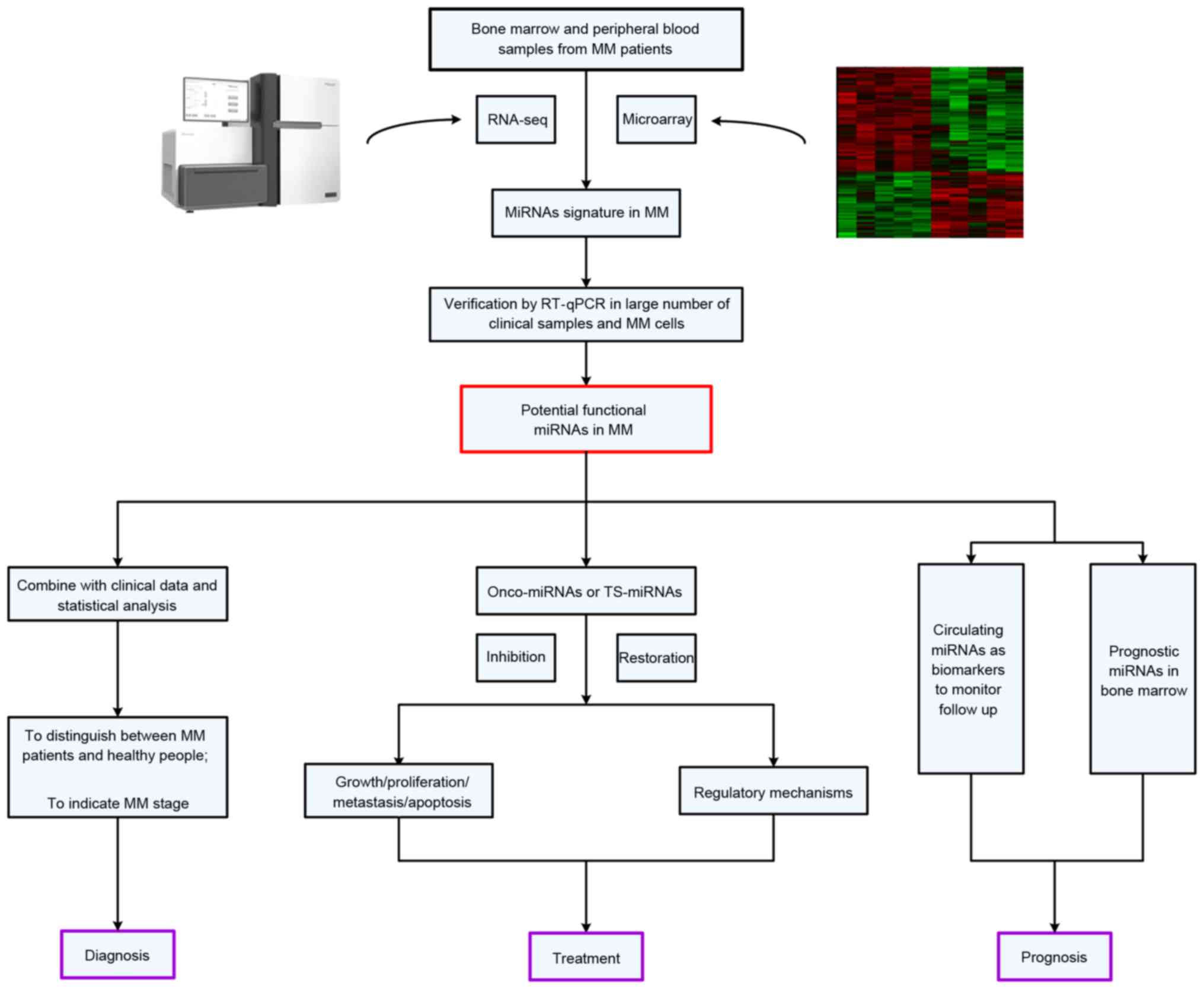
|
1
|
Rebecca RL, Kimberly KD and Jemal A:
Cancer statistics, 2018. CA Cancer J Clin. 68:7–30. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Durie BGM: Patient Handbook: Multiple
Myeloma, Cancer of the Bone Marrow. International Myeloma
Foundation; North Hollywood, CA: 2010
|
|
3
|
Ailawadhi S, Aldoss IT, Yang D, Razavi P,
Cozen W, Sher T and Chanan-Khan A: Outcome disparities in multiple
myeloma: A SEER-based comparative analysis of ethnic subgroups. Br
J Haematol. 158:91–98. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rollig C, Knop S and Bornhauser M:
Multiple myeloma. Lancet. 385:2197–2208. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhao Q, Luo F, Ma J and Yu X: Bone
metastasis-related MicroRNAs: New targets for treatment. Curr
Cancer Drug Targets. 15:716–725. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rossi M, Tagliaferri P and Tassone P:
MicroRNAs in multiple myeloma and related bone disease. Ann Transl
Med. 3:3342015.PubMed/NCBI
|
|
7
|
Rocci A, Hofmeister CC and Pichiorri F:
The potential of miRNAs as biomarkers for multiple myeloma. Expert
Rev Mol Diagn. 14:947–959. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bi CL and Chng WJ: miRNA deregulation in
multiple myeloma. Chin Med J (Engl). 124:3164–3169. 2011.PubMed/NCBI
|
|
9
|
Ghobrial IM: Myeloma as a model for the
process of metastasis: Implications for therapy. Blood. 120:20–30.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cottini F and Anderson K: Novel
therapeutic targets in multiple myeloma. Clin Adv Hematol Oncol.
13:236–248. 2015.PubMed/NCBI
|
|
11
|
Kumar SK, Rajkumar SV, Dispenzieri A, Lacy
MQ, Hayman SR, Buadi FK, Zeldenrust SR, Dingli D, Russell SJ, Lust
JA, et al: Improved survival in multiple myeloma and the impact of
novel therapies. Blood. 111:2516–2520. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kumar SK, Dimopoulos MA, Kastritis E,
Terpos E, Nahi H, Goldschmidt H, Hillengass J, Leleu X, Beksac M,
Alsina M, et al: Natural history of relapsed myeloma, refractory to
immunomodulatory drugs and proteasome inhibitors: A multicenter
IMWG study. Leukemia. 31:2443–2448. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ahmad N, Haider S, Jagannathan S, Anaissie
E and Driscoll JJ: MicroRNA theragnostics for the clinical
management of multiple myeloma. Leukemia. 28:732–738. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kubiczkova L, Kryukov F, Slaby O,
Dementyeva E, Jarkovsky J, Nekvindova J, Radova L, Greslikova H,
Kuglik P, Vetesnikova E, et al: Circulating serum microRNAs as
novel diagnostic and prognostic biomarkers for multiple myeloma and
monoclonal gammopathy of undetermined significance. Haematologica.
99:511–518. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li F, Hao M, Feng X, Zang M, Qin Y, Yi S,
Li Z, Xu Y, Zhou L, Sui W, et al: Downregulated miR-33b is a novel
predictor associated with disease progression and poor prognosis in
multiple myeloma. Leuk Res. 39:793–799. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shukla GC, Singh J and Barik S: MicroRNAs:
Processing, maturation, target recognition and regulatory
functions. Mol Cell Pharmacol. 3:83–92. 2011.PubMed/NCBI
|
|
17
|
Zhang Q, Yan W, Bai Y, Xu H, Fu C, Zheng
W, Zhu Y and Ma J: Synthetic miR-145 mimic inhibits multiple
myeloma cell growth in vitro and in vivo. Oncol Rep. 33:448–456.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sun Y, Pan J, Mao S and Jin J:
IL-17/miR-192/IL-17Rs regulatory feedback loop facilitates multiple
myeloma progression. PLoS One. 9:e1146472014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang Y, Li F, Saha MN, Abdi J, Qiu L and
Chang H: miR-137 and miR-197 induce apoptosis and suppress
tumorigenicity by targeting MCL-1 in multiple myeloma. Clin Cancer
Res. 21:2399–2411. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Macfarlane LA and Murphy PR: MicroRNA:
Biogenesis, function and role in cancer. Curr Genomics. 11:537–561.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rossi M, Amodio N, Di Martino MT,
Tagliaferri P, Tassone P and Cho WC: MicroRNA and multiple myeloma:
From laboratory findings to translational therapeutic approaches.
Curr Pharm Biotechnol. 15:459–467. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Di Martino MT, Amodio N, Tassone P and
Tagliaferri P: Functional analysis of microRNA in multiple myeloma.
Methods Mol Biol. 1375:181–194. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Raimondi L, De Luca A, Morelli E,
Giavaresi G, Tagliaferri P, Tassone P and Amodio N: MicroRNAs:
Novel crossroads between myeloma cells and the bone marrow
microenvironment. Biomed Res Int. 2016:65045932016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Calin GA, Dumitru CD, Shimizu M, Bichi R,
Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al:
Frequent deletions and down-regulation of micro-RNA genes miR15 and
miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci
USA. 99:15524–15529. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pichiorri F, Suh SS, Rocci A, De Luca L,
Taccioli C, Santhanam R, Zhou W, Benson DM Jr, Hofmainster C, Alder
H, et al: Downregulation of p53-inducible microRNAs 192, 194 and
215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma
development. Cancer Cell. 18:367–381. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Misiewicz-Krzeminska I, Sarasquete ME,
Quwaider D, Krzeminski P, Ticona FV, Paíno T, Delgado M, Aires A,
Ocio EM, García-Sanz R, et al: Restoration of microRNA-214
expression reduces growth of myeloma cells through positive
regulation of P53 and inhibition of DNA replication. Haematologica.
98:640–648. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang Q, Wang LQ, Wong KY, Li ZY and Chim
CS: Infrequent DNA methylation of miR-9-1 and miR-9-3 in multiple
myeloma. J Clin Pathol. 68:557–561. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cipolla GA, Park JK, de Oliveira LA,
Lobo-Alves SC, de Almeida RC, Farias TD, Lemos Dde S, Malheiros D,
Lavker RM and Petzl-Erler ML: A 3′UTR polymorphism marks
differential KLRG1 mRNA levels through disruption of a miR-584-5p
binding site and associates with pemphigus foliaceus
susceptibility. Biochim Biophys Acta. 1859:1306–1313. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhao JJ and Carrasco RD: Crosstalk between
microRNA30a/b/c/d/e-5p and the canonical Wnt pathway: Implications
for multiple myeloma therapy. Cancer Res. 74:5351–5358. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Brocke-Heidrich K, Kretzschmar AK, Pfeifer
G, Henze C, Löffler D, Koczan D, Thiesen HJ, Burger R, Gramatzki M
and Horn F: Interleukin-6-dependent gene expression profiles in
multiple myeloma INA-6 cells reveal a Bcl-2 family-independent
survival pathway closely associated with Stat3 activation. Blood.
103:242–251. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Leotta M, Biamonte L, Raimondi L,
Ronchetti D, Di Martino MT, Botta C, Leone E, Pitari MR, Neri A,
Giordano A, et al: A p53-dependent tumor suppressor network is
induced by selective miR-125a-5p inhibition in multiple myeloma
cells. J Cell Physiol. 229:2106–2116. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shen X, Guo Y, Yu J, Qi J, Shi W, Wu X, Ni
H and Ju S: miRNA-202 in bone marrow stromal cells affects the
growth and adhesion of multiple myeloma cells by regulating B
cell-activating factor. Clin Exp Med. 16:307–316. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhou Y, Chen L, Barlogie B, Stephens O, Wu
X, Williams DR, Cartron MA, van Rhee F, Nair B, Waheed S, et al:
High-risk myeloma is associated with global elevation of miRNAs and
overexpression of EIF2C2/AGO2. Proc Natl Acad Sci USA.
107:7904–7909. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hao M, Zang M, Zhao L, Deng S, Xu Y, Qi F,
An G, Qin Y, Sui W, Li F, et al: Serum high expression of miR-214
and miR-135b as novel predictor for myeloma bone disease
development and prognosis. Oncotarget. 7:19589–19600. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pichiorri F, Suh SS, Ladetto M, Kuehl M,
Palumbo T, Drandi D, Taccioli C, Zanesi N, Alder H, Hagan JP, et
al: MicroRNAs regulate critical genes associated with multiple
myeloma pathogenesis. Proc Natl Acad Sci USA. 105:12885–12890.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Unno K, Zhou Y, Zimmerman T, Platanias LC
and Wickrema A: Identification of a novel microRNA cluster
miR-193b-365 in multiple myeloma. Leuk Lymphoma. 50:1865–1871.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Roccaro AM, Sacco A, Thompson B, Leleu X,
Azab AK, Azab F, Runnels J, Jia X, Ngo HT, Melhem MR, et al:
MicroRNAs 15a and 16 regulate tumor proliferation in multiple
myeloma. Blood. 113:6669–6680. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chi J, Ballabio E, Chen XH, Kušec R,
Taylor S, Hay D, Tramonti D, Saunders NJ, Littlewood T, Pezzella F,
et al: MicroRNA expression in multiple myeloma is associated with
genetic subtype, isotype and survival. Biol Direct. 6:232011.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yyusnita, Norsiah, Zakiah I, Chang KM,
Purushotaman VS, Zubaidah Z and Jamal R: MicroRNA (miRNA)
expression profiling of peripheral blood samples in multiple
myeloma patients using microarray. Malays J Pathol. 34:133–143.
2012.PubMed/NCBI
|
|
41
|
Campo S, Allegra A, D'Ascola A, Alonci A,
Scuruchi M, Russo S, Avenoso A, Gerace D, Campo GM and Musolino C:
MiRNome expression is deregulated in the peripheral lymphoid
compartment of multiple myeloma. Br J Haematol. 165:801–813. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang W, Corrigan-Cummins M, Barber EA,
Saleh LM, Zingone A, Ghafoor A, Costello R, Zhang Y, Kurlander RJ,
Korde N, et al: Aberrant Levels of miRNAs in Bone marrow
microenvironment and peripheral blood of myeloma patients and
disease progression. J Mol Diagn. 17:669–678. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hao M, Zang M, Wendlandt E, Xu Y, An G,
Gong D, Li F, Qi F, Zhang Y, Yang Y, et al: Low serum miR-19a
expression as a novel poor prognostic indicator in multiple
myeloma. Int J Cancer. 136:1835–1844. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:2015.doi: 10.7554/eLife.05005. View Article : Google Scholar
|
|
45
|
Betel D, Koppal A, Agius P, Sander C and
Leslie C: Comprehensive modeling of microRNA targets predicts
functional non-conserved and non-canonical sites. Genome Biol.
11:R902010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Anders G, Mackowiak SD, Jens M, Maaskola
J, Kuntzagk A, Rajewsky N, Landthaler M and Dieterich C: doRiNA: A
database of RNA interactions in post-transcriptional regulation.
Nucleic Acids Res. 40:D180–D186. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Shang J, Yao Y, Fan X, Shangguan L, Li J,
Liu H and Zhou Y: miR-29c-3p promotes senescence of human
mesenchymal stem cells by targeting CNOT6 through p53-p21 and
p16-pRB pathways. Biochim Biophys Acta. 1863:520–532. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gulla A, Di Martino MT, Cantafio Gallo ME,
Morelli E, Amodio N, Botta C, Pitari MR, Lio SG, Britti D, Stamato
MA, et al: A 13 mer LNA-i-miR-221 inhibitor restores drug
sensitivity in melphalan-refractory multiple myeloma cells. Clin
Cancer Res. 22:1222–1233. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Di Martino MT, Gullà A, Cantafio ME,
Lionetti M, Leone E, Amodio N, Guzzi PH, Foresta U, Conforti F,
Cannataro M, et al: In vitro and in vivo anti-tumor activity of
miR-221/222 inhibitors in multiple myeloma. Oncotarget. 4:242–255.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhao JJ, Chu ZB, Hu Y, Lin J, Wang Z,
Jiang M, Chen M, Wang X, Kang Y, Zhou Y, et al: Targeting the
miR-221-222/PUMA/BAK/BAX pathway abrogates dexamethasone resistance
in multiple myeloma. Cancer Res. 75:4384–4397. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Galm O, Yoshikawa H, Esteller M, Osieka R
and Herman JG: SOCS-1, a negative regulator of cytokine signaling,
is frequently silenced by methylation in multiple myeloma. Blood.
101:2784–2788. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chen L, Li C, Zhang R, Gao X, Qu X, Zhao
M, Qiao C, Xu J and Li J: miR-17-92 cluster microRNAs confers
tumorigenicity in multiple myeloma. Cancer Lett. 309:62–70. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Peng J, Thakur A, Zhang S, Dong Y, Wang X,
Yuan R, Zhang K and Guo X: Expressions of miR-181a and miR-20a in
RPMI8226 cell line and their potential as biomarkers for multiple
myeloma. Tumour Biol. 36:8545–8552. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Xu S, Cecilia SG, De Veirman K, Vande
Broek I, Leleu X, De Becker A, Van Camp B, Vanderkerken K and Van
Riet I: Upregulation of miR-135b is involved in the impaired
osteogenic differentiation of mesenchymal stem cells derived from
multiple myeloma patients. PLoS One. 8:e797522013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Liang B, Yin JJ and Zhan XR: MiR-301a
promotes cell proliferation by directly targeting TIMP2 in multiple
myeloma. Int J Clin Exp Pathol. 8:9168–9174. 2015.PubMed/NCBI
|
|
56
|
Morelli E, Leone E, Cantafio ME, Di
Martino MT, Amodio N, Biamonte L, Gullà A, Foresta U, Pitari MR,
Botta C, et al: Selective targeting of IRF4 by synthetic
microRNA-125b-5p mimics induces anti-multiple myeloma activity in
vitro and in vivo. Leukemia. 29:2173–2183. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Xu H, Liu C, Zhang Y, Guo X, Liu Z, Luo Z,
Chang Y, Liu S, Sun Z and Wang X: Let-7b-5p regulates proliferation
and apoptosis in multiple myeloma by targeting IGF1R. Acta Biochim
Biophys Sin (Shanghai). 46:965–972. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Amodio N, Leotta M, Bellizzi D, Di Martino
MT, D'Aquila P, Lionetti M, Fabiani F, Leone E, Gullà AM, Passarino
G, et al: DNA-demethylating and anti-tumor activity of synthetic
miR-29b mimics in multiple myeloma. Oncotarget. 3:1246–1258. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Jagannathan S, Vad N, Vallabhapurapu S,
Vallabhapurapu S, Anderson KC and Driscoll JJ: MiR-29b replacement
inhibits proteasomes and disrupts aggresome+autophagosome formation
to enhance the antimyeloma benefit of bortezomib. Leukemia.
29:727–738. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhang YK, Wang H, Leng Y, Li ZL, Yang YF,
Xiao FJ, Li QF, Chen XQ and Wang LS: Overexpression of microRNA-29b
induces apoptosis of multiple myeloma cells through down regulating
Mcl-1. Biochem Biophys Res Commun. 414:233–239. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Amodio N, Bellizzi D, Leotta M, Raimondi
L, Biamonte L, D'Aquila P, Di Martino MT, Calimeri T, Rossi M,
Lionetti M, et al: miR-29b induces SOCS-1 expression by promoter
demethylation and negatively regulates migration of multiple
myeloma and endothelial cells. Cell Cycle. 12:3650–3662. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Amodio N, Di Martino MT, Foresta U, Leone
E, Lionetti M, Leotta M, Gullà AM, Pitari MR, Conforti F, Rossi M,
et al: miR-29b sensitizes multiple myeloma cells to
bortezomib-induced apoptosis through the activation of a feedback
loop with the transcription factor Sp1. Cell Death Dis. 3:e4362012.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Cosco D, Cilurzo F, Maiuolo J, Federico C,
Di Martino MT, Cristiano MC, Tassone P, Fresta M and Paolino D:
Delivery of miR-34a by chitosan/PLGA nanoplexes for the anticancer
treatment of multiple myeloma. Sci Rep. 5:175792015. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Di Martino MT, Campani V, Misso G,
Cantafio Gallo ME, Gullà A, Foresta U, Guzzi PH, Castellano M,
Grimaldi A, Gigantino V, et al: In vivo activity of miR-34a mimics
delivered by stable nucleic acid lipid particles (SNALPs) against
multiple myeloma. PLoS One. 9:e900052014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Di Martino MT, Leone E, Amodio N, Foresta
U, Lionetti M, Pitari MR, Cantafio ME, Gullà A, Conforti F, Morelli
E, et al: Synthetic miR-34a mimics as a novel therapeutic agent for
multiple myeloma: In vitro and in vivo evidence. Clin Cancer Res.
18:6260–6270. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Shen X, Guo Y, Qi J, Shi W, Wu X, Ni H and
Ju S: Study on the association between miRNA-202 expression and
drug sensitivity in multiple myeloma cells. Pathol Oncol Res.
22:531–539. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Gatt ME, Zhao JJ, Ebert MS, Zhang Y, Chu
Z, Mani M, Gazit R, Carrasco DE, Dutta-Simmons J, Adamia S, et al:
MicroRNAs 15a/16-1 function as tumor suppressor genes in multiple
myeloma. Blood. 2010.doi: 10.1182/blood-2009-11-253294. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Sun CY, She XM, Qin Y, Chu ZB, Chen L, Ai
LS, Zhang L and Hu Y: miR-15a and miR-16 affect the angiogenesis of
multiple myeloma by targeting VEGF. Carcinogenesis. 34:426–435.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Tian Z, Zhao JJ, Tai YT, Amin SB, Hu Y,
Berger AJ, Richardson P, Chauhan D and Anderson KC: Investigational
agent MLN9708/2238 targets tumor-suppressor miR33b in MM cells.
Blood. 120:3958–3967. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Min DJ, Ezponda T, Kim MK, Will CM,
Martinez-Garcia E, Popovic R, Basrur V, Elenitoba-Johnson KS and
Licht JD: MMSET stimulates myeloma cell growth through
microRNA-mediated modulation of c-MYC. Leukemia. 27:686–694. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Tessel MA, Benham AL, Krett NL, Rosen ST
and Gunaratne PH: Role for microRNAs in regulating glucocorticoid
response and resistance in multiple myeloma. Horm Cancer.
2:182–189. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Oeztuerk-Winder F, Guinot A, Ochalek A and
Ventura JJ: Regulation of human lung alveolar multipotent cells by
a novel p38a MAPK/miR-17-92 axis. EMBO J. 31:3431–3441. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Ventura A, Young AG, Winslow MM, Lintault
L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone
JR, et al: Targeted deletion reveals essential and overlapping
functions of the miR-17 through 92 family of miRNA clusters. Cell.
132:875–886. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Hirano T, Ishihara K and Hibi M: Roles of
STAT3 in mediating the cell growth, differentiation and survival
signals relayed through the IL-6 family of cytokine receptors.
Oncogene. 19:2548–2556. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Gutierrez NC, Sarasquete ME,
Misiewicz-Krzeminska I, Delgado M, De Las Rivas J, Ticona FV,
Fermiñán E, Martín-Jiménez P, Chillón C, Risueño A, et al:
Deregulation of microRNA expression in the different genetic
subtypes of multiple myeloma and correlation with gene expression
profiling. Leukemia. 24:629–637. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Mott JL, Kobayashi S, Bronk SF and Gores
GJ: mir-29 regulates Mcl-1 protein expression and apoptosis.
Oncogene. 26:6133–6140. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Mraz M, Pospisilova S, Malinova K, Slapak
I and Mayer J: MicroRNAs in chronic lymphocytic leukemia
pathogenesis and disease subtypes. Leuk Lymphoma. 50:506–509. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Amodio N, Stamato MA, Gullà AM, Morelli E,
Romeo E, Raimondi L, Pitari MR, Ferrandino I, Misso G, Caraglia M,
et al: Therapeutic targeting of miR-29b/HDAC4 epigenetic loop in
multiple myeloma. Mol Cancer Ther. 15:1364–1375. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Rossi M, Pitari MR, Amodio N, Di Martino
MT, Conforti F, Leone E, Botta C, Paolino FM, Del Giudice T,
Iuliano E, et al: miR-29b negatively regulates human osteoclastic
cell differentiation and function: Implications for the treatment
of multiple myeloma-related bone disease. J Cell Physiol.
228:1506–1515. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Zhang S, Wu S, Qu X, Zhao M, Xu J,
Jianyong L and Lijuan C: Down-regulation of microRNA-29c is
associated with renal failure in multiple myeloma. Leuk Lymphoma.
55:226–228. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Munker R, Liu CG, Taccioli C, Alder H and
Heerema N: MicroRNA profiles of drug-resistant myeloma cell lines.
Acta Haematol. 123:201–204. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Ma J, Liu S and Wang Y: MicroRNA-21 and
multiple myeloma: Small molecule and big function. Med Oncol.
31:942014. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Zheng P, Guo H, Li G, Han S, Luo F and Liu
Y: PSMB4 promotes multiple myeloma cell growth by activating
NF-κB-miR-21 signaling. Biochem Biophys Res Commun. 458:328–333.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Wang X, Li C, Ju S, Wang Y, Wang H and
Zhong R: Myeloma cell adhesion to bone marrow stromal cells confers
drug resistance by microRNA-21 up-regulation. Leuk Lymphoma.
52:1991–1998. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Jones CI, Zabolotskaya MV, King AJ,
Stewart HJ, Horne GA, Chevassut TJ and Newbury SF: Identification
of circulating microRNAs as diagnostic biomarkers for use in
multiple myeloma. Br J Cancer. 107:1987–1996. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Sevcikova S, Kubiczkova L, Sedlarikova L,
Slaby O and Hajek R: Serum miR-29a as a marker of multiple myeloma.
Leuk Lymphoma. 54:189–191. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Qu X, Zhao M, Wu S, Yu W, Xu J, Xu J, Li J
and Chen L: Circulating microRNA 483-5p as a novel biomarker for
diagnosis survival prediction in multiple myeloma. Med Oncol.
31:2192014. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Li F, Xu Y, Deng S, Li Z, Zou D, Yi S, Sui
W, Hao M and Qiu L: MicroRNA-15a/16-1 cluster located at chromosome
13q14 is down-regulated but displays different expression pattern
and prognostic significance in multiple myeloma. Oncotarget.
6:38270–38282. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Grasedieck S, Sorrentino A, Langer C,
Buske C, Döhner H, Mertens D and Kuchenbauer F: Circulating
microRNAs in hematological diseases: Principles, challenges and
perspectives. Blood. 121:4977–4984. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Chen X, Liang H, Zhang J, Zen K and Zhang
CY: Secreted microRNAs: A new form of intercellular communication.
Trends Cell Biol. 22:125–132. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Seckinger A, Meißner T, Moreaux J, Benes
V, Hillengass J, Castoldi M, Zimmermann J, Ho AD, Jauch A,
Goldschmidt H, et al: miRNAs in multiple myeloma-a survival
relevant complex regulator of gene expression. Oncotarget.
6:39165–39183. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Bergsagel PL, Kuehl WM, Zhan F, Sawyer J,
Barlogie B and Shaughnessy J Jr: Cyclin D dysregulation: An early
and unifying pathogenic event in multiple myeloma. Blood.
106:296–303. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Huang JJ, Yu J, Li JY, Liu YT and Zhong
RQ: Circulating microRNA expression is associated with genetic
subtype and survival of multiple myeloma. Med Oncol. 29:2402–2408.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Rocci A, Hofmeister CC, Geyer S, Stiff A,
Gambella M, Cascione L, Guan J, Benson DM, Efebera YA, Talabere T,
et al: Circulating miRNA markers show promise as new
prognosticators for multiple myeloma. Leukemia. 28:1922–1926. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Wu P, Agnelli L, Walker BA, Todoerti K,
Lionetti M, Johnson DC, Kaiser M, Mirabella F, Wardell C, Gregory
WM, et al: Improved risk stratification in myeloma using a
microRNA-based classifier. Br J Haematol. 162:348–359. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Navarro A, Diaz T, Tovar N, Pedrosa F,
Tejero R, Cibeira MT, Magnano L, Rosiñol L, Monzó M, Bladé J and de
Larrea Fernández C: A serum microRNA signature associated with
complete remission and progression after autologous stem-cell
transplantation in patients with multiple myeloma. Oncotarget.
6:1874–1883. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Zhang L, Pan L, Xiang B, Zhu H, Wu Y, Chen
M, Guan P, Zou X, Valencia CA, Dong B, et al: Potential role of
exosome-associated microRNA panels and in vivo environment to
predict drug resistance for patients with multiple myeloma.
Oncotarget. 7:30876–30891. 2016.PubMed/NCBI
|
|
98
|
Yang WC and Lin SF: Mechanisms of drug
resistance in relapse and refractory multiple myeloma. Biomed Res
Int. 2015:3414302015. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Cantafio Gallo ME, Nielsen BS, Mignogna C,
Arbitrio M, Botta C, Frandsen NM, Rolfo C, Tagliaferri P, Tassone P
and Di Martino MTL: Pharmacokinetics and pharmacodynamics of a
13-mer LNA-inhibitor-miR-221 in mice and non-human primates. Mol
Ther Nucleic Acids. 5:e3362016. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Zhao JJ, Lin J, Zhu D, Wang X, Brooks D,
Chen M, Chu ZB, Takada K, Ciccarelli B, Admin S, et al: miR-30-5p
functions as a tumor suppressor and novel therapeutic tool by
targeting the oncogenic Wnt/β-catenin/BCL9 pathway. Cancer Res.
74:1801–1813. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Guo J, McKenna SL, O'Dwyer ME, Cahill MR
and O'Driscoll CM: RNA interference for multiple myeloma therapy:
Targeting signal transduction pathways. Expert Opin Ther Targets.
20:107–121. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Palagani A, de Beeck KO, Naulaerts S,
Diddens J, Chirumamilla Sekhar C, Van Camp G, Laukens K, Heyninck
K, Gerlo S, Mestdagh P, et al: Ectopic microRNA-150-5p
transcription sensitizes glucocorticoid therapy response in MM1S
multiple myeloma cells but fails to overcome hormone therapy
resistance in MM1R cells. PLoS One. 9:e1138422014. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Li J, Zhang Y, Liu Y, Dai X, Li W, Cai X,
Yin Y, Wang Q, Xue Y, Wang C, et al: Microvesicle-mediated transfer
of microRNA-150 from monocytes to endothelial cells promotes
angiogenesis. J Biol Chem. 288:23586–23596. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Lambert JM, Moshfegh A, Hainaut P, Wiman
KG and Bykov VJ: Mutant p53 reactivation by PRIMA-1MET induces
multiple signaling pathways converging on apoptosis. 29:1329–1338.
2010.
|
|
105
|
Saha MN, Abdi J, Yang Y and Chang H:
miRNA-29a as a tumor suppressor mediates PRIMA-1Met-induced
anti-myeloma activity by targeting c-Myc. Oncotarget. 7:7149–7160.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Besse L, Sedlarikova L, Kryukov F,
Nekvindova J, Radova L, Slaby O, Kuglik P, Almasi M, Penka M,
Krejci M, et al: Circulating serum MicroRNA-130a as a novel
putative marker of extramedullary myeloma. PLoS One.
10:e01372942015. View Article : Google Scholar : PubMed/NCBI
|