth, migration and invasion. Biochem Biophys Res Commun.
391:535–541. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Feng J, Wang J, Chen M, Chen G, Wu Z, Ying
L, Zhuo Q, Zhang J and Wang W: miR-200a suppresses cell growth and
migration by targeting MACC1 and predicts prognosis in
hepatocellular carcinoma. Oncol Rep. 33:713–720. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yao J, Zhou E, Wang Y, Xu F, Zhang D and
Zhong D: microRNA-200a inhibits cell proliferation by targeting
mitochondrial transcription factor A in breast cancer. DNA Cell
Biol. 33:291–300. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cong N, Du P, Zhang A, Shen F, Su J, Pu P,
Wang T, Zjang J, Kang C and Zhang Q: Downregulated microRNA-200a
promotes EMT and tumor growth through the wnt/β-catenin pathway by
targeting the E-cadherin repressors ZEB1/ZEB2 in gastric
adenocarcinoma. Oncol Rep. 29:1579–1587. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Berthois Y, Delfino C, Metellus P, Fina F,
Nanni-Metellus I, Al Aswy H, Pirisi V, Ouafik L and Boudouresque F:
Differential expression of miR200a-3p and miR21 in grade II–III and
grade IV gliomas: Evidence that miR200a-3p is regulated by
O6-methylguanine methyltransferase and promotes
temozolomide responsiveness. Cancer Biol Ther. 15:938–950. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Su Y, He Q, Deng L, Wang J, Liu Q, Wang D,
Huang Q and Li G: MiR-200a impairs glioma cell growth, migration,
and invasion by targeting SIM2-s. Neuroreport. 25(12)2014.7.
PubMed/NCBI
|
|
17
|
Ma CC, Xiong Z, Zhu GN, Wang C, Zong G,
Wang HL, Bian EB and Zhao B: Long non-coding RNA ATB promotes
glioma malignancy by negatively regulating miR-200a. J Exp Clin
Cancer Res. 35:902016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li N, Yang L, Wang H, Yi T, Jia X, Chen C
and Xu P: MiR-130a and miR-374a function as novel regulators of
cisplatin resistance in human ovarian cancer A2780 cells. PLoS One.
10:e01288862015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Elhag R, Mazzio EA and Soliman KF: The
effect of silibinin in enhancing toxicity of temozolomide and
etoposide in p53 and PTEN-mutated resistant glioma cell lines.
Anticancer Res. 35:1263–1269. 2015.PubMed/NCBI
|
|
20
|
Prieto-Vila M, Takahashi RU, Usuba W,
Kohama I and Ochiya T: Drug resistance driven by cancer stem cells
and their niche. Int J Mol Sci. 18:E25742017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang Q, Wang Z, Chu L, Li X, Kan P, Xin X,
Zhu Y and Yang P: The effects and molecular mechanisms of MiR-106a
in multidrug resistance reversal in human glioma U87/DDP and U251/G
cell lines. PLoS One. 10:e01254732015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shi L, Chen J, Yang J, Pan T, Zhang S and
Wang Z: MiR-21 protected human glioblastoma U87MG cells from
chemotherapeutic drug temozolomide induced apoptosis by decreasing
Bax/Bcl-2 ratio and caspase-3 activity. Brain Res. 1352:255–264.
2010. View Article : Google Scholar : PubMed/NCBI
|