Introduction
Esophageal carcinoma (EC) is the sixth most common
type of cancer in China (1). Although
the contributions of environmental factors, lifestyles and trace
elements have been extensively investigated, the etiology of
esophageal cancer has not yet been established (2–4).
In 1982, Syrjänen (5)
first proposed that esophageal cancer is associated with HPV
infection. Since then, numerous studies have supported this
hypothesis (6–10).
In a previous study, HPV18 was detected and
localized in human chromosome 8 in the esophageal squamous cell
carcinoma EC109 cell line, which supported the hypothesis that HPV
infection is a pathogenic factor in the development of esophageal
cancer (11). Moreover, chronic
high-risk HPV infection and the integration of elements of the
viral genome into the human chromosome have proven to be pathogenic
factors for cervical malignant transformation and lesions (12,13).
Integrated and episomal HPV may be distinguished by
amplifying papillomavirus oncogene transcripts (APOT) using the
polymerase chain reaction (PCR), and Klaes et al (14) proposed that the location of HPV genome
implantation in the human chromosome is an appropriate molecular
tag for pathological lesions and the development of cancer.
Nevertheless, few reports (11–14) have
described the HPV integration site. The objective of the present
study was to assess HPV18 infection and its location in the human
genome in the setting of EC.
Materials and methods
Sample collection
Fresh surgically resected tissue samples from 189
patients who were pathologically diagnosed with esophageal
carcinoma were obtained between March 2013 and December 2015;
written informed consent was obtained from each specimen donor. All
specimen donors were treated in the Pathology Department of
Tangshan People's Hospital (Hebei, China); all patients came from
the Tangshan area. The mean age was 58 years (range, 40–76 years),
of which 136 were male and 53 female. All samples were esophageal
squamous cell carcinoma, of which 30 were well differentiated, 104
were moderately differentiated and 55 were poorly differentiated.
The samples were separated into 98 early-stage, 63 middle-stage and
28 late-stage specimens, according to the sixth and seventh
editions of International Union Against Cancer classification
(15); all fresh samples were stored
at −80°C prior to the experiments.
Cells and materials
The following media were used in the present study:
Dulbecco's modified Eagle medium (DMEM) with antibiotics (100 U/ml
penicillin/streptomycin), fetal bovine serum, trypsin, and these
reagents were provided by BioTek China, Beijing, China. QIAamp DNA
Mini kit, Qiagen RNeasy Mini kit and Qiaquick Gel Extraction kit
were provided by Qiagen GmbH, Hilden, Germany. Takara Ex Taq kit
(including buffer, MgCl2, dNTPs, Taq DNA polymerase) was
provided by Takara Bio, Inc., Otsu, Japan. The 293 cell line
obtained from National Institute for Viral Disease Control and
Prevention, Chinese Center for Disease Control and Prevention
(Beijing, China).
Cell culture conditions
The 293 cell line were cultivated using 10 ml media
(DMEM with 100 U/ml penicillin-Streptomycin and 10% fetal bovine
serum) in a 10 cm petri dish and maintained at 37°C in humidified
atmosphere, containing 5% CO2/and 95% air. Cell media
were changed every 2–3 days.
Cell collection and DNA
extraction
The 293 cells were washed twice using phosphate
buffer saline (PBS) when the 293 cells were allowed to reach 90%
confluence. Subsequently, cells were digested using 2 ml 0.25%
trypsin (including 0.02% EDTA), which was terminated using 2 ml
fresh cell medium, and transferred to 10 ml flasks. Cells were
centrifuged at 3,000 × g for 5 min at 4°C and the supernatant was
removed. DNA extraction was performed using QIAamp DNA Mini kit,
according to the manufacturer's protocol.
Plasmid
The plasmid HPV18/pBR322 (National Institute for
Viral Disease Control and Prevention, Chinese Center for Disease
Control and Prevention, China) DNA, containing the whole HPV18
genome, and DNA from the 293 cell line were stored at −20°C.
DNA extraction
DNA was extracted from each tissue specimen (~25 mg)
using the QIAamp DNA Mini kit according to the manufacturer's
protocol, and each sample was eluted with ~50 µl sterilized
distilled water. The extracted DNA was stored at −20°C until
further processing.
Detection of specimen quality and HPV
DNA
The quality of the DNA from each tissue sample was
assessed using PCR with β-actin primers (16). DNA from the 293 cell line was used as
a control to ensure specimen quality.
HPV DNA for each specimen was analyzed by PCR using
the primers My09/11 for HPV L1 (My09, 5′-CGTCCMARRGGAWACTGATC-3′;
MY11, 5′-GCMCAGGGWCATAAYAATGG-3′, PCR product ~450 bp) (17) and HPV18 E6-specific primers (forward,
5′-GCGCTTTGAGGATCCAACAC-3′ and reverse, 5′-ATTCAACGGTTTCTGGCAC-3′;
the PCR product generated was ~335 bp). The HPV18 E6 primers were
designed on the basis of the genomic sequence of HPV18 in GenBank
(accession number X05015.1).
Each PCR amplification was performed in a volume of
25 µl, containing 1X Ex Buffer (MgCl2-free), 2.5 mM
MgCl2, 0.2 mM dNTPs, 1 U Ex Taq DNA polymerase, and 5
pmol each of the aforementioned forward and reverse primers. A
total of 100 ng extracted DNA template and control DNA were added
to the reaction mixture. The following thermal cycling profile was
employed: 95°C for 5 min, followed by 31 cycles of 95°C for 30 sec,
55°C for 30 sec and 72°C for 30 sec. The last cycle was followed by
a final extension step of 5 min at 72°C.
Sequence analysis products of
type-specific HPV18
To confirm the specificity of the HPV18 primers, the
HPV18 E6-positive PCR products were purified using a Qiaquick Gel
Extraction kit according to the manufacturer's protocol. The
products were then ligated into the pMD-18T vector prior to being
subjected to sequence analysis by Beijing Rui Bo Xing ke Biological
Technology company using DNAman software v6.0 (www.shinegene.org.cn/q2.html).
Reverse transcription (RT)
Total RNA was extracted from each sample using a
Qiagen RNeasy Mini kit according to the manufacturer's protocol. RT
was performed in a total volume of 25 µl consisting of 4 µl 5X
first-strand buffer, 1 µl dNTPs (0.2 mM), 1 µl RNase inhibitor (40
U), 100 ng total RNA, 1 µl (dT)17-p3 (10 pmol primer,
GACTCGAGTCGACATCGATTTTTTTTTTTTTTTTT), 1 µl SuperScript reverse
transcriptase (200 U) and 9.2 µl RNase-free water. The RNA was
reverse-transcribed at 42°C for 50 min and deactivated at 70°C for
15 min, and the resultant product was stored at 4°C.
To verify that each tissue sample was harboring HPV
with detectable HPV18 E7 mRNA, human GAPDH was employed as a
reference gene for RT-PCR using the following primers: GAPDH
forward, 5′-CATCACCATCTTCCAGGA-3′ and reverse,
5′-GTCTACCACCCTATTGCA-3′ (13). This
approach guaranteed that the integrity of all mRNA extracted from
each sample was sufficient for the detection of viral-cell fusion
transcripts.
Detection of viral-cell fusion
transcripts
The first PCR was prepared according to the method
of Klaes et al (14), in a
volume of 25 µl using HPV18 E7-specific forward primers for p1-18
(5′-TAGAAAGCTCAGCAGACGACC-3′) in HPV18 gene sequence 816…836 and
reverse primers for p3 (5′-GACTCGAGTCGACATCG-3′); the reaction
system consisted of 1X Buffer, 10 pM primers, 2.5 mM
MgCl2, 0.2 mM dNTPs, 100 ng cDNA product and 1 U Ex Taq
DNA polymerase. PCR was performed according to the following steps:
95°C for 5 min, followed by 30 cycles at 95°C for 1 min, 56°C for 1
min and 72°C for 3 min. The final cycle consisted of 72°C for 5
min, and the sample was stored at 4°C.
Nested PCR was performed under identical conditions
(except for the annealing temperature, which was 67°C) and the
primers: the HPV18 E7-specific forward primer p2-18
(5′-ACGACCTTCGAGCATTCCAGCAG-3′) in HPV18 gene sequence 831…853 and
the reverse primer (dT)17-p3; 5 µl PCR product from the
first round was used as the template. The two primers span bp
814–835 (p1-18) and bp 830–853 (p2-18) in the HPV18 genome. To
ensure the specificity of these primers, a negative control (293
cell DNA template) was added to each reaction system.
Integration sites analysis
The HPV18 E7 PCR products for p2-18 were cloned into
the pMD-18T vector. Subsequently, Beijing Rui Bo Xing ke Biological
Technology sequenced. The results were blasted using the National
Center for Biotechnology Information (blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome),
HPV18 integration sites in human chromosome were determined for
every specimen.
Results
Detection of specimen quality and HPV
DNA
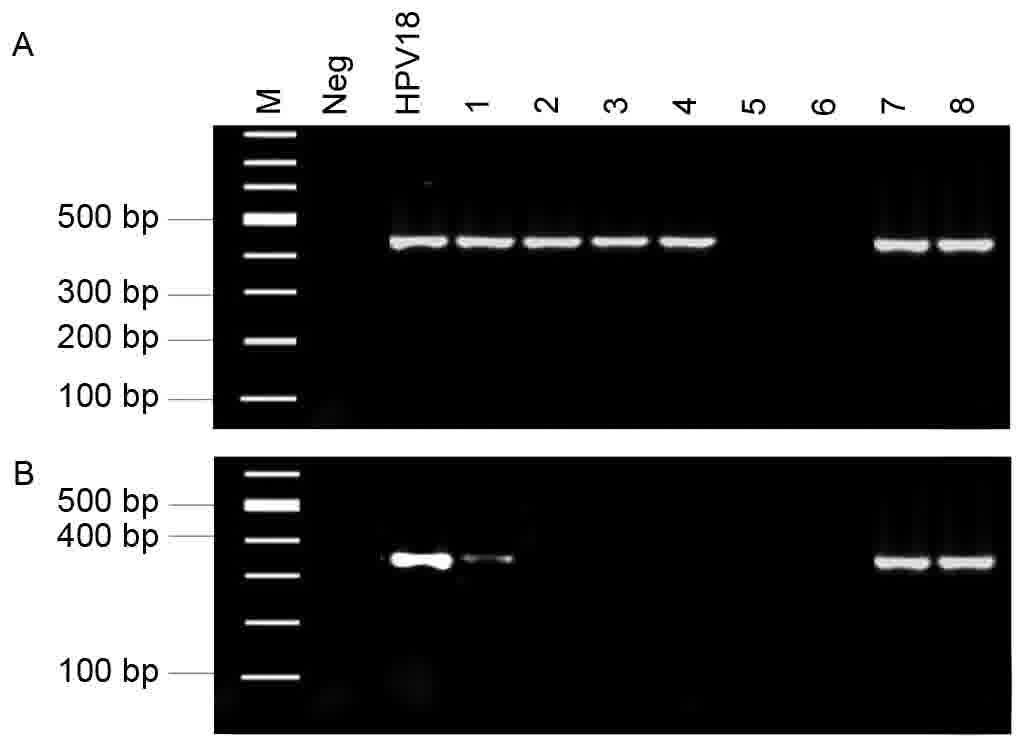
β-actin expression in a total of 189 samples was
investigated to evaluate the DNA quality of the tissue specimens. A
290-bp β-actin product was detected in each sample. This result
suggested that the DNA from each specimen was of high quality and
satisfied the requirements for further experiments. Of the 189
specimens, 168 were positive for HPV DNA, as assessed using MY09/11
primers (Fig. 1A); 33 of these
specimens were positive for HPV18 (Fig.
1B), assessed using the HPV18 E6-specific primers.
Sequence analysis for type-specific
HPV18 products
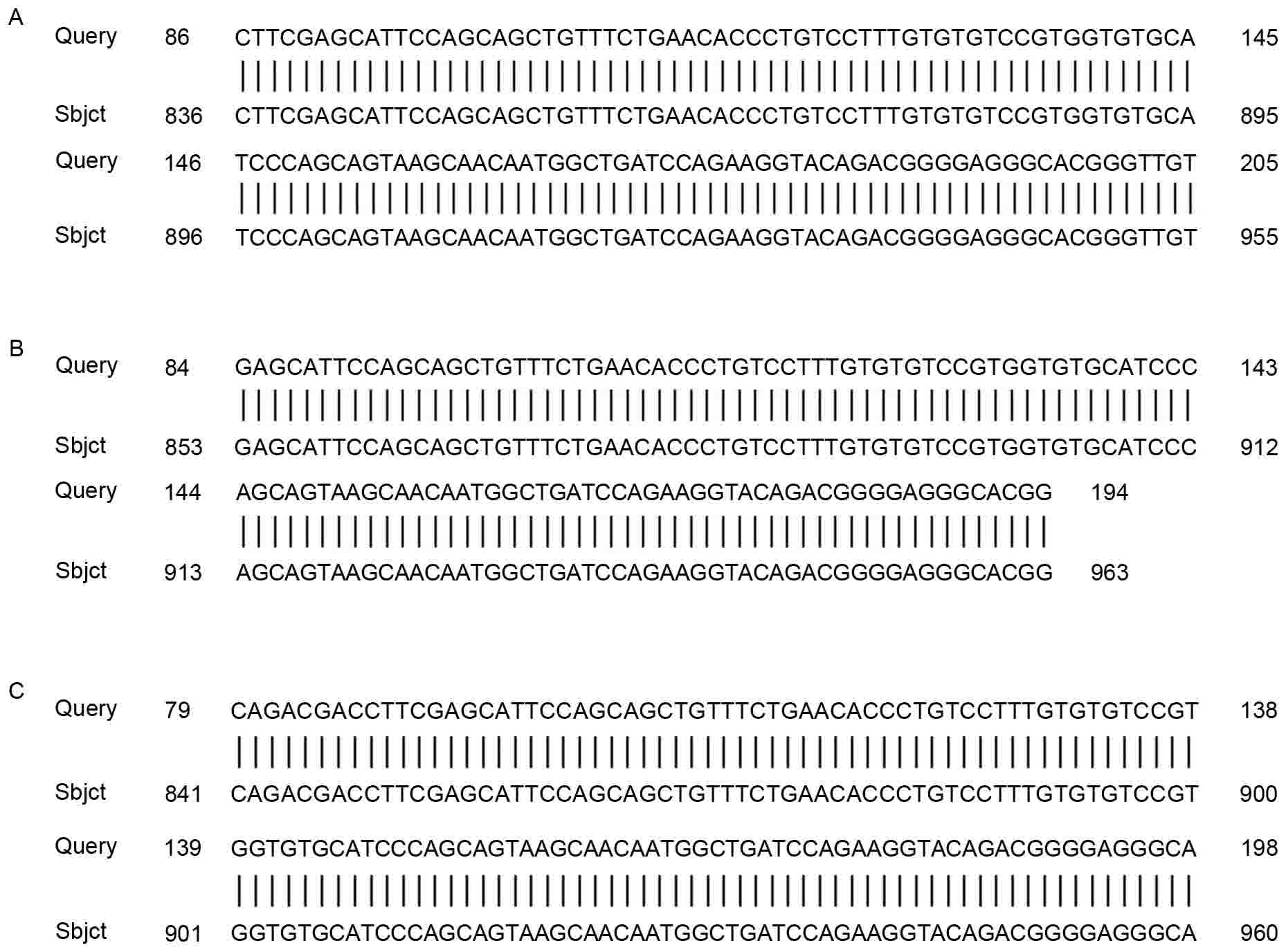
The sequencing results of samples positive for HPV18
were analyzed with the DNA man software and compared with the HPV18
sequence provided in GenBank (X05015.1). The results of this
analysis verified that four of the HPV18 E6-positive samples
harbored mutations in the viral DNA: Two samples contained one
mutation; the other two contained two mutations (Fig. 2).
HPV18 integration-derived transcripts
in HPV18 E6-positive esophageal cancer samples
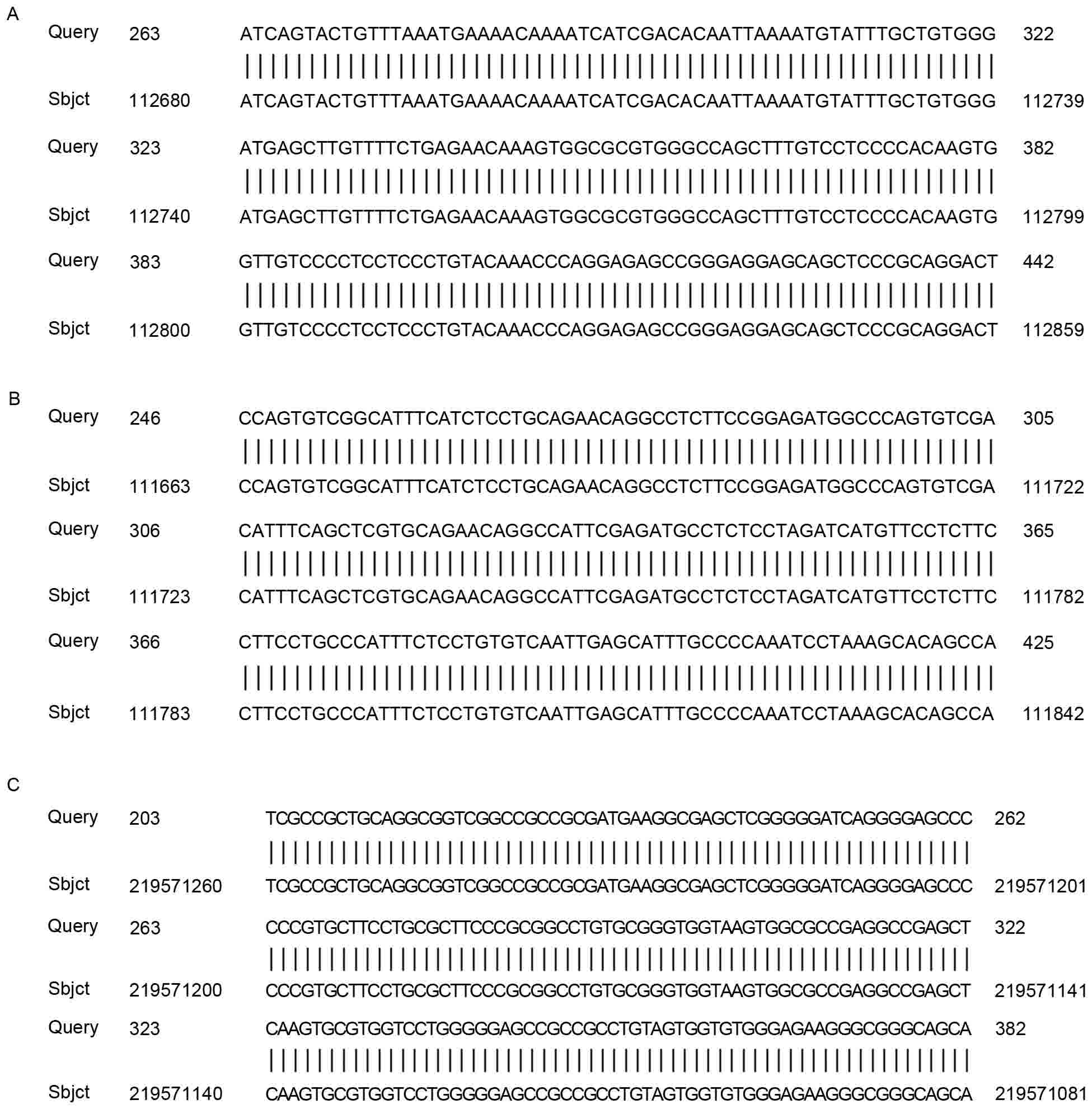
HPV18 integration sites were identified in all HPV18
E6-positive specimens. A ~600 bp HPV18 E7 PCR product was
identified in three samples using p2-18 primers. The HPV18 E7 PCR
product obtained (using primer p2-18) from the three samples was
ligated into the pMD-18T vector, and sequence analysis of the HPV18
integration sites was performed. The results of this sequencing
analysis indicated that the HPV18 E7 gene PCR product, amplified
using the p2-18 primer in these three samples, was part of the
HPV18 E7-E1 sequence (Fig. 3A-C).
Other sequences from two samples were similar to sequences from
human chromosome 5 (Fig. 4A and B)
and human chromosome 2 (Fig. 4C).
Discussion
The HPV positivity rate of 168/189 observed in the
present study is atypically high, comparing with previous studies
in esophageal cancer (10,18). This result indicated that the
prevalence of HPV infection for esophageal cancer in the Tangshan
area, and, taken in combination with previous studies (6–10),
evidence indicates HPV may serve an etiological role in esophageal
cancer. A total of 17.5% of 189 samples were positive for HPV18.
The sequence analysis of PCR products positive for HPV18 E6 DNA
identified two samples with one mutation in the viral DNA and two
samples with two mutations. The results indicated that the samples
harbored HPV18, and the high rates of HPV18 infection indicated
that HPV18 has an etiological role in the development of esophageal
carcinoma. Only part of HPV genome integrates into the human
genome, and the HPV genome is usually cleaved at gene E1 or E2
(14,19,20). The
integration of HPV DNA, including the E6 and E7 genes, into the
human genome generally includes the 5′ ends of the viral gene and
the 3′ ends of the human cell sequences (14,19,21,22).
Viral-host cell-gene interconnection resulting from HPV DNA
integration mediates DNA replication and recombination, with HPV E6
and E7 oncogene amplification being involved in tumorigenesis
(23). HPV integration sites may be
used as individualized biomarkers for cancer tumorigenesis
(24). Chronic HPV infection results
in the integration of the viral genome, which alters the expression
patterns of viral oncogenes and integration-associated host-cell
genes; this integration may have a vital function in the
progression of cervical damage and cancer tumorigenesis (25).
Applying APOT analysis to 33 HPV18-positive samples,
of which three HPV18-positive samples of integration-derived
transcripts were: Two cases of late-stage, poorly differentiated
cancer in men (aged 49 and 57 years); one case of early-stage
moderately differentiated cancer in a woman (aged 71 years). Three
patient samples is too few to be able to indicate that HPV18
integration is associated with sex, age, degree of differentiation
or pathological stage of esophageal carcinoma, supporting the
hypothesis that HPV-infected cells undergo preferred selective
outgrowth in pre-neoplastic lesions that express the
human-genome-integrated viral oncogenes E6 and E7. HPV18
integration was identified in esophageal cancer cells from three
patient specimens, integrating into chromosomes 2 and 5. Taken
together with the integration of HPV into the genome of EC109
cells, these results indicated that HPV randomly integrates into
the host cell and that the HPV viral genome was cleaved at the E1
and E2 open reading frames prior to integration. The E2 gene serves
as a pivotal modulator for E6 and E7 gene expression in the viral
life cycle (26,27). In a great proportion of HPV-infected
patients, HPV E2 restrains E6 and E7 gene transcription, which aids
the regulation of cellular proliferation (28). Cleavage of the E2 gene increased HPV
E6 and E7 gene expression, which disrupts the cell cycle and leads
to aberrant cellular proliferation (29–31). The
HPV E6 and E7 genes usually integrate into the host cell genome and
require longer incubation periods for the viral DNA replication and
recombination to produce a variety of genetic changes in the viral
and human genome (32,33). The expression levels of E6 and E7
simultaneously increase, which results in human chromosomal
instability and the development of malignant tumors (34).
The high prevalence of HPV18 infection suggested
that HPV18 infection is a pathogenic factor for esophageal
carcinoma progression. The integration of HPV18 DNA into the host
cell genome suggests that persistent HPV infection has a function
in esophageal epithelial cell malignant transformation and
carcinogenesis.
Acknowledgements
The present study was supported by the Project of
Science and Technology for Overseas Scholars in Hebei Province
(grant no. C201400559), the Project of North China University
Science and Technology (grant no. SP201506), the Hebei Education
Department (grant no. ZD2016003), and Beijing Natural Science
Foundation (grant no. 5162003).
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhao J, He YT, Zheng RS, Zhang SW and Chen
WQ: Analysis of esophageal cancer time trends in China, 1989–2008.
Asian Pac J Cancer Prev. 13:4613–4617. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sun X, Chen W, Chen Z, Wen D, Zhao D and
He Y: Population-based casecontrol study on risk factors for
esophageal cancer in five high-risk areas in China. Asian Pac J
Cancer Prev. 11:1631–1636. 2010.PubMed/NCBI
|
|
3
|
Zhang HZ, Jin GF and Shen HB:
Epidemiologic differences in esophageal cancer between Asian and
Western populations. Chin J Cancer. 31:281–286. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Esophageal cancer: Epidemiology,
pathogenesis and prevention. Nat Clin Pract Gastroenterol Hepatol.
5:517–526. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Syrjänen KJ: Histological changes
identical to those of condylomatous lesions found in esophageal
squamous cell carcinomas. Arch Geschwulstforsch. 52:283–292.
1982.PubMed/NCBI
|
|
6
|
Dong HC, Cui XB, Wang LH, Li M, Shen YY,
Zhu JB, Li CF, Hu JM, Li SG, Yang L, et al: Type-specific detection
of human papillomaviruses in Kazakh esophageal squamous cell
carcinoma by genotyping both E6 and L1 genes with MALDI-TOF mass
spectrometry. Int J Clin Exp Pathol. 8:13156–13165. 2015.PubMed/NCBI
|
|
7
|
Türkay DÖ, Vural Ç, Sayan M and Gürbüz Y:
Detection of human papillomavirus in esophageal and
gastroesophageal junction tumors: A retrospective study by
real-time polymerase chain reaction in an instutional experience
from Turkey and review of literature. Pathol Res Pract. 212:77–82.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ludmir EB, Stephens SJ, Palta M, Willett
CG and Czito BG: Human papillomavirus tumor infection in esophageal
squamous cell carcinoma. J Gastrointest Oncol. 6:287–295.
2015.PubMed/NCBI
|
|
9
|
Georgantis G, Syrakos T, Agorastos T,
Miliaras S, Gagalis A, Tsoulfas G, Spanos K and Marakis G:
Detection of human papillomavirus DNA in esophageal carcinoma in
Greece. World J Gastroenterol. 21:2352–2357. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu HY, Zhou SL, Ku JW, Zhang DY, Li B,
Han XN, Fan ZM, Cui JL, Lin HL, Guo ET, et al: Prevalence of human
papillomavirus infection in esophageal and cervical cancers in the
high incidence area for the two diseases from 2007 to 2009 in
Linzhou of Henan Province, Northern China. Arch Virol.
159:1393–1401. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang K, Li JT, Li SY, Zhu LH, Zhou L and
Zeng Y: Integration of human papillomavirus 18 DNA in esophageal
carcinoma 109 cells. World J Gastroenterol. 17:4242–4246. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang R, Shen C, Zhao L, Wang J, McCrae M,
Chen X and Lu F: Dysregulation of host cellular genes targeted by
human papillomavirus (HPV) integration contributes to HPV-related
cervical carcinogenesis. Int J Cancer. 138:1163–1174. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu F, Cao M, Shi Q, Chen H, Wang Y and Li
X: Integration of the full-length HPV16 genome in cervical cancer
and Caski and Siha cell lines and the possible ways of HPV
integration. Virus Genes. 50:210–220. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Klaes R, Woerner SM, Ridder R, Wentzensen
N, Duerst M, Schneider A, Lotz B, Melsheimer P and von Knebel
Doeberitz M: Detection of high-risk cervical intraepithelial
neoplasia and cervical cancer by amplification of transcripts
derived from integrated papillomavirus oncogenes. Cancer Res.
59:6132–6136. 1999.PubMed/NCBI
|
|
15
|
Reeh M, Nentwich MF, von Loga K, Schade J,
Uzunoglu FG, Koenig AM, Bockhorn M, Rosch T, Izbicki JR and
Bogoevski D: An attempt at validation of the Seventh edition of the
classification by the International UnionAgainst Cancer for
esophageal carcinoma. Ann Thorac Surg. 93:890–896. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee DC, Cheung CY, Law AH, Mok CK, Peiris
M and Lau AS: p38 mitogen- activated protein kinase-dependent
hyperinduction of tumor necrosis factor alpha expression in
response to avian influenza virus H5N1. J Virol. 79:10147–10154.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Karlsen F, Kalantari M, Jenkins A,
Pettersen E, Kristensen G, Holm R, Johansson B and Hagmar B: Use of
multiple PCR sets for optimal detection of human papillomavirus. J
Clin Microbiol. 34:2095–2100. 1996.PubMed/NCBI
|
|
18
|
Syrjänen KJ: HPV infections and
oesophageal cancer. J Clin Pathol. 55:721–728. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kadaja M, Sumerina A, Verst T, Ojarand M,
Ustav E and Ustav M: Genomic instability of the host cell induced
by the human papillomavirus replication machinery. EMBO J.
26:2180–2191. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Johansson C, Somberg M, Li X, Winquist
Backström E, Fay J, Ryan F, Pim D, Banks L and Schwartz S: HPV-16
E2 contributes to induction of HPV-16 late gene expression by
inhibiting early polyadenylation. EMBO J. 31:3212–3227. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Venuti A and Marcante ML: Presence of
human papillomavirus type 18 DNA in vulvar carcinomas and its
integration into the cell genome. J Gen Virol. 70:1587–1592. 1989.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Klimov E, Vinokourova S, Moisjak E,
Rakhmanaliev E, Kobseva V, Laimins L, Kisseljov F and Sulimova G:
Human papilloma viruses and cervical tumours: Mapping of
integration sites and analysis of adjacent cell ularsequences. BMC
Cancer. 2:242002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Akagi K, Li J, Broutian TR, Padilla-Nash
H, Xiao W, Jiang B, Rocco JW, Teknos TN, Kumar B, Wangsa D, et al:
Genome-wide analysis of HPV integration in human cancers reveals
recurrent, focal genomic instability. Genome Res. 24:185–199. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xu B, Chotewutmontri S, Wolf S, Klos U,
Schmitz M, Dürst M and Schwarz E: Multiplex identification of human
papillomavirus 16 DNA integration sites in cervical carcinomas.
PLoS One. 8:e666932013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lu X, Lin Q, Lin M, Duan P, Ye L, Chen J,
Chen X, Zhang L and Xue X: Multiple-integrations of HPV16 genome
and altered transcription of viral oncogenes and cellular genes are
associated with the development of cervical cancer. PLoS One.
9:e975882014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Steger G and Corbach S: Dose-dependent
regulation of the early promoter of human papillomavirus type 18 by
the viral E2 protein. J Virol. 71:50–58. 1997.PubMed/NCBI
|
|
27
|
Nishimura A, Ono T, Ishimoto A, Dowhanick
JJ, Frizzell MA, Howley PM and Sakai H: Mechanisms of human
papillomavirus E2-mediated repression of viral oncogene expression
and cervical cancer cell growth inhibition. J Virol. 74:3752–3760.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wells SI, Aronow BJ, Wise TM, Williams SS,
Couget JA and Howley PM: Transcriptome signature of irreversible
senescence in human papillomavirus-positive cervical cancer cells.
Proc Natl Acad Sci USA. 100:7093–7098. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bergner S, Halec G, Schmitt M, Aubin F,
Alonso A and Auvinen E: Individual and complementary effects of
human papillomavirus oncogenes on epithelial cell proliferation and
differentiation. Cells Tissues Organs. 201:97–108. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ekalaksananan T, Jungpol W, Prasitthimay
C, Wongjampa W, Kongyingyoes B and Pientong C: Polymorphisms and
functional analysis of the intact human papillomavirus16 e2 gene.
Asian Pac J Cancer Prev. 15:10255–10262. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Scheffner M, Romanczuk H, Münger K,
Huibregtse JM, Mietz JA and Howley PM: Functions of human
papillomavirus proteins. Curr Top Microbiol Immunol. 186:83–99.
1994.PubMed/NCBI
|
|
32
|
Alp Avcı G: Genomic organization and
proteins of human papillomavirus. Mikrobiyol Bul. 46:507–515.
2012.(In Turkish). PubMed/NCBI
|
|
33
|
Kessis TD, Connolly DC, Hedrick L and Cho
KR: Expression of HPV16 E6 or E7 increases integration of foreign
DNA. Oncogene. 13:427–431. 1996.PubMed/NCBI
|
|
34
|
Hillemanns P and Wang X: Integration of
HPV-16 and HPV-18 DNA in vulvar intraepithelial neoplasia. Gynecol
Oncol. 100:276–282. 2006. View Article : Google Scholar : PubMed/NCBI
|