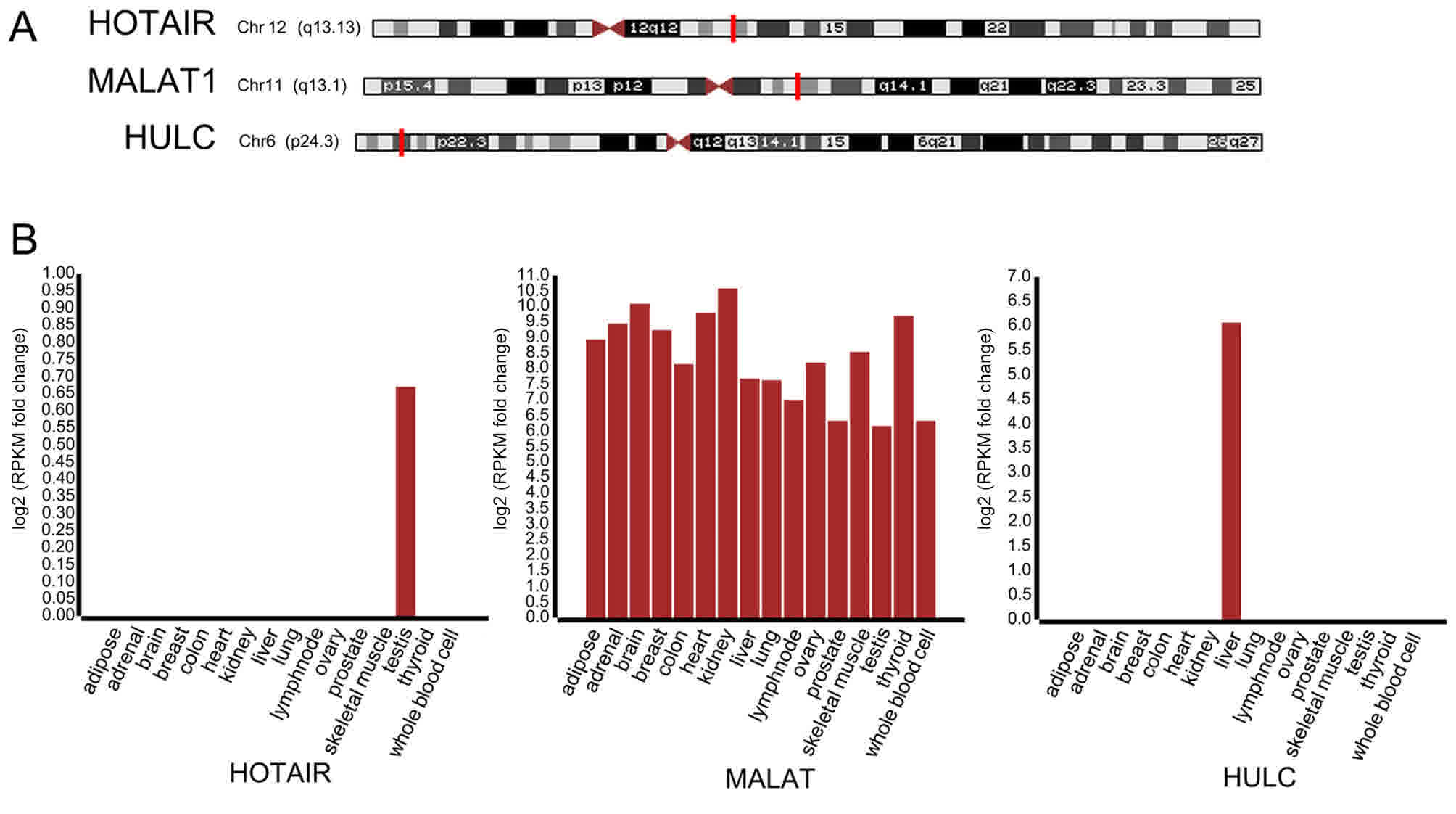
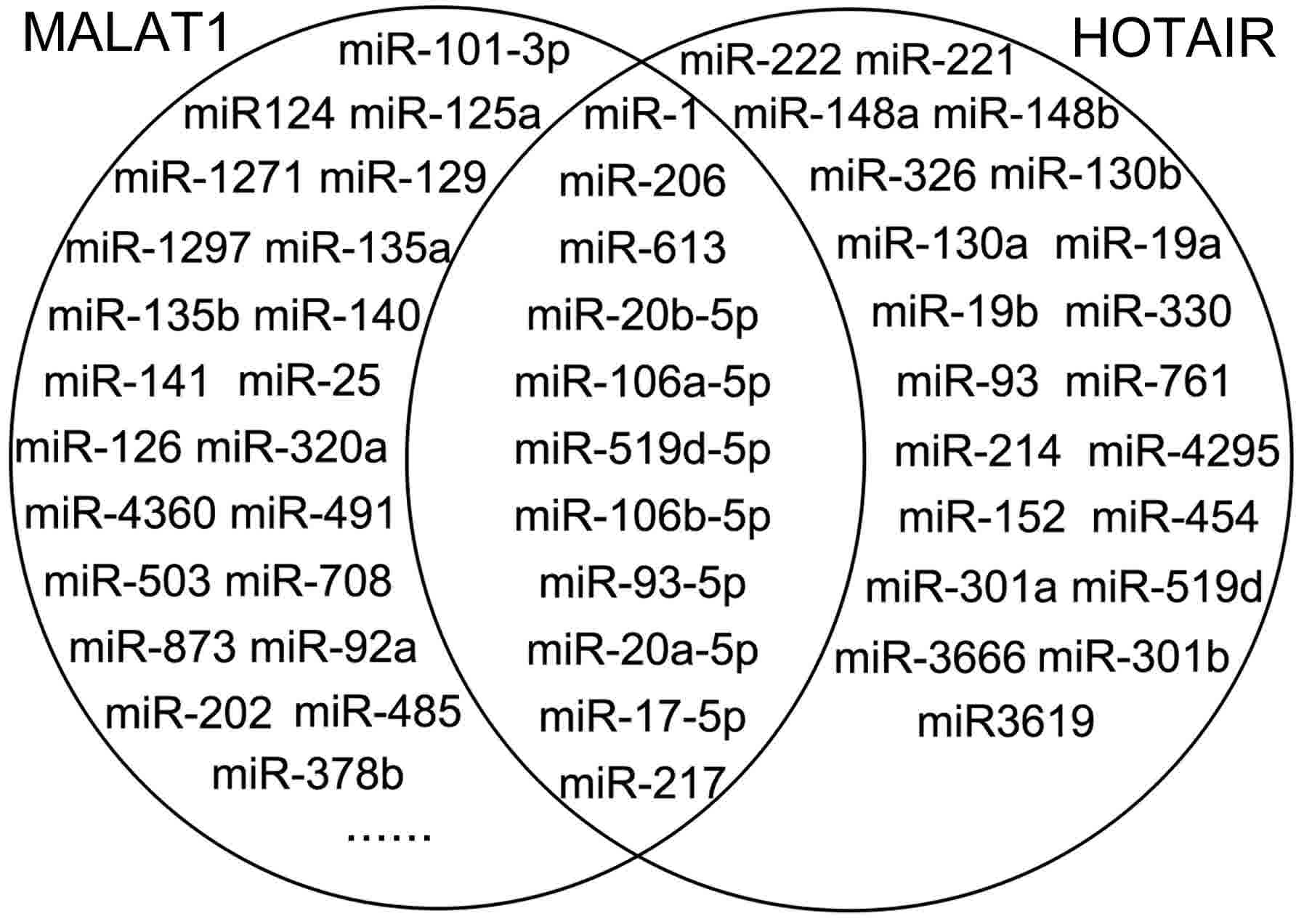
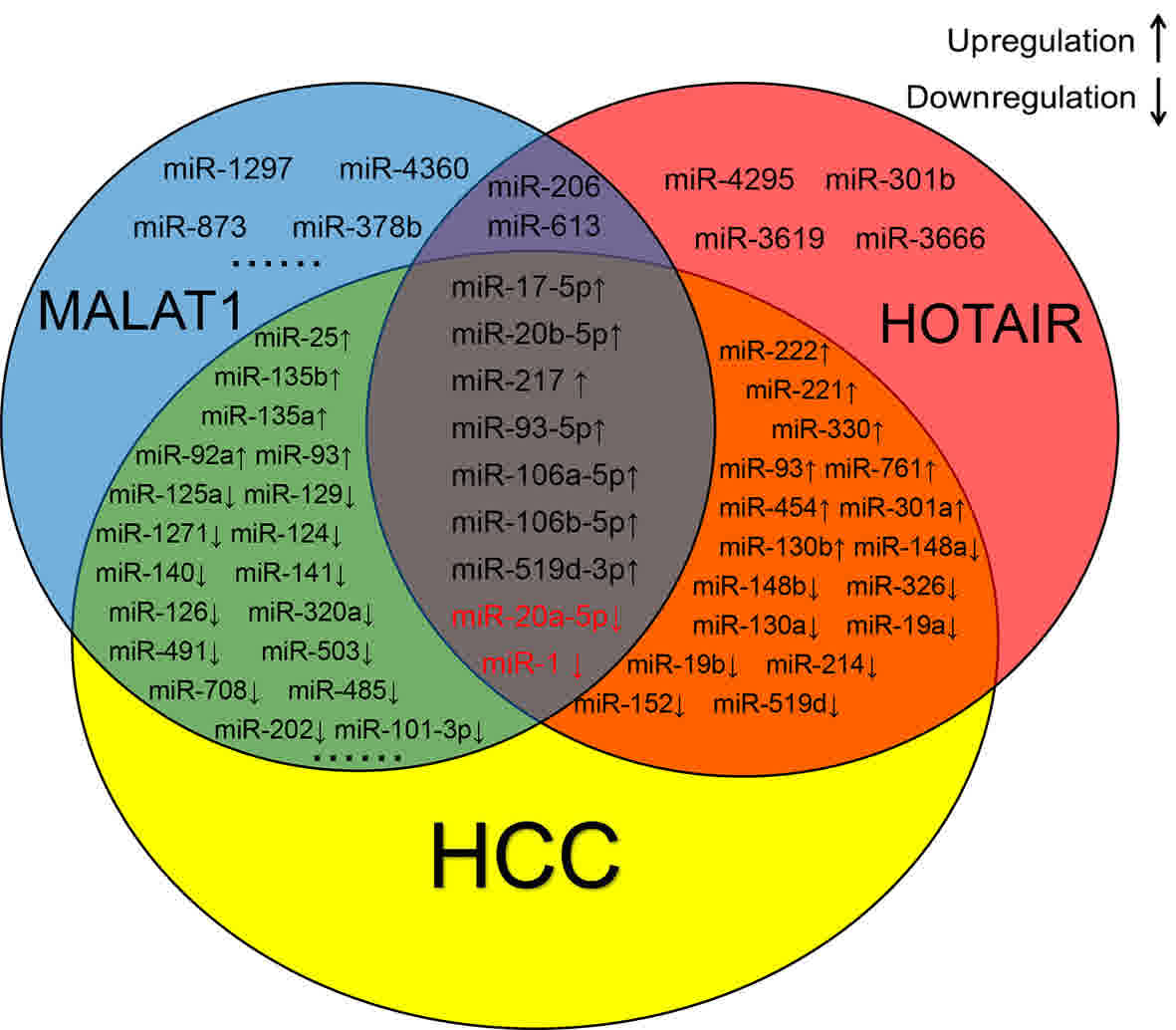
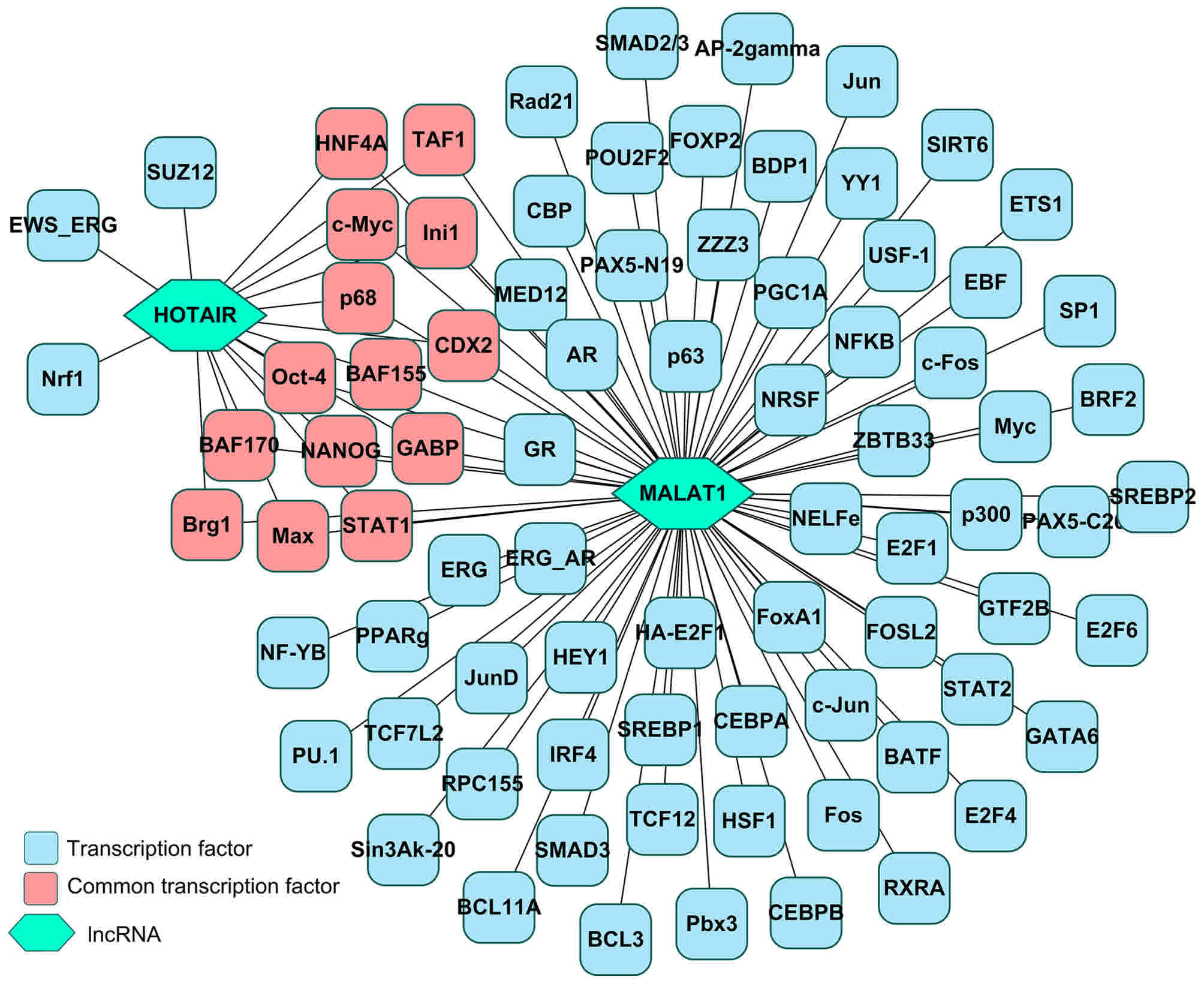
|
1
|
JTorre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang X, Xie X, Xiao YF, Xie R, Hu CJ, Tang
B, Li BS and Yang SM: The emergence of long non-coding RNAs in the
tumorigenesis of hepatocellular carcinoma. Cancer Lett.
360:119–124. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Anwar SL and Lehmann U: DNA methylation,
microRNAs, and their crosstalk as potential biomarkers in
hepatocellularcarcinoma. World J Gastroenterol. 20:7894–7913. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Roderburg C and Luedde T: The role of the
gut microbiome in the development and progression of liver
cirrhosis and hepatocellular carcinoma. Gut Microbes. 5:441–445.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chew CL, Conos SA, Unal B and Tergaonkar
V: Noncoding RNAs: Master regulators of inflammatory signaling.
Trends Mol Med. 24:66–84. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Weidle UH, Birzele F, Kollmorgen G and
Rüger R: Long Non-coding RNAs and their Role in Metastasis. Cancer
Genomics Proteomics. 14:143–160. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bassett AR, Akhtar A, Barlow DP, Bird AP,
Brockdorff N, Duboule D, Ephrussi A, Ferguson-Smith AC, Gingeras
TR, Haerty W, et al: Considerations when investigating lncRNA
function in vivo. Elife. 3:e030582014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Quinn JJ and Chang HY: Unique features of
long non-coding RNA biogenesis and function. Nat Rev Genet.
17:47–62. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Marchese FP, Raimondi I and Huarte M: The
multidimensional mechanisms of long noncoding RNA function. Genome
Biol. 18:2062017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zheng P, Yin Z, Wu Y, Xu Y, Luo Y and
Zhang TC: LncRNA HOTAIR promotes cell migration and invasion by
regulating MKL1 via inhibition miR206 expression in HeLa cells.
Cell Commun Signal. 16:52018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gutschner T, Richtig G, Haemmerle M and
Pichler M: From biomarkers to therapeutic targets-the promises and
perils of long non-coding RNAs in cancer. Cancer Metastasis Rev.
37:83–105. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
YiRen H, YingCong Y, Sunwu Y, Keqin L,
Xiaochun T, Senrui C, Ende C, XiZhou L and Yanfan C: Long noncoding
RNA MALAT1 regulates autophagy associated chemoresistance via
miR-23b-3p sequestration in gastric cancer. Mol Cancer. 16:1742017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Qiu L, Tang Q, Li G and Chen K: Long
non-coding RNAs as biomarkers and therapeutic targets: Recent
insights into hepatocellular carcinoma. Life Sci. 191:273–282.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu YR, Tang RX, Huang WT, Ren FH, He RQ,
Yang LH, Luo DZ, Dang YW and Chen G: Long noncoding RNAs in
hepatocellular carcinoma: Novel insights into their mechanism.
World J Hepatol. 7:2781–2791. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
He JH, Han ZP, Liu JM, Zhou JB, Zou MX, Lv
YB, Li YG and Cao MR: Overexpression of long non-coding RNA MEG3
inhibits proliferation of hepatocellular carcinoma Huh7 cells via
negative modulation of miRNA-664. J Cell Biochem. 118:3713–3721.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Malakar P, Shilo A, Mogilevsky A, Stein I,
Pikarsky E, Nevo Y, Benyamini H, Elgavish S, Zong X, Prasanth KV
and Karni R: Long Noncoding RNA MALAT1 promotes hepatocellular
carcinoma development by SRSF1 upregulation and mTOR activation.
Cancer Res. 77:1155–1167. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Quek XC, Thomson DW, Maag JL, Bartonicek
N, Signal B, Clark MB, Gloss BS and Dinger ME: lncRNAdb v2.0:
Expanding the reference database for functional long noncoding
RNAs. Nucleic Acids Res. 43:(Database Issue). D168–D173. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42:(Database Issue). D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li Y, Qiu C, Tu J, Geng B, Yang J, Jiang T
and Cui Q: HMDD v2.0: A database for experimentally supported human
microRNA and disease associations. Nucleic Acids Res. 42:(Database
Issue). D1070–D1074. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang JH, Li JH, Jiang S, Zhou H and Qu LH:
ChIPBase: A database for decoding the transcriptional regulation of
long non-coding RNA and microRNA genes from ChIP-Seq data. Nucleic
Acids Res. 41:(Database Issue). D177–D187. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dweep H, Gretz N and Sticht C: miRWalk
database for miRNA-target interactions. Methods Mol Biol.
1182:289–305. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Su WH, Chao CC, Yeh SH, Chen DS, Chen PJ
and Jou YS: OncoDB. HCC: An integrated oncogenomic database of
hepatocellular carcinoma revealed aberrant cancer target genes and
loci. Nucleic Acids Res. 35:(Database Issue). D727–D731. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar
|
|
27
|
Sandelin A, Wasserman WW and Lenhard B:
ConSite: Web-based prediction of regulatory elements using
cross-species comparison. Nucleic Acids Res. 32:(Web Server Issue).
W249–W252. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lin Z, Hu Y, Lai S, Xue M, Lin J, Qian Y,
Zhuo W, Chen S, Si J and Wang L: Long Noncoding RNA: Its partners
and their roles in cancer. Neoplasma. 62:846–854. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen G, Wang H, Xie S, Ma J and Wang G:
STAT1 negatively regulates hepatocellular carcinoma cell
proliferation. Oncol Rep. 29:2303–2310. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yao D, Peng S and Dai C: The role of
hepatocyte nuclear factor 4alpha in metastatic tumor formation of
hepatocellular carcinoma and its close relationship with the
mesenchymal-epithelial transition markers. BMC Cancer. 13:4322013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chang TS, Wu YC, Chi CC, Su WC, Chang PJ,
Lee KF, Tung TH, Wang J, Liu JJ, Tung SY, et al: Activation of
IL6/IGFIR confers poor prognosis of HBV-related hepatocellular
carcinoma through induction of OCT4/NANOG expression. Clin Cancer
Res. 21:201–210. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhu R, Wong KF, Lee NP, Lee KF and Luk JM:
HNF1α and CDX2 transcriptional factors bind to cadherin-17 (CDH17)
gene promoter and modulate its expression in hepatocellular
carcinoma. J Cell Biochem. 111:618–626. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kitagawa N, Ojima H, Shirakihara T,
Shimizu H, Kokubu A, Urushidate T, Totoki Y, Kosuge T, Miyagawa S
and Shibata T: Downregulation of the microRNA biogenesis components
and its association with poor prognosis inhepatocellular carcinoma.
Cancer Sci. 104:543–551. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Endo M, Yasui K, Zen Y, Gen Y, Zen K,
Tsuji K, Dohi O, Mitsuyoshi H, Tanaka S, Taniwaki M, et al:
Alterations of the SWI/SNF chromatin remodelling subunit-BRG1 and
BRM in hepatocellular carcinoma. Liver Int. 33:105–117. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mauleon I, Lombard MN, Muñoz-Alonso MJ,
Cañelles M and Leon J: Kinetics of myc-max-mad gene expression
during hepatocyte proliferation in vivo: Differential regulation of
mad family and stress-mediated induction of c-myc. Mol Carcinog.
39:85–90. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang D, Qi J, Liu R, Dai B, Ma W, Zhan Y
and Zhang Y: c-Myc plays a key role in TADs-induced apoptosis and
cell cycle arrest in human hepatocellular carcinoma cells. Am J
Cancer Res. 5:1076–1088. 2015.PubMed/NCBI
|
|
37
|
Hou X, Peng JX, Hao XY, Cai JP, Liang LJ,
Zhai JM, Zhang KS, Lai JM and Yin XY: DNA methylation profiling
identifies EYA4 gene as a prognostic molecular marker in
hepatocellular carcinoma. Ann Surg Oncol. 21:3891–3899. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tu H, Wei G, Cai Q, Chen X, Sun Z, Cheng
C, Zhang L, Feng Y, Zhou H, Zhou B and Zeng T: MicroRNA-212
inhibits hepatocellular carcinoma cell proliferation and induces
apoptosis by targeting FOXA1. Onco Targets Ther. 8:2227–2235.
2015.PubMed/NCBI
|
|
39
|
Abdou AG, Abd-Elwahed M, Badr M, Helmy M,
Soliman EA and Maher D: The differential immunohistochemical
expression of p53, c-Jun, c-Myc, and p21 between HCV-related
hepatocellular carcinoma with and without cirrhosis. Appl
Immunohistochem Mol Morphol. 24:75–87. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Du T and Zamore PD: Beginning to
understand microRNA function. Cell Res. 17:6612007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhou ZJ, Dai Z, Zhou SL, Hu ZQ, Chen Q,
Zhao YM, Shi YH, Gao Q, Wu WZ, Qiu SJ, Zhou J and Fan J: HNRNPAB
induces epithelial-mesenchymal transition and promotes metastasis
of hepatocellularcarcinoma by transcriptionally activating SNAIL.
Cancer Res. 74:2750–2762. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Struhl K: Transcriptional noise and the
fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol.
14:103–105. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Caley DP, Pink RC, Trujillano D and Carter
DR: Long noncoding RNAs, chromatin, and development.
ScientificWorldJournal. 10:90–102. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Vizoso M and Esteller M: The activatory
long non-coding RNA DBE-T reveals the epigenetic etiology of
facioscapulohumeral muscular dystrophy. Cell Res. 22:1413–1415.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ishigaki S, Masuda A, Fujioka Y, Iguchi Y,
Katsuno M, Shibata A, Urano F, Sobue G and Ohno K:
Position-dependent FUS-RNA interactions regulate alternative
splicing events and transcriptions. Sci Rep. 2:5292012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yang Z, Zhou L, Wu LM, Lai MC, Xie HY,
Zhang F and Zheng SS: Overexpression of long non-coding RNA HOTAIR
predicts tumor recurrence in hepatocellular carcinoma patients
following liver transplantation. Ann Surg Oncol. 18:1243–1250.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Geng YJ, Xie SL, Li Q, Ma J and Wang GY:
Large intervening non-coding RNA HOTAIR is associated with
hepatocellular carcinoma progression. J Int Med Res. 39:2119–2128.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Niinuma T, Suzuki H, Nojima M, Nosho K,
Yamamoto H, Takamaru H, Yamamoto E, Maruyama R, Nobuoka T, Miyazaki
Y, et al: Upregulation of miR-196a and HOTAIR drive malignant
character in gastrointestinal stromal tumors. Cancer Res.
72:1126–1136. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Tano K and Akimitsu N: Long non-coding
RNAs in cancer progression. Front Genet. 3:2192012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Kiss T: Small nucleolar RNA-guided
post-transcriptional modification of cellular RNAs. EMBO J.
20:3617–3622. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY,
Zhang F, Wu LM, Chen LM and Zheng SS: Long non-coding RNA MALAT-1
overexpression predicts tumor recurrence of hepatocellular
carcinoma after liver transplantation. Med Oncol. 29:1810–1816.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Du Y, Kong G, You X, Zhang S, Zhang T, Gao
Y, Ye L and Zhang X: Elevation of highly up-regulated in liver
cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell
proliferation via down-regulating p18. J Biol Chem.
287:26302–26311. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Takeshita F, Patrawala L, Osaki M,
Takahashi RU, Yamamoto Y, Kosaka N, Kawamata M, Kelnar K, Bader AG,
Brown D and Ochiya T: Systemic delivery of synthetic microRNA-16
lnhibits the growth of metastatic prostate tumors via dowregulation
of multiple cell-cycle genes. Mol Ther. 18:181–187. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Fu WM, Zhu X, Wang WM, Lu YF, Hu BG, Wang
H, Liang WC, Wang SS, Ko CH, Waye MM, et al: Hotair mediates
hepatocarcinogenesis through suppressing miRNA-218 expression and
activating P14 and P16 signaling. J Hepatol. 63:886–895. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Cui M, Xiao Z, Wang Y, Zheng M, Song T,
Cai X, Sun B, Ye L and Zhang X: Long noncoding RNA HULC modulates
abnormal lipid metabolism in hepatoma cells through an
miR-9-mediated RXRA signaling pathway. Cancer Res. 75:846–857.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Tang Y, Jin X, Xiang Y, Chen Y, Shen CX,
Zhang YC and Li YG: The lncRNA MALAT1 protects the endothelium
against ox-LDL-induced dysfunction via upregulating the expression
of the miR-22-3p target genes CXCR2 and AKT. FEBS Lett.
589:3189–3196. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wan Z, Pan H, Liu S, Zhu J, Qi W, Fu K,
Zhao T and Liang J: Downregulation of SNAIL sensitizes
hepatocellular carcinoma cells to TRAIL-induced apoptosis by
regulating the NF-κB pathway. Oncol Rep. 33:1560–1566. 2015.
View Article : Google Scholar : PubMed/NCBI
|