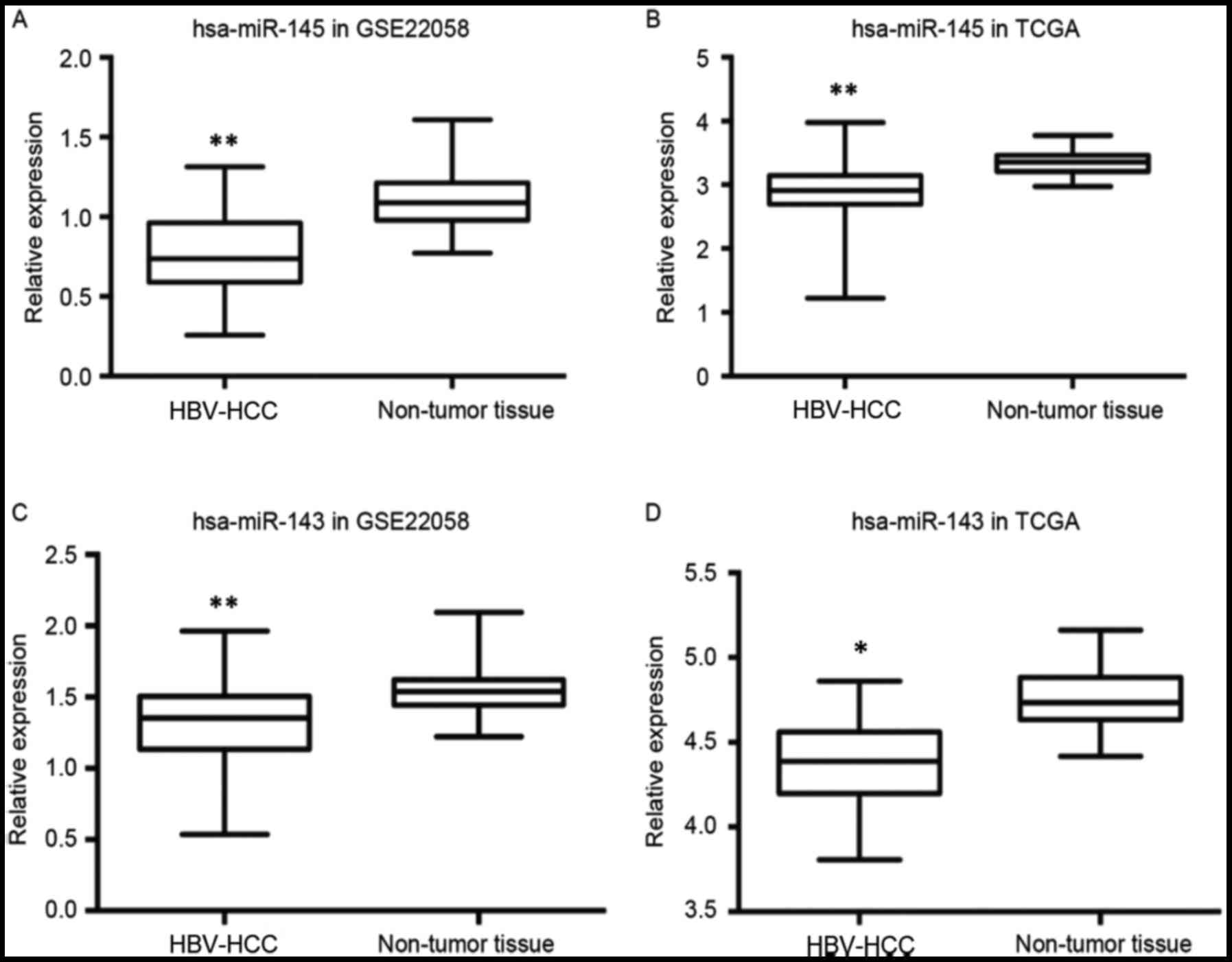
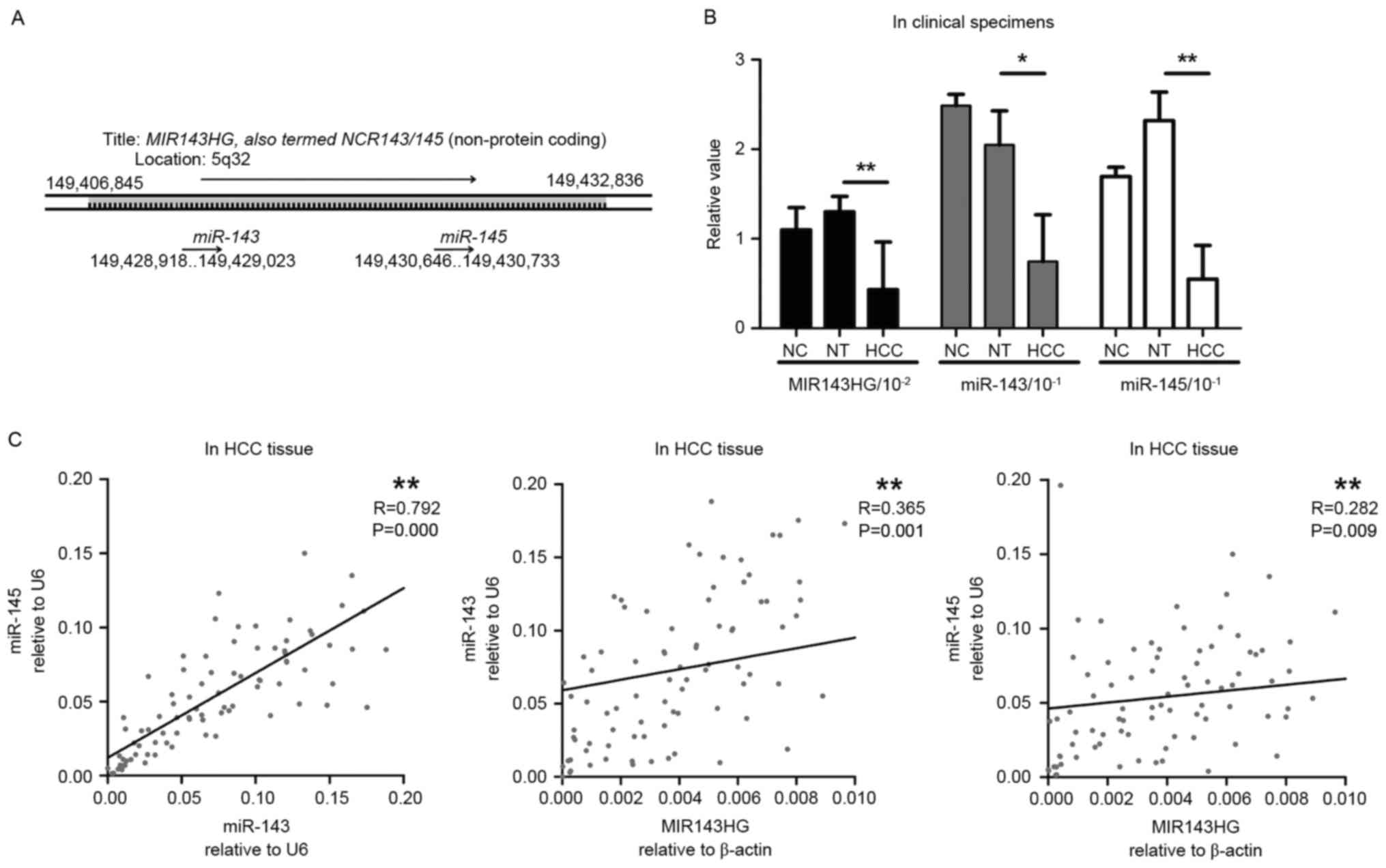
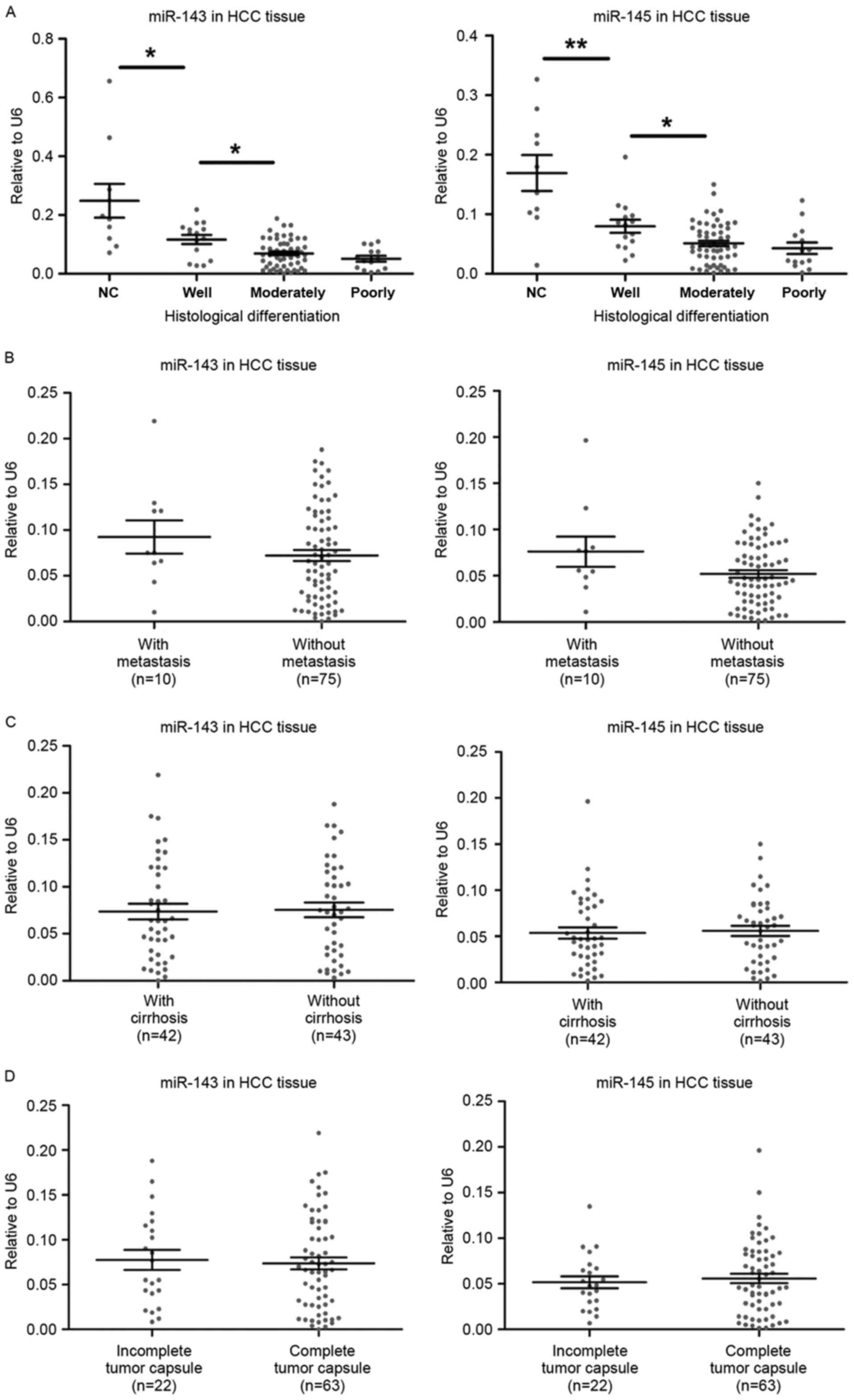
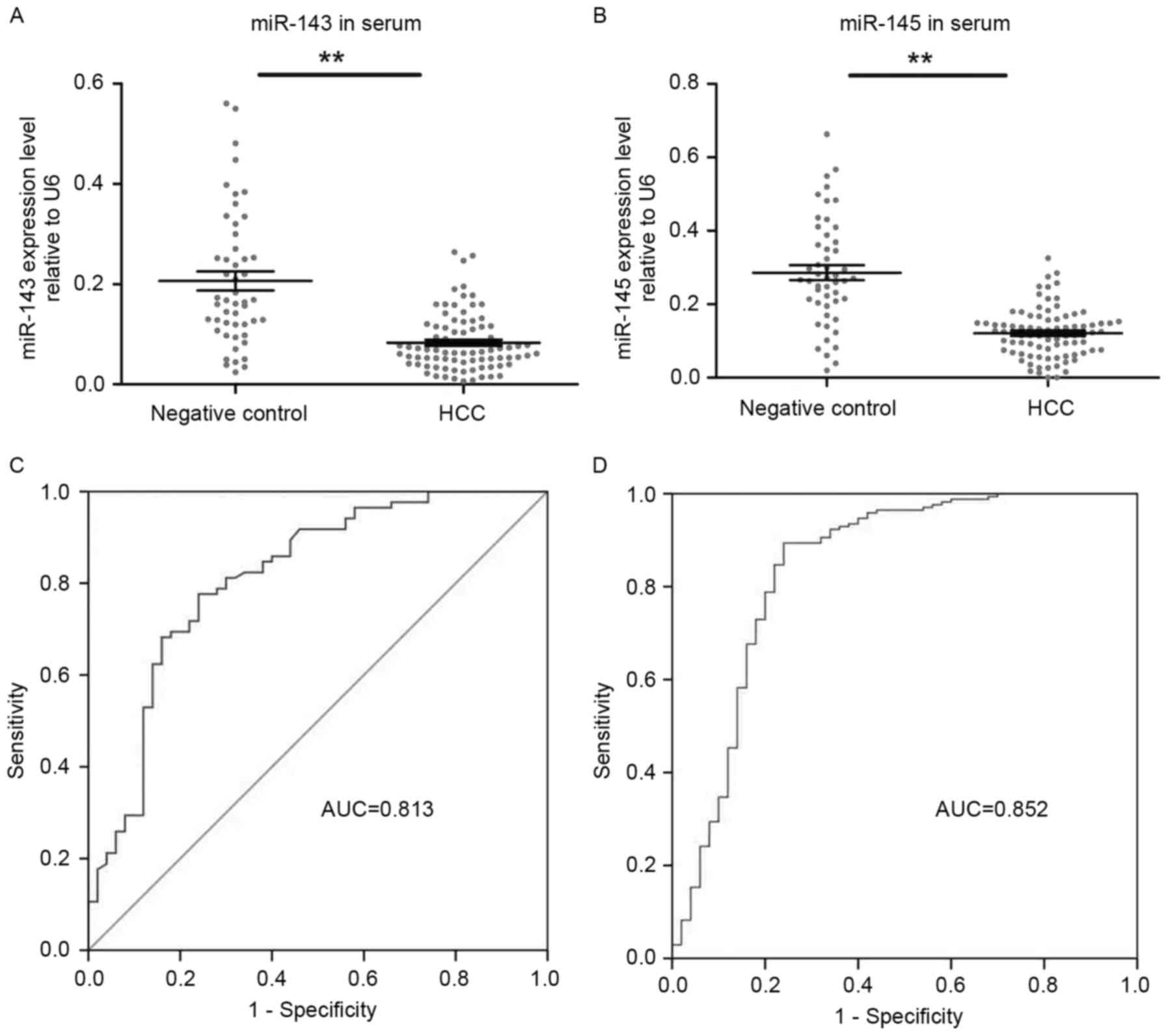
|
1
|
Bréchot C, Gozuacik D, Murakami Y and
Paterlini-Bréchot P: Molecular bases for the development of
hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC).
Semin Cancer Biol. 10:211–231. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhao Q, Li T, Qi J, Liu J and Qin C: The
miR-545/374a cluster encoded in the Ftx IncRNA is overexpressed in
HBV-related hepatocellular carcinoma and promotes tumorigenesis and
tumor progression. PLoS One. 9:e1097822014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dimitrova N, Gocheva V, Bhutkar A, Resnick
R, Jong RM, Miller KM, Bendor J and Jacks T: Stromal expression of
miR-143/145 promotes neoangiogenesis in lung cancer development.
Cancer Discov. 6:188–201. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Callegari E, Gramantieri L, Domenicali M,
D'Abundo L, Sabbioni S and Negrini M: MicroRNAs in liver cancer: A
model for investigating pathogenesis and novel therapeutic
approaches. Cell Death Differ. 22:46–57. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu H, Zhu L, Liu B, Yang L, Meng X, Zhang
W, Ma Y and Xiao H: Genome-wide microRNA profiles identify miR-378
as a serum biomarker for early detection of gastric cancer. Cancer
Lett. 316:196–203. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Iio A, Nakagawa Y, Hirata I, Naoe T and
Akao Y: Identification of non-coding RNAs embracing
microRNA-143/145 cluster. Mol Cancer. 9:1362010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang X, Liu S, Hu T, Liu S, He Y and Sun
S: Up-regulated MicroRNA-143 transcribed by nuclear factor kappa B
enhances hepatocarcinoma metastasis by repressing fibronectin
expression. Hepatology. 50:490–499. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fan X, Chen X, Deng W, Zhong G, Cai Q and
Lin T: Up-regulated microRNA-143 in cancer stem cells
differentiation promotes prostate cancer cells metastasis by
modulating FNDC3B expression. BMC Cancer. 13:612013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ma Q, Jiang Q, Pu Q, Zhang X, Yang W, Wang
Y, Ye S, Wu S, Zhong G, Ren J, et al: MicroRNA-143 inhibits
migration and invasion of human non-small-cell lung cancer and its
relative mechanism. Int J Biol Sci. 9:680–692. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ni Y, Meng L, Wang L, Dong W, Shen H, Wang
G, Liu Q and Du J: MicroRNA-143 functions as a tumor suppressor in
human esophageal squamous cell carcinoma. Gene. 517:197–204. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Qian X, Yu J, Yin Y, He J, Wang L, Li Q,
Zhang LQ, Li CY, Shi ZM, Xu Q, et al: MicroRNA-143 inhibits tumor
growth and angiogenesis and sensitizes chemosensitivity to
oxaliplatin in colorectal cancers. Cell Cycle. 12:1385–1394. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang R, Wang L and Yang AG: Is
microRNA-143 really a turncoat of tumor suppressor MicroRNA in
hepatitis B virus-related hepatocellular carcinoma? Hepatology.
50:987–988. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Noh JH, Chang YG, Kim MG, Jung KH, Kim JK,
Bae HJ, Eun JW, Shen Q, Kim SJ, Kwon SH, et al: MiR-145 functions
as a tumor suppressor by directly targeting histone deacetylase 2
in liver cancer. Cancer Lett. 335:455–462. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang Y, Hu C, Cheng J, Chen B, Ke Q, Lv Z,
Wu J and Zhou Y: MicroRNA-145 suppresses hepatocellular carcinoma
by targeting IRS1 and its downstream Akt signaling. Biochem Biophys
Res Commun. 446:1255–1260. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Edgar R, Domrachev M and Lash AE: Gene
expression omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Burchard J, Zhang C, Liu AM, Poon RT, Lee
NP, Wong KF, Sham PC, Lam BY, Ferguson MD, Tokiwa G, et al:
microRNA-122 as a regulator of mitochondrial metabolic gene network
in hepatocellular carcinoma. Mol Syst Biol. 6:4022010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shi R and Chiang VL: Facile means for
quantifying microRNA expression by real-time PCR. Biotechniques.
39:519–525. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Schetter AJ, Leung SY, Sohn JJ, Zanetti
KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, et
al: MicroRNA expression profiles associated with prognosis and
therapeutic outcome in colon adenocarcinoma. JAMA. 299:425–436.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sun HH, Vaynblat A and Pass HI: Diagnosis
and prognosis-review of biomarkers for mesothelioma. Ann Transl
Med. 5:2442017. View Article : Google Scholar : PubMed/NCBI
|