Introduction
Multiple myeloma (MM) is the second most common
cause of hematological malignancy-associated mortality in America,
second only to non Hodgkin's lymphoma (1). The incidence of MM in Asia is
0.5–1/100,000, whereas the incidence in Africa and America is
10–12/100,000 (2). Generally, the
median age of patients diagnosed with MM is 69 years old, and two
thirds of patients are male (3). Over
the previous two decades, the median survival time of patients with
MM has increased from 3 to 6 years due to improvements in available
treatments (4). However, the majority
of patients have only a several-year remission and will eventually
relapse or become refractory, with MM accompanied by severe
multiple systemic lesions (5,6).
The pathogenesis of MM includes multistep
carcinogenic events. Monoclonal gammopathy of undetermined
significance (MGUS), the pre-malignant condition of MM, is
typically followed by smoldering myeloma and finally develops into
MM (7). A previous report indicated
that patients with MGUS progress to MM or other associated
malignant tumors at a rate of 1% per year (8). Therefore, it is important to accurately
classify the different stages of MM to identify early stage and
high-risk patients to enable timely clinical intervention (9).
At present, the treatment for MM includes autologous
stem cell transplantation (ASCT) and the combined application of
multiple chemotherapeutic drugs (10,11).
However, there are a number of limitations with present therapeutic
strategies. Firstly, patients with MM have differing responses to
the same standardized treatment and drug-resistance may be induced.
Secondly, although disease-free survival has been prolonged in
patients with MM, the majority relapse eventually and the condition
is more complicated following relapse (12). Therefore, uncovering more effective
biomarkers and therapeutic agents for MM is desirable (13–15).
MicroRNAs (miRNA/miRs) are small (~22 nucleotides)
non-coding RNAs that are associated with the initiation and
progression of tumors by regulating ~30% genes at a
post-transcriptional level (16).
Accumulating evidence has demonstrated that miRNAs may be
associated with regulating cellular apoptosis, proliferation,
differentiation, metabolism, invasion and migration in vitro
(17–19). The regulatory mechanisms of miRNAs in
tumor cells have been studied extensively and the majority of the
results concur that mature miRNAs are loaded into the RNA-induced
silencing complex, which results in the degradation or
translational inhibition of their targets depending on perfect or
partial base complementarity with the 3′ untranslated region (UTR)
of genes (20,21). This mode of interaction between miRNAs
and their target genes in tumor cells means that they possess the
potential to become novel therapeutic agents via the knockdown
onco-miRs or the restoration of tumor-suppressive (TS)-miRs
(22). Additionally, aberrant miRNA
expression profiling in MM may be used as a biomarker for tumor
classification, grading and clinical outcomes prediction in
addition to providing the rationale for clinical individual therapy
(7,23).
miRNAs deregulated in MM
MM is a heterogeneous malignancy with complex
genetic abnormalities, including the presence of hypodiploidy, gene
mutations, chromosome translocations, amplifications and deletions
(24). Emerging evidence demonstrates
that the expression of miRNA may be affected by numerous genetic
diversities, including genomic alterations (25), transcriptional regulation (26), epigenetic regulation (27,28), RNA
editing and sequence variations in miRNA binding sites, including
in SNPs (29).
Dysregulated miRNAs in MM often serve similar
functions in pathological processes as oncogenes or tumor
suppressor genes via the activation of multiple signaling pathways
associated with MM, including the nuclear factor-κB (NF-κB)
signaling pathway (30), interleukin
(IL)6/signal transducer and activator of transcription (STAT)3
signaling pathway (31), tumor
protein P53 (P53)/mouse double minute 2 homolog signaling pathway
(32) and phosphatidylinositide
3-kinases (PI3K)/protein kinase B (AKT) signaling pathway (33). During the occurrence and development
of MM, the regulatory mechanisms of these miRNAs also provide the
theoretical foundation for clinical-associated application research
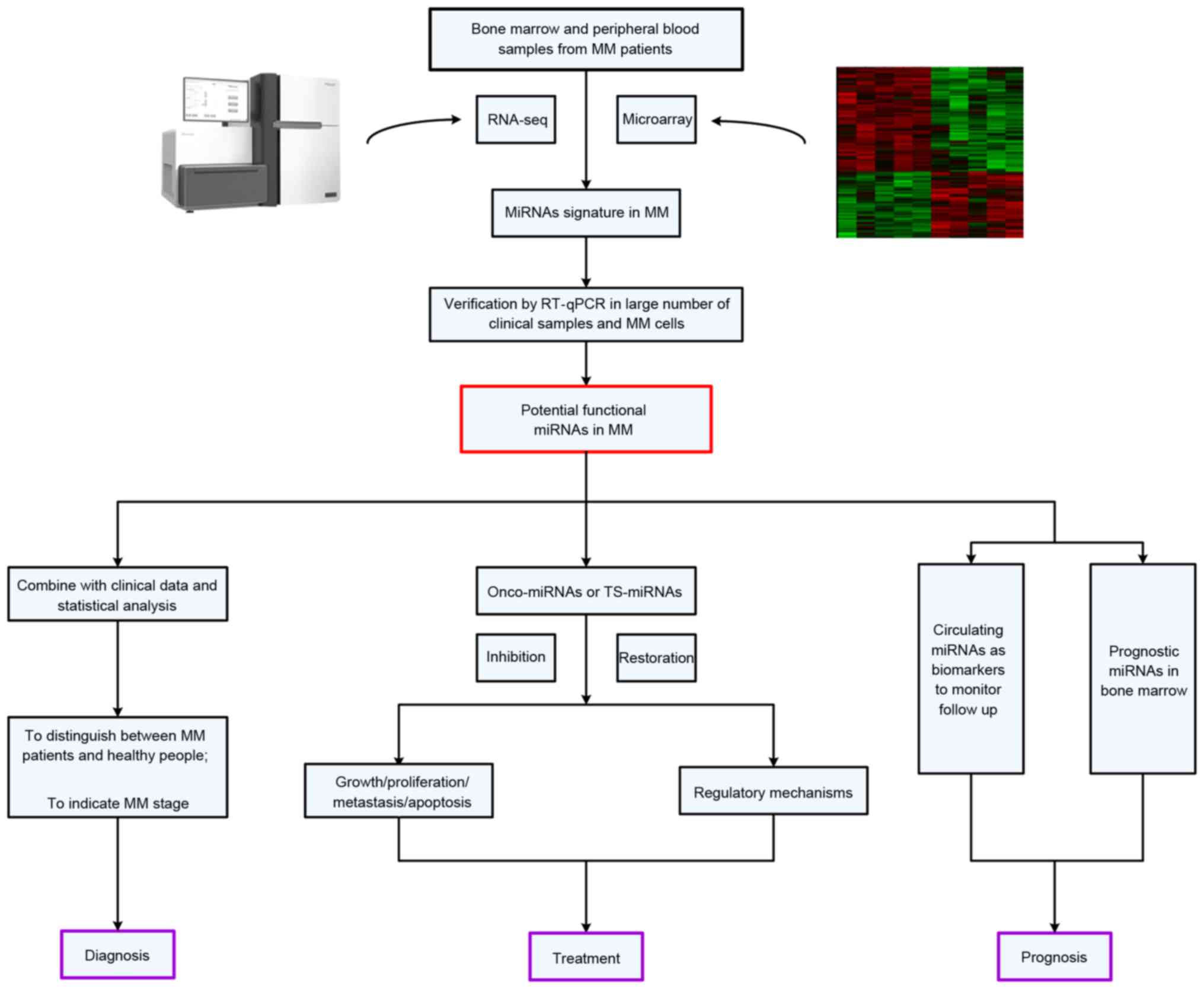
in the future (Fig. 1).
miRNA expression profiling analyses in
MM
Increasingly, evidence suggests that aberrant miRNA
expression is a hallmark in patients with MM, and that normal PCs
have distinct miRNA expression profiles with malignant PCs. Zhou
et al (34) profiled the miRNA
expression pattern of syndecan-1 (CD138)+ cells isolated from 52
newly diagnosed patients with MM and two healthy donors (HDs), and
revealed an elevated total miRNA level in malignant PCs. Microarray
data analyses demonstrated that 39 miRNAs including miR-18,
miR-92a, miR-181a, miR-181b, miR-221 and miR-222 were consistently
expressed at higher levels in samples from newly diagnosed cases
compared with HDs, whereas only miR-370 was downregulated in
MM.
Although MM is a type of cancer that originates from
malignant PCs in bone marrow, it also exerts considerable influence
on ectopic miRNA expression profiles in body fluid, including
serum, plasma, urine etc. Hao et al (35) performed a miRNA expression profile
analysis on the serum samples from seven newly diagnosed
symptomatic patients with MM and five HDs using the miRCURYTM LNA
Array, and the results indicated that amongst all 1,891 miRNAs, 4
miRNAs were upregulated and 23 were downregulated. miR-214 (fold
change of 4.80), miR-135b (fold change of 3.60), miR-132 (fold
change of 0.43) and miR-92a (fold change of 0.49) among them were
selected to be further validated in a large cohort of 108 newly
diagnosed patients with MM and 44 HDs by RT-qPCR assay due to their
critical function in regulating the differentiation of osteoclasts
and osteoblasts as previously reported. Results confirmed that the
level of miR-214 (2.34 vs. 0.23, P=0.0005) and miR-135b (1.83 vs.
−0.18, P=0.0022) were significantly increased in patients with MM
compared with HDs. Furthermore, the receiver operating
characteristic (ROC) analysis revealed that miR-214 and miR-135b
may offer a powerful diagnostic tool for the identification of bone
disease related to MM with high sensitivity and specificity.
In general, these previous studies disclosed a
series of abnormally expressing miRNAs in patients with MM compared
with HDs by high-throughput screening technologies, summarized in
Table I (10,35–43).
However, the results from these studies do not appear to be
consistent. This discrepancy may partly be due to the differences
in the platforms used for microarray technologies, the number of
miRNAs analyzed, and the types and sizes of the samples included
and the statistical methods designed.
 | Table I.High throughput screening of miRNAs
with abnormal expression in MM. |
Table I.
High throughput screening of miRNAs
with abnormal expression in MM.
| Type of sample | Sample size | Platform | Dysregulated miRs
in MM validated by RT-qPCR | Refs. |
|---|
| CD138+ PCs, MM cell
lines | 41 MM cell lines,
10 untreated MM, 4 N | Academic miRNA
microarray | Up: miR-32,
miR-17-5, miR-19a, miR-19b, miR-20a, miR-92, miR-106a, miR-106b,
miR-93, miR-25, miR-181a/181b | (36) |
| CD138+ PCs, MM cell
lines | 7 MM cell lines, 10
untreated MM, 2 N | miRCURY™LNA
microarray | Up: miR-365,
miR-193 | (37) |
| CD138+ PCs | 15
relapsed/refractory MM, 4 N | Liquid phase
Luminex microbead miRNA profiling | Up: miR-181a/181b,
miR-382, miR-222, miR-221 Down: miR-15a, miR-16 | (38) |
| CD138+ PCs | 52 untreated MM, 2
N | Agilent miRNA
microarray V2 | Up: miR-15b,
miR-16, miR-19b, miR-21, miR-22, miR-29c, let-7a, let-7d,
let-7f | (10) |
| CD138+ PCs | 33 untreated MM,5
MGUS, 9 N | µRNA
microarray | Up: miR-21, miR-16,
miR-221, miR-200b, miR-19a, miR-342; Down: miR-15a | (39) |
| Serum | 14 untreated MM, 21
follow-up MM, 7 N | Agilent miRNA 15K
array | Up: let-7b,
miR-106a Down: miR-98, miR-16, let-7c, let-7i | (40) |
| Peripheral
mononuclear cells | 5 untreated MM, 5
N | Applied Biosystems
miProfile miRNA qPCR arrays | Up: miR-16-1,
miR-21, miR-24-1, miR-28, miR-33a, miR-101-1, miR-124-1, miR-125a,
miR-125b-1, miR-129-1, miR-139, miR-145, miR-149, miR-202, miR-212,
miR-221, miR-410, miR-424, miR-1297 Down: miR-23a, miR-520e | (41) |
| Extracellular
supernatant fluid of BM | 20
relapsed/refractory MM, 8 N | Agilent miRNA
microarray v3 | Down: let-7a,
let-7b, miR-15a, miR-16, miR-20a, miR-106b, miR-223 | (42) |
| Serum | 7 untreated MM, 5
N | miRCURY™LNA
microarray | Up: miR-214-3p,
miR-135b-5p, miR-4254, miR-3658 miR-33b; Down: miR-19a,
miR-92a | (43) |
| Serum | 7 untreated MM, 5
N | miRCURY™LNA
microarray | Up: miR-214,
miR-135b, miR-132; Down: miR-92a | (35) |
In addition to being associated with clinical
pathological parameters, a number of these deregulated miRNAs may
be involved in the pathogenesis of MM. Global abnormal miRNA
expression profiling provide the basis for further investigations
on the specific functions of a single miRNA in the pathogenesis of
MM. Firstly, further in vitro and in vivo experiments
on the function of miRNAs are required to validate their biological
function in MM. Subsequently, it is important to determine the
potential mRNA targets of the deregulated miRNAs and investigate
the underlying mechanism. In previous years, numerous bioinformatic
softwares using different computational algorithms have been well
developed to predict miRNA targets including TargetScan (http://www.targetscan.org) (44), miRanda (http://www.microRNA.org) (45) and PicTar (http://pictar.mdc-berlin.de/) (46). A number of studies selected the
intersection of a number of different software predicted target
genes in order to conduct in-depth research. This putative binding
relationship should be validated using dual-luciferase reporter
assays, the change of luciferase activity will decide whether this
miRNA may bind directly to the 3′UTR of its target gene (47).
Functional studies of dysregulated
miRNAs in MM
Following the identification of dysregulated miRNAs
in MM, an increasing number of studies have emphasized how these
small molecules function in the process of transformation from
normal PCs to malignant PCs. These studies demonstrate that miRNAs
may exert an important function in regulating cell processes
(including apoptosis, proliferation, migration and the cell cycle)
in MM by directly binding to the corresponding target genes.
Table II (18,36,48–55)
and Table III (18,27,33,38,56–71)
summarize all deregulated miRNAs and their target genes in MM. The
understanding of this mode of interaction in MM will lay the
theoretical foundation for the clinical miRNA-based therapy.
 | Table II.Principal oncogenic microRNAs with
upregulation in MM. |
Table II.
Principal oncogenic microRNAs with
upregulation in MM.
| MicroRNAs | Cell processes | Validated target
genes | Refs. |
|---|
| miR-221/222 | Cell apoptosis,
cell proliferation | p27Kip1, p57Kip2,
PUMA, PTEN | (48–50) |
| miR-17-92
cluster | Cell apoptosis,
cell proliferation | BIM, SOCS1,
IL-17RA, IL-17RE, IL-17RC | (18,36,51,52) |
| miR-181a/b | Cell apoptosis,
cell proliferation | PCAF | (36,53) |
| miR-106b-25
cluster | Cell apoptosis,
cell proliferation | PCAF | (36) |
| miR-32 | Cell apoptosis,
cell proliferation | PCAF | (36) |
| miR-135b | Cell
differentiation | SMAD5 | (54) |
| miR-125a | Cell apoptosis,
cell proliferation | P53 | (36) |
| miR-301a | Cell apoptosis,
cell proliferation | TIMP2 | (55) |
 | Table III.Principal tumor suppressor microRNAs
downregulated in MM. |
Table III.
Principal tumor suppressor microRNAs
downregulated in MM.
| MicroRNAs | Cell processes | Validated target
genes | Refs. |
|---|
| miR-125b | Cell apoptosis,
cell proliferation | IRF4 | (56) |
| Let-7b | Cell apoptosis,
cell proliferation | IGF-1R | (57) |
| miR-29 family | Cell apoptosis,
cell proliferation, cell migration | DNMT3A, DNMT3B,
PSME4, Sp1, CDK6, MCL-1 | (58–62) |
| miR-34a | Cell apoptosis,
cell proliferation | NOTCH1, BCL2,
CDK6 | (63–65) |
| miR-202 | Cell apoptosis,
cell proliferation | BAFF | (33,66) |
| miR-15a/16 | Cell apoptosis,
cell proliferation, cell migration, cell cycle, angiogenesis | FGFR1, PIK3α,
PI3KC2A, MDM4, VEGF | (38,67,68) |
| miR-214 | Cell apoptosis,
cell proliferation, cell cycle | PSMD10, ASF1B | (27) |
| miR-192, 194,
215 | Cell apoptosis,
cell proliferation, cell cycle | MDM2, IL-17R | (18) |
| miR-33b | Cell apoptosis,
cell proliferation, cell migration | PIM-1 | (69) |
| miR-126 | Cell
proliferation | C-MYC | (70) |
| miR-130b | Cell apoptosis,
cell proliferation | GR-α | (71) |
miR-17-92 cluster
miR-17-92 cluster, located in chromosome 13q31.3,
including miR-18a, miR-20a, miR-92, miR-17 and miR-19a/b, is
activated by the oncogene MYC proto-oncogene, BHLH transcription
factor (C-MYC) and its expression is upregulated in a variety of
types of cancer (72). It has
previously been verified that an abnormally elevated expression of
the miR-17-92 cluster is involved in the malignant progression of
MM (52,73). Pichiorri et al (36) reported that the miR-17-92 cluster was
significantly increased in the malignant PCs of patients with MM
compared with normal ones of HDs. Additionally, the cluster members
miR-19a and miR-19b were able to downregulate the protein
expression of suppressor of cytokine signaling 1 (SOCS1), and then
promote the proliferation of MM cells. It is well acknowledged that
SOCS1 is a negative regulator of the signaling pathways mediated by
IL6, and the precise mechanism is partially due to the fact that
its decreased expression may induce the phosphorylation of signal
transducer and activator of transcription 3 (STAT3), eventually
resulting in unlimited growth of tumor cells (31,74).
Furthermore, it has been revealed that miR-19 targeted the BCL2
like 11 gene and downregulated its protein expression, resulting in
reduced apoptosis and the increased proliferation of malignant PCs.
Although this cluster has a recognized carcinogenic effect in
tumors, studies have also revealed that it may function as a tumor
suppressor gene in inhibiting the proliferation in breast cancer
cells. Additionally, Gutierrez et al (75) reported that the expression of miR-20a,
miR-18a and miR-19b were downregulated in patients with MM with
retinoblastoma gene deletion. It was suggested that this cluster
may serve a different function in different subtypes of MM. It is
worth mentioning that no abnormal expression of the miR-17-92
cluster is present in MGUS, indicating that the miR-17-92 cluster
may participate in the progression from MGUS to MM and has the
potential to be used to distinguish MGUS from MM.
miR-29 family
The miR-29 family includes miR-29a, miR-29b and
miR-29c, have been reported to possess a significant tumor
inhibitory effect and are downregulated in hematological
malignancies via regulating cell proliferation, differentiation and
apoptosis (76,77). Amongst a variety of miRNAs, the miR-29
family represents a prototypical example of epi-miRNAs by targeting
epigenetic regulators including DNA methyltransferases (DNMTs). In
MM, miR-29b was revealed to target DNMTs, resulting in the
demethylation of SOCS1 and an increase of its protein expression.
SOCS1 inhibited the phosphorylation of its receptor, then inhibited
the activation of STAT3 by binding with Janus Kinase, which finally
resulted in the decreased proliferation of MM cells via the
suppression of the transcription of downstream genes (61). In addition, miR-29b may also
negatively regulate the PI3K/AKT signaling pathway. miR-29b
inhibited the phosphorylation of AKT and re-activated suppressed
apoptosis-promoting proteins, including P53, BCL2 associated
agonist of cell death, and caspase-9 by reducing the interaction of
AKT and its substrate glycogen synthase kinase 3β, which led to the
limitation of cell proliferation and an increase of cell apoptosis
(62). Because of these significant
tumor inhibitory effects, miRNA-29b has potential in clinical
application as a micromolecular nucleic acid drug. There have been
numerous in vivo studies that combine clinical routine
chemotherapy drugs with miRNAs. The results concluded that
miRNA-29b additionally possesses a strong tumor inhibitory effect
in vivo (78). In addition,
this miRNA family is considered to be associated with complications
in MM. Rossi et al (79)
revealed that miR-29b expression declined alongside osteoclast
differentiation, and its negative regulation of osteoclast activity
may overcome the strong pro-osteoclastic stimuli provided by MM
cells. Another previous study indicated that the expression of
miR-29c is negatively correlated with the severity of renal failure
and the expression level of β2-microglobulin (β2-M), but the
precise mechanism remains yet to be fully understood (80).
miR-181a/b
miR-181a/b, located in chromosome 1q32.1, is
reported to be upregulated in the malignant PCs of MGUS and MM.
This upregulation suggests that miR-181a/b may be involved in the
primary pathological course of MM (34,38). In
MM, the P53 protein may inhibit proliferation and promote the
apoptosis of tumor cells. The underlying mechanism may be
attributed in part to the fact that P53 may arrest the cell cycle
at the G1/S point and accelerating the DNA repairing process.
Generally, the P53 gene mutation is observed in the progression
from MM to plasma cell leukemia. miR-181a/b serves an important
function in regulating P53. A number of studies have revealed that
miR-181a/b may negatively regulate the expression of P-300-CBP
associated factor (PCAF), antagonize the positive effect of PCAF on
P53, and eventually result in the decreased expression of P53
(36). Additionally, miR-181a/b may
work as a histone acetyltransferase to keep P53 at a low level or
partially inactivated by controlling its stability through human
double minute 2. Furthermore, miR-181a/b is abnormally expressed in
two drug-resistance MM cell lines (U266 Dox resistant and 8226 Dox
resistant) (81), suggesting that
miR-181a/b may participate in the drug-resistance course of MM, but
the underlying mechanisms remain unclear.
miR-21
miR-21, located in chromosome 17q23.2, is one of the
most important onco-miRNAs in MM and its expression is
significantly elevated in the malignant PCs of MM (82). miR-21 is closely associated with the
bone marrow microenvironment. IL6, secreted by bone marrow
mesenchymal stem cells (BMSCs) may induce an elevated expression of
miR-21 through activating the STAT3 signaling pathway. Zheng et
al (83) revealed that, in MM
cells, miR-21 expression was positively correlated with an
abnormally increased expression of proteasome subunit β4 which may
promote the growth and proliferation of MM cells by stimulating the
NFκB-miR-21 signaling pathway. miR-21 is not only associated with
the malignant behavior of MM cells, but is additionally involved in
the drug-resistance behavior of MM cells. It is partly due to the
fact that miR-21 may target oncogene ras homolog gene family member
B and stimulate the NFκB signaling pathway, and then overcomes BMSC
induced drug-resistance of MM. Increasingly, evidence suggests that
miR-21 may become a novel therapeutic target for patients with MM
with severe drug-resistance (84).
miRNAs as potential diagnostic biomarkers of
MM
Bone marrow biopsy is the gold standard for the
clinical diagnosis of MM. However, this traditional diagnostic
method is invasive and is a painful procedure for patients. It is
urgent to identify a more sensitive, convenient and noninvasive
biomarker to apply in the clinical diagnosis of MM. ROC curve
analysis provides a regular way to assess the value of diagnosis.
Here, studies evaluating the diagnostic values of miRNAs have been
summarized in Table IV (14,35,43,85–88).
 | Table IV.Studies evaluating the diagnostic
values of miRNAs using ROC analysis. |
Table IV.
Studies evaluating the diagnostic
values of miRNAs using ROC analysis.
| Name | Sample | Sensitivity
(%) | Specificity
(%) | AUC | Cut-off value | Refs. |
|---|
| miR-720 | MM/MG vs. N | 87.2 | 92.3 | 0.9112 | 5,773.0000 | (85) |
| miR-1308 | MM/MG vs. N | 82.1 | 92.3 | 0.8920 | 405,400.0000 | (85) |
|
miR-720+miR-1308 | MM vs. MG | 97.4 | 92.3 | 0.9862 | 83.9000 | (85) |
|
miR-1246+miR-1308 | MM vs. MG | 79.2 | 66.7 | 0.7250 | 6.4000 | (85) |
| miR-29a | MM vs. N | 88.0 | 70.0 | 0.8320 | 0.0103 | (86) |
| miR-483 | MM vs. N | 50.0 | 90.0 | 0.7450 | 12.6900 | (87) |
| miR-20a | MM vs. N | 63.0 | 85.0 | 0.7400 | 478.9000 | (87) |
| miR-34a+Let-7e | MM vs. N | 80.6 | 86.7 | 0.8980 | ND | (14) |
|
miR-19a+miR-4254 | MM vs. N | 91.7 | 90.5 | 0.9500 | ND | (43) |
| miR-15a | MM vs. N | 100.0 | 73.0 | 0.8640 | 2.3500 | (88) |
| miR-16-1 | MM vs. N | 78.9 | 56.7 | 0.6640 | 3.1300 | (88) |
| miR-214 | MM with BD vs. MM
without BD | 97.0 | 86.0 | 0.7670 | ND | (35) |
| miR-135b | MM with BD vs. MM
without BD | 100.0 | 73.0 | 0.9070 | ND | (35) |
miRNAs may distinguish patients with
MM from HDs
Extracellular free miRNAs are protected by tiny
vesicles, exosomes, particles and apoptosis bodies, or bind to
proteins in serum, plasma, saliva, urine and other body fluids to
avoid being degraded (89). This
feature means that miRNAs possess the potential to be indicators of
MM in clinical applications (90).
Jones et al (85) obtained a
series of miRNAs whose expression differed between patients with MM
and HDs through gene array analysis of the peripheral serum in
patients with MM and conducted a large-scale validation for three
miRNAs (miR-720, miR-1308 and miR-1246) with a relatively large
difference of expression between them. ROC curve analysis revealed
that serum miR-720 yielded an area under the curve (AUC) of 0.9112
(P<0.001) with 87.2% sensitivity and 92.3% specificity for
discriminating patients with MGUS and MM from healthy controls. A
combination of miR-1308 and miR-720 provided a more powerful
diagnostic effect with the AUC rising to 0.9862, with a sensitivity
of 97.4% and specificity of 92.3%. Another previous study indicated
that miR-142-5p, miR-660 and miR-29a were upregulated in the serum
of patients with MM (86). Further
ROC curve analysis demonstrated that serum miR-29a expression
possessed a potent ability in discriminating patients with MM from
HDs with an AUC of 0.8320, with 88.0% sensitivity and 70.0%
specificity. Qu et al (87)
discovered that serum miR-483-5p and miR-20a possessed a
considerable diagnostic efficacy yielding an AUC of 0.7450
(sensitivity 58.0%, specificity 90.0%) and 0.7400 (sensitivity
63.0%, specificity 85.0%), respectively. Furthermore, a previous
study concluded that the combined application of the serum
expression of miR-19a with miR-4254 had a significant diagnostic
value with an AUC of 0.9500, with a sensitivity of 91.7% and
specificity of 90.5% (43).
These studies utilizing ROC analysis suggest that
miRNAs possess considerable diagnostic efficacy as serological
markers for MM, and have advantages over traditional diagnostic
methods, including convenience and noninvasiveness. Despite the
fact that the preliminary results are positive, larger samples are
required for further validation. Additionally, the methods of
specimen collection, RNA extraction and data analysis varied
between different studies, and it is necessary to set up a unified
standard in the process of testing.
Correlation between miRNAs and
clinical parameters of MM
miRNAs are closely associated with the occurrence
and development of MM and their expression levels are significantly
correlated with a number of common laboratory biomarkers of MM
(91). Therefore, miRNAs may be used
as indicators to reflect the severity of MM. It was identified that
serum miR-214 and miR-135b expression levels had the ability to
distinguish between patients with MM with or without bone disease
and may reflect the severity of bone lesions (35). The AUC of miR-214 was 0.767 with 97%
sensitivity and 86% specificity, and the serum level of miR-135b
was a powerful diagnostic tool in the identification of MM
associated bone diseases with an AUC of 0.907, sensitivity of 100%
and specificity of 73%. Additionally, these two miRNAs were
positively correlated with the severity of bone disease. Kubiczkova
et al (14) revealed that
serum miR-744, miR-130a, let-7d, and let-7e levels were positively
correlated with hemoglobin levels in patients with MM and the
expression levels of miR-744, miR-130a, let-7d, and let-7e were
positively correlated with the thrombocyte count and significantly
negatively correlated with creatinine and β-2 microglobulin levels.
Furthermore, the expression levels of miR-744, let-7d and let-7e
were positively associated with albumin levels and miR-744, let-7e
were negatively associated with C-reactive protein. Only the let-7e
expression was negatively correlated with monoclonal immunoglobulin
levels.
miRNAs possess advantages in sensitivity and
specificity of detection compared with conventional hematological
auxiliary diagnostic indicators. Furthermore, one abnormally
expressed miRNA may have a correlation with multiple conventional
hematological parameters (14). This
quality will increase their convenience for clinical application. A
number of miRNAs were also significantly associated with
imagological examination. A previous study investigated the ability
of miR-214 and miR-135b to distinguish between patients with MM
with and without bone disease using the ROC analysis and it was
revealed that they possess greater potential as an alternative of
imageological examination due to the fact that they are more
convenient and economical (35).
miRNAs as potential prognostic biomarkers of
MM
MM is a hematological tumor with obvious
heterogeneity. Patients with different stages and classifications
vary in therapeutic strategies and clinical outcomes. Clinical
research on the prognosis of MM has been continuously progressing.
Presently, commonly used indicators for prognosis of MM are usually
based on cytogenetic abnormities including Translocation/Cyclin D
(TC), International Staging System (ISS) and fluorescence
hybridization (FISH) of malignant PCs, and gene expression
profiling (GEP) (92). These
classification methods have clinical effectiveness to some extent,
but cannot accurately reflect all genetic mutations during the
process of MM. miRNAs may regulate almost all cellular processes
with a more comprehensive reflection of the dysfunctional state of
patients with MM. Therefore, miRNAs are more likely to become novel
effective biomarkers for predicting the prognosis of MM patients.
Table V (14,15,35,43,87,88,93,94)
summarizes studies evaluating the prognostic values of miRNAs using
survival analysis.
 | Table V.Studies evaluating the prognostic
values of miRNAs using survival analysis. |
Table V.
Studies evaluating the prognostic
values of miRNAs using survival analysis.
| Name | P-value | HR (95% CI) | Method | Refs. |
|---|
| RFS |
|
|
|
|
|
miR-20a | 0.0100 | ND | K-M | (93) |
|
miR-148a | 0.0200 | ND | K-M | (93) |
| PFS |
|
|
|
|
|
miR-25 | 0.0340 | 0.920 (0.840,
0.990) | Univariate Cox
regression | (94) |
|
miR-483 | 0.0250 | ND | K-M | (87) |
|
miR-33b | 0.0160 | ND | Univariate Cox
regression | (15) |
|
miR-19 | 0.0030 | 2.787 (1.421,
5.468) | Multivariable Cox
regression | (43) |
|
miR-15a | 0.0080 | 0.260 (0.090,
0.710) | Multivariable Cox
regression | (88) |
|
miR-214 | 0.0150 | ND | K-M | (35) |
| OS |
|
|
|
|
|
miR-25 | 0.0005 | 0.810 (0.720,
0.910) | Univariate Cox
regression | (94) |
|
miR-25 | 0.0130 | 0.760 (0.620,
0.940) | Multivariable Cox
regression | (94) |
|
miR-744 | 0.0001 | 0.670 (0.548,
0.819) | Univariate Cox
regression | (14) |
|
Let-7e | 0.0020 | 0.611 (0.450,
0.829) | Univariate Cox
regression | (14) |
|
miR-33b | 0.0330 | ND | Univariate Cox
regression | (15) |
|
miR-19 | 0.0230 | 2.995 (1.167,
7.690) | Multivariable Cox
regression | (43) |
|
miR-15a | 0.0350 | 0.280 (0.080,
0.930) | Multivariable Cox
regression | (88) |
|
miR-214 | 0.0020 | ND | K-M | (35) |
Correlation of miRNAs and conventional
prognostic biomarkers of MM
Wu et al (95)
analyzed miRNA expression profiling in CD138+ cells from the bone
marrow tissues of patients with MM and followed them up. A distinct
upregulation of the miRNA cluster miR-99b/let-7e/miR-125a on 19q
was identified in TC 4p16 cases, and additionally
miR-150/miR-155/miR-34a were upregulated in the MAF subgroup. An
outcome scale model was established based on the combined
application of miR-17 and miR-886-5p, and patients were divided
into high-, moderate- and low-risk groups (P=0.001) according to
their overall survival (OS). The prediction accuracy of the
combined application of these two miRNAs is 98.46%, much greater
than that of the outcome scale approach based on ISS/FISH
(P=0.0004). This novel outcome scale mode based on miRNAs was also
able to identify that patients with MM carrying the t(4;14)
mutation and patients carrying the t(4;14) mutation with a lower
expression of these two miRNAs had a longer OS (71 months).
Additionally, Huang et al (93) conducted FISH tests in malignant PCs
and then detected miRNA expression levels in plasma of patients
with MM. The expression level of miR-99b in the plasma of patients
with MM was revealed to be significantly correlated with t(4;14;
immunoglobulin heavy locus; fibroblast growth factor receptor 3)
translocation and the low expression of miR-221 was significant
associated with del(13q14). Qu et al (87) observed the significantly association
between plasma miR-483-5p expression levels and the ISS stage.
Furthermore, Kubiczkova et al (14) revealed that serum miR-744, let-7d, and
let-7e expression levels were positively correlated with ISS
staging in patients with MM and serum let-7e expression level was
significantly associated with del (13q14) obtained from the FISH
tests of MM PCs.
These results indicated that miRNAs have a huge
potential to function as prognostic indicators. Furthermore, as
they may be detected more conveniently, circulating miRNAs may
offer an advantage over the traditional FISH detection of malignant
PCs in bone marrow.
Survival analyses of miRNAs in MM
In addition to being associated with conventional
prognostic markers, long-term follow-up studies may reveal the
association between miRNA expression and the life expectancy of
patients with MM directly. Huang et al (93) assessed the association between plasma
miRNAs and the life expectancy of patients using Kaplan-Meier
survival curves. Results demonstrated that high expression levels
of plasma miR-20a and miR-148a in patients with MM were
significantly correlated with relapse-free survival (P=0.01 and
P=0.02, respectively). Rocci et al (94) selected ten miRNAs which had stable
serum expression for a follow-up study. Results indicated that
patients with MM with higher levels of miR-25 [hazard ratio
(HR)=0.81; P=0.0005], miR-16 (HR=0.87; P=0.008) and miR-30a
(HR=0.86, P=0.016) in their serum had a longer OS than those with a
lower expression of these miRNAs. Furthermore, serum miR-25
expression level was significantly correlated with progression-free
survival (PFS; P=0.034). Additionally, it was concluded that miR-25
and miR-16 were independent prognostic indicators for the OS of
newly diagnosed patients with MM using Cox regression analysis. Hao
et al (35) reported that the
high expression of serum miR-21 may be an effective indicator of
the poor outcomes in patients with MM. Patients with high
expression level of serum miR-214 had significant shortened PFS
(P=0.015) and OS (P=0.002) compared with those with a low
expression level.
miRNAs have proved to be effective indicators in
survival analysis and risk evaluation. The next step of these
studies should focus on validating these biomarkers in larger
samples and then combining utilization with conventional prognostic
markers including ISS, FISH and GEP to improve the accuracy of risk
stratification and outcome prediction.
miRNAs used for predicting of
therapeutic response in MM
Patients with MM have varied therapeutic responses
and a number of patients are resistant to one or more chemotherapy
drugs, which bring a number of difficulties to clinical treatment.
It is imperative to discover novel clinical indicators that may
effectively predict the therapeutic efficacy. ASCT is a recognized
effective therapy for MM, but its therapeutic efficacy remains
different for different individuals. Navarro et al (96) investigated the difference of serum
miRNA expression profiling in patients with MM prior to and
following performing ASCT. Results indicated that patients with a
high expression of serum miR-19b and miR-331 had significantly
prolonged PFS subsequent to receiving ASCT (P<0.001 and P=0.001,
respectively) and combined application of these two miRNAs may be a
more effective predicting indicator for PFS following ASCT (HR=5.3,
P=0.033). Hao et al (43)
identified that patients with a low expression of serum miR-19a had
an improved therapeutic response to bortezomib (BZ) and
significantly extended PFS and OS following BZ treatment (P=0.002),
whereas patients with a high expression of serum miR-19a had no
obvious efficacy and no improvement in survival analysis. Li et
al (88) investigated whether
miR-15a downregulated in bone marrow tissues may influence the
response of patients to different therapies. It was revealed that
therapy based on thalidomide and BZ may not remarkably improve the
PFS and OS of patients with a low-expression of miR-15a. Another
previous study on miRNA expression profiling analysis associated
with serum exosomes revealed that the expression levels of serum
exosome derived miR-16-5p, miR-15a-5p, miR-20a-5p and miR-17-5p had
increased 3.91, 1.83, 2.96 and 1.97 times, respectively, in the
BZ-effective group compared with those in the BZ-resistant group
(97).
These previous studies suggest that different miRNA
expression profiling are significantly associated with the varied
therapeutic responses of patients and may be used as predicting
indicators to monitor therapeutic responses in patients with MM. In
addition, these miRNAs may also provide an important basis for
individual treatment.
Dynamics of miRNA levels during MM
progression
As deregulated miRNA expression in patients with
MGUS and MM was observed at the time of diagnosis, the further aim
for researchers was to check if this profiling changed during
disease progression. Kubiczkova et al (14) collected serum samples at the time of
diagnosis and relapse (following 2 lines of treatment), and
revealed that almost all miRNAs in MM samples were significantly
different from HDs (miR-744: FC=0.270, miR-130a: FC=0.487, miR-34a:
FC=10.083, let-7d: FC=0.243, let-7e: FC=0.300). Furthermore, a
significant increase of miR-34a (P<0.0001, FC=3.560) and
decrease of let-7d (P=0.0182, FC=0.460) were revealed in relapsed
samples compared with samples at the time of first diagnosis.
Yyusnita et al (40) claimed
that four miRNAs (miR-494, 130a, let-7i, let-7c) in peripheral
blood samples were exclusively expressed in novel MM cases whereas
nine (miR-148a, 1225, 423, 484, 99a, 106a, 224, 638, let-7b) were
exclusively expressed in follow-up MM cases. Hao et al
(43) indicated that miR-19a and
miR-4254 expression levels were closer to the levels in HDs when in
a remission phase. However, downregulated miR-19a and upregulated
miR-4254 levels returned to diagnostic levels during relapse. These
results demonstrated that the expression of miRNAs varied in
patients with different stages of MM and had utility in monitoring
MM progression.
miRNA-based therapeutic strategies in
MM
With the exception of ASCT, traditional therapies of
MM are usually based on the combined treatment of multiple
chemotherapy drugs, including immunomodulatory drugs (lenalidomide
and thalidomide), proteasome inhibitors (BZ), alkylating agents
(melphalan and cyclophosphamide), and steroid hormones
(dexamethasone). In spite of advances in present therapies, a low
remission rate and drug-resistance towards the traditional
chemotherapy regimen in patients with MM often result in
difficulties in the clinical cure of this disease. Multiple
signaling pathways may enhance the growth, proliferation, survival
and migration of MM cells, and additionally induce the
drug-resistance of MM cells in the pathological process of MM
(98). miRNA-based therapy may
selectively downregulate genes involved in the pathological
processes of MM. In addition to targeting genes effectively, this
therapy has the advantage of being safer. Gallo Cantafio et
al (99) studied the
pharmacokinetics and pharmacodynamics of the
13-merLNA-inhibitor-miR-221. The results of this previous study
indicated that LNA-i-miR-221 harbored a short half-life, optimal
tissue bioavailability and minimal urine excretion in mice and
monkeys and revealed no toxicity in the pilot monkey study.
Therefore, miRNA-based therapy is a promising novel therapeutic
method for MM (100,101).
miRNAs as novel drugs used in MM
miRNAs may regulate the growth of tumor cells
through suppressing target gene expression. This therapeutic
strategy has proved effective in vivo by an increasing
number of studies. Di Martino et al (65) confirmed that miR-34a had a significant
tumor inhibitory effect on severe combined immunodeficient (SCID)
mice. First, miR-34a was proved to downregulate oncogenic
expression, including BCL2, apoptosis regulator, cyclin dependent
kinase 6 and Notch 1, at mRNA and protein levels in vitro.
MM cells overexpressed miR-34a demonstrated growth inhibition, low
colony formation activity and increased apoptosis. Next, in in
vivo studies, researchers conducted a xenograft tumor
experiment through transfecting MM cells with lentiviral miR-34a
over-expression vector then injecting these cells in SCID mice.
Lentiviral vector-transduced MM xenografts with constitutive
miR-34a expression demonstrated high growth inhibition in SCID mice
with a significant inhibition of tumor formation (P<0.05) and
the decrease of the tumor average size (P=0.008). Subsequently, in
order to investigate the anti-tumor effect of lipid-formulated
miR-34a, miR-34a or miR-NC were administered into neutral lipid
emulsion (NLE) particles and make a systemic delivery of
oligonucleotides in mice. Results indicated that, after 21 days,
tumors were inhibited in 50% of mice treated with formulated
miR-34a mimics. In addition, a prolongation of survival time
(P=0.0009) was observed in mice treated with miR-34a mimics
compared with control groups. Additionally, a biopolymeric 3D
scaffold constructed by MM cells and BMSCs was implanted into SCID
mice to simulate a bone marrow microenvironment-dependent effect on
MM cells. The significant inhibition of tumor growth (P<0.01)
and prolonged survival (P=0.041) were observed in mice with miR-34a
overexpression, which indicated that miR-34a may overcome the
protective function of BMSCs for MM cells and suppress tumors.
Therapeutic effects of miRNAs were dependent on
different vectors, including lentivirus, liposome and NLE.
Furthermore, compared with traditional chemotherapeutic drugs,
miRNAs have the obvious therapeutic efficacy in vivo without
toxic reaction, release of inflammatory factors or other drug side
effects.
Advantages of combined application of
miRNAs and chemotherapy drugs
Previous studies indicated that the combined use of
chemotherapy drugs and miRNAs may improve the therapeutic efficacy
compared with a single application of one miRNA or chemotherapy
drug and reduced drug-resistance of traditional chemotherapy
drugs.
Glucocorticoid (GC) is frequently included in the
chemotherapeutic regimen for lymphoid malignant tumors because of
its capability for killing lymphoid cells. In the long-term,
high-dose GC will result in the reduction of glucocorticoid
receptors (GR), and then induce resistance to GC. However, low-dose
GC does not have enough therapeutic efficacy. Palagani et al
(102) investigated the possibility
of improving this situation using a miRNA combined application with
GC. The results of this previous study confirmed that the miR-150
synthetic vector combined with low-dose GC had a synergistic
therapeutic effect on MM cells by markedly raising the cell
sensitivity for GC-induced death. Furthermore, miR-150 may induce
the specific response of GR via indirectly regulating the mRNAs of
the proteins interacting with GR, including hormone receptors,
molecular chaperones, unfolded protein stress and transcriptional
factors, so that resistance to GC is unlikely to occur. In
addition, miR-150 in tiny vesicles may also be considered to be a
monitoring biomarker of therapeutic response to GC (103). Zhao et al (50) reported that mir-221/222 family
expression was elevated in GC-resistance cell lines MM1R and
further study verified that this miRNA family reduced the
drug-resistance of MM cells to dexamethasone (Dex) and promoted the
survival of MM cells by BCL2 binding component 3/BCL2
antagonist/killer 1/BCL2 associated X, apoptosis regulator
signaling pathway. Therefore, miR-221/222 antagonists may be
promising therapeutic targets for reversing the drug-resistance of
MM cells to Dex. Subsequently to the construction of a xenograft
tumor model in SCID mice, mice treated with miR-221/222 antagonist
and Dex had significantly longer survival than the mice treated
only with Dex (P<0.05). In addition, Gulla et al
(48) reported that the inhibition of
miR-221/222 may restore melphalan sensitivity in MM and induced
apoptosis of MM cells in vitro. It was also revealed
LNA-i-miR-221 may perform anti-MM activity by systemic delivery
in vivo. They demonstrated the rationale of the combined use
of LNA-i-miR-221 and melphalan in drug-refractory stage of patients
with MM.
BZ may inhibit the formation of proteasome, so that
misfolded and short-lived proteins may not be eliminated, leading
to the death of tumor cells eventually. At present, BZ has been
widely used in the standard treatment regimen of MM, but BZ has a
number of side effects. For example, the enhanced accumulation of
polyubiquitin in cells results in an increase of protein
aggregation and autophagosome to eliminate excessive polyubiquitin.
This autophagy behavior eventually contributes to the occurrence of
drug-resistance in tumor cells. Therefore, it is warranted to seek
a type of chemotherapy drug that may eliminate proteasomes in cells
without increased protective autophagy behavior. Jagannathan et
al (59) identified that the
increased expression of miR-29b reinforced the BZ-induced
aggregation of polyubiquitin without inducing the formation of a
protein autophagosome. miR-29b may result in proteasome degradation
and cell death by targeting proteasome activator subunit 4, and
this pattern does not affect the accumulation of ubiquitinated
proteins and is different from the mechanism of BZ-induced
proteasome degradation. Additionally, the combined application of
miRNA with BZ may decrease drug-resistance to BZ. A previous study
concerning miR-29b and BZ disclosed that miR-29b with transcription
factor Sp1 transcription factor may increase the sensitivity of MM
cells to BZ and led to the increased apoptosis of MM cells through
the PI3K-AKT signaling pathway (62).
The group treated with BZ and miR-29b had a significantly enhanced
effect on cell apoptosis, compared with the control group
(P<0.01). Furthermore, Wang et al (84) reported that miR-21 combined with BZ,
Dex, and doxorubicin (Dox) had a synergistic effect in killing MM
cells and were demonstrated to be more effective than the
application of any drug alone.
PRIMA-1Met is a novel micromolecular
chemotherapy drug (104). The
therapeutic efficacy of the combination of PRIMA-1Met
with miR-29a for MM treatment was previously studied (105). miR-29a was considered to be a
TS-miRNA and served an important function in the
PRIMA-1Met induced cell apoptosis by targeting the C-MYC
gene. Further xenograft tumor experiments on SCID mice verified
that miR-29a combined with PRIMA-1Met may significantly
inhibit the growth of tumors and the extended survival time of SCID
mice, which provided a novel therapeutic strategy for the treatment
of MM.
Preliminary basic studies of miRNAs in the treatment
of MM have made progress, but its clinical application remains in
its infancy and faces several challenges. Tumor formation and
treatment duration in animal models are markedly different from
those in human beings. It remains unclear if miRNAs exert a similar
effect and long-term function in human beings. Furthermore, vectors
that deliver miRNAs in vivo remain to be improved. It should
be ensured that during the process of delivering, carriers are safe
and produce no toxic or other side effects, and transported nucleic
acid molecules are stable without degradation by endogenic
nuclease, which requires more effective chemical modification
technology to improve current vectors for clinical application.
Conclusion
Basic studies of tumors has always been an area of
concentrated research, but the transformation from preliminary
basic studies to clinical application is a problem at present. A
number of researchers put forward the concept of ‘Theragnostics’,
representing cutting-edge, multi-disciplinary strategies that
combine diagnostics with therapeutics in order to generate
personalized therapies and improve the outcomes of tumor patients
(13). Application of theragnostics
in patients with clinical cancer may raise their survival rate,
with a more accurate diagnosis of cancer and optimized choice of
treatment regimen.
Circulating miRNAs possess potential to become novel
biomarkers obtained in a non-invasive manner (106), but the origin they derive from is
still unclear. Furthermore, which is the best detection source out
of whole blood, plasma, serum and exosomes remains controversial
(89). Kubiczkova et al
(14) discussed the derivation of
several miRNAs in the exosomal and exosome-depleted supernatant of
six newly diagnosed patients with MM. Concentration of miR-744,
miR-130a, let-7d and let-7e (all P<0.05) were revealed to be
significantly higher in the exosome pellet compared with the
exosome-depleted supernatant. Previous studies considered that
exosomal miRNA profiling was superior for detecting pathology in
secretory cell types. In addition, there remains a lack of
standardized protocols for sample collection, small RNA extraction
and data-analytical methods when quantifying miRNAs.
miRNAs and their target genes represent basic
networks, regulating a variety of cellular functions and that are
also the rationale of miRNA-based therapeutic strategies. However,
there are multiple miRNAs targeting the same gene, thus interfering
with a single miRNA may not produce enough effects;
correspondingly, one miRNA may work on multiple targets, which will
result in unknown side effects in the long term. Furthermore, in
the delivery system, it is necessary to ascertain the right way to
inject these vectors to achieve the highest efficacy in the safest
manner.
Acknowledgements
Not applicable.
Funding
The present review was supported by the National
Nature Science Foundation of China (grant nos. 81301498 and
81271920), the Project of Jiangsu Provincial Department of Health
and Family Planning Commission (grant no. H201526), the Project of
the Jiangsu Provincial Six Talent Peaks (grant no. WS-066), the
Social Science and Technology Innovation and Demonstration Project
in Nantong (grant no. HS2014059) and the Innovation Project of
Graduate Students of Nantong University (grant no. YKC15099).
Availability of data and materials
Not applicable.
Authors' contributions
BZ wrote the initial draft and designed the outline
of the manuscript. HCo, SJ and XS designed the outline and revised
the manuscript. HCh revised and expanded the manuscript. YZ and XL
contributed to the acquisition and analysis of data for the work.
All authors have seen and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Rebecca RL, Kimberly KD and Jemal A:
Cancer statistics, 2018. CA Cancer J Clin. 68:7–30. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Durie BGM: Patient Handbook: Multiple
Myeloma, Cancer of the Bone Marrow. International Myeloma
Foundation; North Hollywood, CA: 2010
|
|
3
|
Ailawadhi S, Aldoss IT, Yang D, Razavi P,
Cozen W, Sher T and Chanan-Khan A: Outcome disparities in multiple
myeloma: A SEER-based comparative analysis of ethnic subgroups. Br
J Haematol. 158:91–98. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rollig C, Knop S and Bornhauser M:
Multiple myeloma. Lancet. 385:2197–2208. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhao Q, Luo F, Ma J and Yu X: Bone
metastasis-related MicroRNAs: New targets for treatment. Curr
Cancer Drug Targets. 15:716–725. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rossi M, Tagliaferri P and Tassone P:
MicroRNAs in multiple myeloma and related bone disease. Ann Transl
Med. 3:3342015.PubMed/NCBI
|
|
7
|
Rocci A, Hofmeister CC and Pichiorri F:
The potential of miRNAs as biomarkers for multiple myeloma. Expert
Rev Mol Diagn. 14:947–959. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bi CL and Chng WJ: miRNA deregulation in
multiple myeloma. Chin Med J (Engl). 124:3164–3169. 2011.PubMed/NCBI
|
|
9
|
Ghobrial IM: Myeloma as a model for the
process of metastasis: Implications for therapy. Blood. 120:20–30.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cottini F and Anderson K: Novel
therapeutic targets in multiple myeloma. Clin Adv Hematol Oncol.
13:236–248. 2015.PubMed/NCBI
|
|
11
|
Kumar SK, Rajkumar SV, Dispenzieri A, Lacy
MQ, Hayman SR, Buadi FK, Zeldenrust SR, Dingli D, Russell SJ, Lust
JA, et al: Improved survival in multiple myeloma and the impact of
novel therapies. Blood. 111:2516–2520. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kumar SK, Dimopoulos MA, Kastritis E,
Terpos E, Nahi H, Goldschmidt H, Hillengass J, Leleu X, Beksac M,
Alsina M, et al: Natural history of relapsed myeloma, refractory to
immunomodulatory drugs and proteasome inhibitors: A multicenter
IMWG study. Leukemia. 31:2443–2448. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ahmad N, Haider S, Jagannathan S, Anaissie
E and Driscoll JJ: MicroRNA theragnostics for the clinical
management of multiple myeloma. Leukemia. 28:732–738. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kubiczkova L, Kryukov F, Slaby O,
Dementyeva E, Jarkovsky J, Nekvindova J, Radova L, Greslikova H,
Kuglik P, Vetesnikova E, et al: Circulating serum microRNAs as
novel diagnostic and prognostic biomarkers for multiple myeloma and
monoclonal gammopathy of undetermined significance. Haematologica.
99:511–518. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li F, Hao M, Feng X, Zang M, Qin Y, Yi S,
Li Z, Xu Y, Zhou L, Sui W, et al: Downregulated miR-33b is a novel
predictor associated with disease progression and poor prognosis in
multiple myeloma. Leuk Res. 39:793–799. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shukla GC, Singh J and Barik S: MicroRNAs:
Processing, maturation, target recognition and regulatory
functions. Mol Cell Pharmacol. 3:83–92. 2011.PubMed/NCBI
|
|
17
|
Zhang Q, Yan W, Bai Y, Xu H, Fu C, Zheng
W, Zhu Y and Ma J: Synthetic miR-145 mimic inhibits multiple
myeloma cell growth in vitro and in vivo. Oncol Rep. 33:448–456.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sun Y, Pan J, Mao S and Jin J:
IL-17/miR-192/IL-17Rs regulatory feedback loop facilitates multiple
myeloma progression. PLoS One. 9:e1146472014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang Y, Li F, Saha MN, Abdi J, Qiu L and
Chang H: miR-137 and miR-197 induce apoptosis and suppress
tumorigenicity by targeting MCL-1 in multiple myeloma. Clin Cancer
Res. 21:2399–2411. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Macfarlane LA and Murphy PR: MicroRNA:
Biogenesis, function and role in cancer. Curr Genomics. 11:537–561.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rossi M, Amodio N, Di Martino MT,
Tagliaferri P, Tassone P and Cho WC: MicroRNA and multiple myeloma:
From laboratory findings to translational therapeutic approaches.
Curr Pharm Biotechnol. 15:459–467. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Di Martino MT, Amodio N, Tassone P and
Tagliaferri P: Functional analysis of microRNA in multiple myeloma.
Methods Mol Biol. 1375:181–194. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Raimondi L, De Luca A, Morelli E,
Giavaresi G, Tagliaferri P, Tassone P and Amodio N: MicroRNAs:
Novel crossroads between myeloma cells and the bone marrow
microenvironment. Biomed Res Int. 2016:65045932016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Calin GA, Dumitru CD, Shimizu M, Bichi R,
Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al:
Frequent deletions and down-regulation of micro-RNA genes miR15 and
miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci
USA. 99:15524–15529. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pichiorri F, Suh SS, Rocci A, De Luca L,
Taccioli C, Santhanam R, Zhou W, Benson DM Jr, Hofmainster C, Alder
H, et al: Downregulation of p53-inducible microRNAs 192, 194 and
215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma
development. Cancer Cell. 18:367–381. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Misiewicz-Krzeminska I, Sarasquete ME,
Quwaider D, Krzeminski P, Ticona FV, Paíno T, Delgado M, Aires A,
Ocio EM, García-Sanz R, et al: Restoration of microRNA-214
expression reduces growth of myeloma cells through positive
regulation of P53 and inhibition of DNA replication. Haematologica.
98:640–648. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang Q, Wang LQ, Wong KY, Li ZY and Chim
CS: Infrequent DNA methylation of miR-9-1 and miR-9-3 in multiple
myeloma. J Clin Pathol. 68:557–561. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cipolla GA, Park JK, de Oliveira LA,
Lobo-Alves SC, de Almeida RC, Farias TD, Lemos Dde S, Malheiros D,
Lavker RM and Petzl-Erler ML: A 3′UTR polymorphism marks
differential KLRG1 mRNA levels through disruption of a miR-584-5p
binding site and associates with pemphigus foliaceus
susceptibility. Biochim Biophys Acta. 1859:1306–1313. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhao JJ and Carrasco RD: Crosstalk between
microRNA30a/b/c/d/e-5p and the canonical Wnt pathway: Implications
for multiple myeloma therapy. Cancer Res. 74:5351–5358. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Brocke-Heidrich K, Kretzschmar AK, Pfeifer
G, Henze C, Löffler D, Koczan D, Thiesen HJ, Burger R, Gramatzki M
and Horn F: Interleukin-6-dependent gene expression profiles in
multiple myeloma INA-6 cells reveal a Bcl-2 family-independent
survival pathway closely associated with Stat3 activation. Blood.
103:242–251. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Leotta M, Biamonte L, Raimondi L,
Ronchetti D, Di Martino MT, Botta C, Leone E, Pitari MR, Neri A,
Giordano A, et al: A p53-dependent tumor suppressor network is
induced by selective miR-125a-5p inhibition in multiple myeloma
cells. J Cell Physiol. 229:2106–2116. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shen X, Guo Y, Yu J, Qi J, Shi W, Wu X, Ni
H and Ju S: miRNA-202 in bone marrow stromal cells affects the
growth and adhesion of multiple myeloma cells by regulating B
cell-activating factor. Clin Exp Med. 16:307–316. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhou Y, Chen L, Barlogie B, Stephens O, Wu
X, Williams DR, Cartron MA, van Rhee F, Nair B, Waheed S, et al:
High-risk myeloma is associated with global elevation of miRNAs and
overexpression of EIF2C2/AGO2. Proc Natl Acad Sci USA.
107:7904–7909. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hao M, Zang M, Zhao L, Deng S, Xu Y, Qi F,
An G, Qin Y, Sui W, Li F, et al: Serum high expression of miR-214
and miR-135b as novel predictor for myeloma bone disease
development and prognosis. Oncotarget. 7:19589–19600. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pichiorri F, Suh SS, Ladetto M, Kuehl M,
Palumbo T, Drandi D, Taccioli C, Zanesi N, Alder H, Hagan JP, et
al: MicroRNAs regulate critical genes associated with multiple
myeloma pathogenesis. Proc Natl Acad Sci USA. 105:12885–12890.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Unno K, Zhou Y, Zimmerman T, Platanias LC
and Wickrema A: Identification of a novel microRNA cluster
miR-193b-365 in multiple myeloma. Leuk Lymphoma. 50:1865–1871.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Roccaro AM, Sacco A, Thompson B, Leleu X,
Azab AK, Azab F, Runnels J, Jia X, Ngo HT, Melhem MR, et al:
MicroRNAs 15a and 16 regulate tumor proliferation in multiple
myeloma. Blood. 113:6669–6680. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chi J, Ballabio E, Chen XH, Kušec R,
Taylor S, Hay D, Tramonti D, Saunders NJ, Littlewood T, Pezzella F,
et al: MicroRNA expression in multiple myeloma is associated with
genetic subtype, isotype and survival. Biol Direct. 6:232011.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yyusnita, Norsiah, Zakiah I, Chang KM,
Purushotaman VS, Zubaidah Z and Jamal R: MicroRNA (miRNA)
expression profiling of peripheral blood samples in multiple
myeloma patients using microarray. Malays J Pathol. 34:133–143.
2012.PubMed/NCBI
|
|
41
|
Campo S, Allegra A, D'Ascola A, Alonci A,
Scuruchi M, Russo S, Avenoso A, Gerace D, Campo GM and Musolino C:
MiRNome expression is deregulated in the peripheral lymphoid
compartment of multiple myeloma. Br J Haematol. 165:801–813. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang W, Corrigan-Cummins M, Barber EA,
Saleh LM, Zingone A, Ghafoor A, Costello R, Zhang Y, Kurlander RJ,
Korde N, et al: Aberrant Levels of miRNAs in Bone marrow
microenvironment and peripheral blood of myeloma patients and
disease progression. J Mol Diagn. 17:669–678. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hao M, Zang M, Wendlandt E, Xu Y, An G,
Gong D, Li F, Qi F, Zhang Y, Yang Y, et al: Low serum miR-19a
expression as a novel poor prognostic indicator in multiple
myeloma. Int J Cancer. 136:1835–1844. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:2015.doi: 10.7554/eLife.05005. View Article : Google Scholar
|
|
45
|
Betel D, Koppal A, Agius P, Sander C and
Leslie C: Comprehensive modeling of microRNA targets predicts
functional non-conserved and non-canonical sites. Genome Biol.
11:R902010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Anders G, Mackowiak SD, Jens M, Maaskola
J, Kuntzagk A, Rajewsky N, Landthaler M and Dieterich C: doRiNA: A
database of RNA interactions in post-transcriptional regulation.
Nucleic Acids Res. 40:D180–D186. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Shang J, Yao Y, Fan X, Shangguan L, Li J,
Liu H and Zhou Y: miR-29c-3p promotes senescence of human
mesenchymal stem cells by targeting CNOT6 through p53-p21 and
p16-pRB pathways. Biochim Biophys Acta. 1863:520–532. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gulla A, Di Martino MT, Cantafio Gallo ME,
Morelli E, Amodio N, Botta C, Pitari MR, Lio SG, Britti D, Stamato
MA, et al: A 13 mer LNA-i-miR-221 inhibitor restores drug
sensitivity in melphalan-refractory multiple myeloma cells. Clin
Cancer Res. 22:1222–1233. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Di Martino MT, Gullà A, Cantafio ME,
Lionetti M, Leone E, Amodio N, Guzzi PH, Foresta U, Conforti F,
Cannataro M, et al: In vitro and in vivo anti-tumor activity of
miR-221/222 inhibitors in multiple myeloma. Oncotarget. 4:242–255.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhao JJ, Chu ZB, Hu Y, Lin J, Wang Z,
Jiang M, Chen M, Wang X, Kang Y, Zhou Y, et al: Targeting the
miR-221-222/PUMA/BAK/BAX pathway abrogates dexamethasone resistance
in multiple myeloma. Cancer Res. 75:4384–4397. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Galm O, Yoshikawa H, Esteller M, Osieka R
and Herman JG: SOCS-1, a negative regulator of cytokine signaling,
is frequently silenced by methylation in multiple myeloma. Blood.
101:2784–2788. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chen L, Li C, Zhang R, Gao X, Qu X, Zhao
M, Qiao C, Xu J and Li J: miR-17-92 cluster microRNAs confers
tumorigenicity in multiple myeloma. Cancer Lett. 309:62–70. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Peng J, Thakur A, Zhang S, Dong Y, Wang X,
Yuan R, Zhang K and Guo X: Expressions of miR-181a and miR-20a in
RPMI8226 cell line and their potential as biomarkers for multiple
myeloma. Tumour Biol. 36:8545–8552. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Xu S, Cecilia SG, De Veirman K, Vande
Broek I, Leleu X, De Becker A, Van Camp B, Vanderkerken K and Van
Riet I: Upregulation of miR-135b is involved in the impaired
osteogenic differentiation of mesenchymal stem cells derived from
multiple myeloma patients. PLoS One. 8:e797522013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Liang B, Yin JJ and Zhan XR: MiR-301a
promotes cell proliferation by directly targeting TIMP2 in multiple
myeloma. Int J Clin Exp Pathol. 8:9168–9174. 2015.PubMed/NCBI
|
|
56
|
Morelli E, Leone E, Cantafio ME, Di
Martino MT, Amodio N, Biamonte L, Gullà A, Foresta U, Pitari MR,
Botta C, et al: Selective targeting of IRF4 by synthetic
microRNA-125b-5p mimics induces anti-multiple myeloma activity in
vitro and in vivo. Leukemia. 29:2173–2183. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Xu H, Liu C, Zhang Y, Guo X, Liu Z, Luo Z,
Chang Y, Liu S, Sun Z and Wang X: Let-7b-5p regulates proliferation
and apoptosis in multiple myeloma by targeting IGF1R. Acta Biochim
Biophys Sin (Shanghai). 46:965–972. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Amodio N, Leotta M, Bellizzi D, Di Martino
MT, D'Aquila P, Lionetti M, Fabiani F, Leone E, Gullà AM, Passarino
G, et al: DNA-demethylating and anti-tumor activity of synthetic
miR-29b mimics in multiple myeloma. Oncotarget. 3:1246–1258. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Jagannathan S, Vad N, Vallabhapurapu S,
Vallabhapurapu S, Anderson KC and Driscoll JJ: MiR-29b replacement
inhibits proteasomes and disrupts aggresome+autophagosome formation
to enhance the antimyeloma benefit of bortezomib. Leukemia.
29:727–738. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhang YK, Wang H, Leng Y, Li ZL, Yang YF,
Xiao FJ, Li QF, Chen XQ and Wang LS: Overexpression of microRNA-29b
induces apoptosis of multiple myeloma cells through down regulating
Mcl-1. Biochem Biophys Res Commun. 414:233–239. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Amodio N, Bellizzi D, Leotta M, Raimondi
L, Biamonte L, D'Aquila P, Di Martino MT, Calimeri T, Rossi M,
Lionetti M, et al: miR-29b induces SOCS-1 expression by promoter
demethylation and negatively regulates migration of multiple
myeloma and endothelial cells. Cell Cycle. 12:3650–3662. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Amodio N, Di Martino MT, Foresta U, Leone
E, Lionetti M, Leotta M, Gullà AM, Pitari MR, Conforti F, Rossi M,
et al: miR-29b sensitizes multiple myeloma cells to
bortezomib-induced apoptosis through the activation of a feedback
loop with the transcription factor Sp1. Cell Death Dis. 3:e4362012.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Cosco D, Cilurzo F, Maiuolo J, Federico C,
Di Martino MT, Cristiano MC, Tassone P, Fresta M and Paolino D:
Delivery of miR-34a by chitosan/PLGA nanoplexes for the anticancer
treatment of multiple myeloma. Sci Rep. 5:175792015. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Di Martino MT, Campani V, Misso G,
Cantafio Gallo ME, Gullà A, Foresta U, Guzzi PH, Castellano M,
Grimaldi A, Gigantino V, et al: In vivo activity of miR-34a mimics
delivered by stable nucleic acid lipid particles (SNALPs) against
multiple myeloma. PLoS One. 9:e900052014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Di Martino MT, Leone E, Amodio N, Foresta
U, Lionetti M, Pitari MR, Cantafio ME, Gullà A, Conforti F, Morelli
E, et al: Synthetic miR-34a mimics as a novel therapeutic agent for
multiple myeloma: In vitro and in vivo evidence. Clin Cancer Res.
18:6260–6270. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Shen X, Guo Y, Qi J, Shi W, Wu X, Ni H and
Ju S: Study on the association between miRNA-202 expression and
drug sensitivity in multiple myeloma cells. Pathol Oncol Res.
22:531–539. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Gatt ME, Zhao JJ, Ebert MS, Zhang Y, Chu
Z, Mani M, Gazit R, Carrasco DE, Dutta-Simmons J, Adamia S, et al:
MicroRNAs 15a/16-1 function as tumor suppressor genes in multiple
myeloma. Blood. 2010.doi: 10.1182/blood-2009-11-253294. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Sun CY, She XM, Qin Y, Chu ZB, Chen L, Ai
LS, Zhang L and Hu Y: miR-15a and miR-16 affect the angiogenesis of
multiple myeloma by targeting VEGF. Carcinogenesis. 34:426–435.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Tian Z, Zhao JJ, Tai YT, Amin SB, Hu Y,
Berger AJ, Richardson P, Chauhan D and Anderson KC: Investigational
agent MLN9708/2238 targets tumor-suppressor miR33b in MM cells.
Blood. 120:3958–3967. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Min DJ, Ezponda T, Kim MK, Will CM,
Martinez-Garcia E, Popovic R, Basrur V, Elenitoba-Johnson KS and
Licht JD: MMSET stimulates myeloma cell growth through
microRNA-mediated modulation of c-MYC. Leukemia. 27:686–694. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Tessel MA, Benham AL, Krett NL, Rosen ST
and Gunaratne PH: Role for microRNAs in regulating glucocorticoid
response and resistance in multiple myeloma. Horm Cancer.
2:182–189. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Oeztuerk-Winder F, Guinot A, Ochalek A and
Ventura JJ: Regulation of human lung alveolar multipotent cells by
a novel p38a MAPK/miR-17-92 axis. EMBO J. 31:3431–3441. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Ventura A, Young AG, Winslow MM, Lintault
L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone
JR, et al: Targeted deletion reveals essential and overlapping
functions of the miR-17 through 92 family of miRNA clusters. Cell.
132:875–886. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Hirano T, Ishihara K and Hibi M: Roles of
STAT3 in mediating the cell growth, differentiation and survival
signals relayed through the IL-6 family of cytokine receptors.
Oncogene. 19:2548–2556. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Gutierrez NC, Sarasquete ME,
Misiewicz-Krzeminska I, Delgado M, De Las Rivas J, Ticona FV,
Fermiñán E, Martín-Jiménez P, Chillón C, Risueño A, et al:
Deregulation of microRNA expression in the different genetic
subtypes of multiple myeloma and correlation with gene expression
profiling. Leukemia. 24:629–637. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Mott JL, Kobayashi S, Bronk SF and Gores
GJ: mir-29 regulates Mcl-1 protein expression and apoptosis.
Oncogene. 26:6133–6140. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Mraz M, Pospisilova S, Malinova K, Slapak
I and Mayer J: MicroRNAs in chronic lymphocytic leukemia
pathogenesis and disease subtypes. Leuk Lymphoma. 50:506–509. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Amodio N, Stamato MA, Gullà AM, Morelli E,
Romeo E, Raimondi L, Pitari MR, Ferrandino I, Misso G, Caraglia M,
et al: Therapeutic targeting of miR-29b/HDAC4 epigenetic loop in
multiple myeloma. Mol Cancer Ther. 15:1364–1375. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Rossi M, Pitari MR, Amodio N, Di Martino
MT, Conforti F, Leone E, Botta C, Paolino FM, Del Giudice T,
Iuliano E, et al: miR-29b negatively regulates human osteoclastic
cell differentiation and function: Implications for the treatment
of multiple myeloma-related bone disease. J Cell Physiol.
228:1506–1515. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Zhang S, Wu S, Qu X, Zhao M, Xu J,
Jianyong L and Lijuan C: Down-regulation of microRNA-29c is
associated with renal failure in multiple myeloma. Leuk Lymphoma.
55:226–228. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Munker R, Liu CG, Taccioli C, Alder H and
Heerema N: MicroRNA profiles of drug-resistant myeloma cell lines.
Acta Haematol. 123:201–204. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Ma J, Liu S and Wang Y: MicroRNA-21 and
multiple myeloma: Small molecule and big function. Med Oncol.
31:942014. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Zheng P, Guo H, Li G, Han S, Luo F and Liu
Y: PSMB4 promotes multiple myeloma cell growth by activating
NF-κB-miR-21 signaling. Biochem Biophys Res Commun. 458:328–333.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Wang X, Li C, Ju S, Wang Y, Wang H and
Zhong R: Myeloma cell adhesion to bone marrow stromal cells confers
drug resistance by microRNA-21 up-regulation. Leuk Lymphoma.
52:1991–1998. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Jones CI, Zabolotskaya MV, King AJ,
Stewart HJ, Horne GA, Chevassut TJ and Newbury SF: Identification
of circulating microRNAs as diagnostic biomarkers for use in
multiple myeloma. Br J Cancer. 107:1987–1996. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Sevcikova S, Kubiczkova L, Sedlarikova L,
Slaby O and Hajek R: Serum miR-29a as a marker of multiple myeloma.
Leuk Lymphoma. 54:189–191. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Qu X, Zhao M, Wu S, Yu W, Xu J, Xu J, Li J
and Chen L: Circulating microRNA 483-5p as a novel biomarker for
diagnosis survival prediction in multiple myeloma. Med Oncol.
31:2192014. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Li F, Xu Y, Deng S, Li Z, Zou D, Yi S, Sui
W, Hao M and Qiu L: MicroRNA-15a/16-1 cluster located at chromosome
13q14 is down-regulated but displays different expression pattern
and prognostic significance in multiple myeloma. Oncotarget.
6:38270–38282. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Grasedieck S, Sorrentino A, Langer C,
Buske C, Döhner H, Mertens D and Kuchenbauer F: Circulating
microRNAs in hematological diseases: Principles, challenges and
perspectives. Blood. 121:4977–4984. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Chen X, Liang H, Zhang J, Zen K and Zhang
CY: Secreted microRNAs: A new form of intercellular communication.
Trends Cell Biol. 22:125–132. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Seckinger A, Meißner T, Moreaux J, Benes
V, Hillengass J, Castoldi M, Zimmermann J, Ho AD, Jauch A,
Goldschmidt H, et al: miRNAs in multiple myeloma-a survival
relevant complex regulator of gene expression. Oncotarget.
6:39165–39183. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Bergsagel PL, Kuehl WM, Zhan F, Sawyer J,
Barlogie B and Shaughnessy J Jr: Cyclin D dysregulation: An early
and unifying pathogenic event in multiple myeloma. Blood.
106:296–303. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Huang JJ, Yu J, Li JY, Liu YT and Zhong
RQ: Circulating microRNA expression is associated with genetic
subtype and survival of multiple myeloma. Med Oncol. 29:2402–2408.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Rocci A, Hofmeister CC, Geyer S, Stiff A,
Gambella M, Cascione L, Guan J, Benson DM, Efebera YA, Talabere T,
et al: Circulating miRNA markers show promise as new
prognosticators for multiple myeloma. Leukemia. 28:1922–1926. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Wu P, Agnelli L, Walker BA, Todoerti K,
Lionetti M, Johnson DC, Kaiser M, Mirabella F, Wardell C, Gregory
WM, et al: Improved risk stratification in myeloma using a
microRNA-based classifier. Br J Haematol. 162:348–359. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Navarro A, Diaz T, Tovar N, Pedrosa F,
Tejero R, Cibeira MT, Magnano L, Rosiñol L, Monzó M, Bladé J and de
Larrea Fernández C: A serum microRNA signature associated with
complete remission and progression after autologous stem-cell
transplantation in patients with multiple myeloma. Oncotarget.
6:1874–1883. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Zhang L, Pan L, Xiang B, Zhu H, Wu Y, Chen
M, Guan P, Zou X, Valencia CA, Dong B, et al: Potential role of
exosome-associated microRNA panels and in vivo environment to
predict drug resistance for patients with multiple myeloma.
Oncotarget. 7:30876–30891. 2016.PubMed/NCBI
|
|
98
|
Yang WC and Lin SF: Mechanisms of drug
resistance in relapse and refractory multiple myeloma. Biomed Res
Int. 2015:3414302015. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Cantafio Gallo ME, Nielsen BS, Mignogna C,
Arbitrio M, Botta C, Frandsen NM, Rolfo C, Tagliaferri P, Tassone P
and Di Martino MTL: Pharmacokinetics and pharmacodynamics of a
13-mer LNA-inhibitor-miR-221 in mice and non-human primates. Mol
Ther Nucleic Acids. 5:e3362016. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Zhao JJ, Lin J, Zhu D, Wang X, Brooks D,
Chen M, Chu ZB, Takada K, Ciccarelli B, Admin S, et al: miR-30-5p
functions as a tumor suppressor and novel therapeutic tool by
targeting the oncogenic Wnt/β-catenin/BCL9 pathway. Cancer Res.
74:1801–1813. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Guo J, McKenna SL, O'Dwyer ME, Cahill MR
and O'Driscoll CM: RNA interference for multiple myeloma therapy:
Targeting signal transduction pathways. Expert Opin Ther Targets.
20:107–121. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Palagani A, de Beeck KO, Naulaerts S,
Diddens J, Chirumamilla Sekhar C, Van Camp G, Laukens K, Heyninck
K, Gerlo S, Mestdagh P, et al: Ectopic microRNA-150-5p
transcription sensitizes glucocorticoid therapy response in MM1S
multiple myeloma cells but fails to overcome hormone therapy
resistance in MM1R cells. PLoS One. 9:e1138422014. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Li J, Zhang Y, Liu Y, Dai X, Li W, Cai X,
Yin Y, Wang Q, Xue Y, Wang C, et al: Microvesicle-mediated transfer
of microRNA-150 from monocytes to endothelial cells promotes
angiogenesis. J Biol Chem. 288:23586–23596. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Lambert JM, Moshfegh A, Hainaut P, Wiman
KG and Bykov VJ: Mutant p53 reactivation by PRIMA-1MET induces
multiple signaling pathways converging on apoptosis. 29:1329–1338.
2010.
|
|
105
|
Saha MN, Abdi J, Yang Y and Chang H:
miRNA-29a as a tumor suppressor mediates PRIMA-1Met-induced
anti-myeloma activity by targeting c-Myc. Oncotarget. 7:7149–7160.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Besse L, Sedlarikova L, Kryukov F,
Nekvindova J, Radova L, Slaby O, Kuglik P, Almasi M, Penka M,
Krejci M, et al: Circulating serum MicroRNA-130a as a novel
putative marker of extramedullary myeloma. PLoS One.
10:e01372942015. View Article : Google Scholar : PubMed/NCBI
|