Introduction
Colorectal cancer (CRC) is a common gastrointestinal
malignancy. It is the third most common cancer worldwide and
accounts for >600,000 mortalities annually (1,2). The
5-year relative survival rate for patients with CRC is <65%
(3). Early diagnosis and treatment
are crucial to increase the survival rate of patients with CRC.
However, there is a lack of highly sensitive and specific
biomarkers for detection and prognosis of CRC.
CRC is a complex disease that is caused by genetic
and epigenetic alterations (4,5).
Epigenetic alterations are stable changes in gene expression,
without altering the underlying DNA sequence (6). DNA methylation is a common epigenetic
alteration and serves a key function in the regulation of gene
activity. Hypermethylation in gene promoter regions may lead to
transcriptional silencing (7).
Silencing of tumor suppressor genes is a crucial mechanism involved
in carcinogenesis (8). Gene
methylation may be reversible and may be modified by environmental
factors (9). It is reasonable to
hypothesize that methylated genes may be attractive candidates for
the detection and prevention of cancer. Previous studies suggested
several aberrantly methylated tumor suppressor genes to be used as
biomarkers for the early detection and prognosis of CRC (10,11).
However, their diagnostic sensitivity and specificity remains
unsatisfactory.
Smooth muscle protein 22α (SM22α), also known as
transgelin or TAGLN, is an actin-binding protein that is abundantly
expressed in smooth muscle cells in vertebrates (12). Although the biological function of
SM22α remains unclear, it has been suggested to regulate muscle
fiber contractility, cell differentiation, cell migration, tissue
invasion and tumor suppression (13–16).
Accumulating evidence suggests that SM22α may act as a tumor
suppressor. Previous studies reported decreased expression of SM22α
in several types of human cancer, including lung, prostate, renal
and breast cancer (17–20). The function of SM22α was also reported
to be associated with increased apoptosis of prostate cancer cells
(21). Furthermore, it was
demonstrated that SM22α may suppress the expression of matrix
metalloproteinase 9 (MMP-9), which serves an important function in
tumor progression (22). A previous
study demonstrated that SM22α expression was significantly
decreased in CRC (23). However, the
molecular mechanism underlying the downregulation of SM22α in CRC
remains to be elucidated.
In the present study, the methylation status and
protein expression of SM22α is examined in CRC and adjacent normal
tissue. To the best of our knowledge, this is the first study to
investigate the association between the protein expression and
methylation level of SM22α in CRC tissues and their adjacent normal
tissues. The aim of the present study is to investigate the
function of SM22α in the pathogenesis of CRC, and to identify
candidate biomarkers for the detection and prognosis of CRC.
Materials and methods
Tissue extraction
A total of 78 CRC tissues and adjacent normal
tissues were obtained from the Department of General Surgery of the
Fourth Hospital of Hebei Medical University (Hebei, China) between
October 2013 and November 2014. The mean age of the patients was 62
years (range, 42–78 years). A total of 45 cases were male and 33
were female. Postoperative pathological examination confirmed the
diagnosis of CRC. No patients received chemotherapy, radiotherapy
or immunotherapy prior to surgery. Adjacent normal tissues were
collected ≥10 cm away from the edge of the tumor. CRC tissues and
corresponding adjacent normal tissues were snap frozen in liquid
nitrogen within 30 min after their removal and stored at −80°C.
Following surgery, the patients were followed-up every 3 months.
Overall survival was defined as the time from the date of surgery
to the date of mortality due to CRC. Disease-free survival was
defined as the time from the primary surgical treatment to the date
of tumor recurrence or the last follow-up. At the time of the last
follow-up, 18 (23.08%) had succumbed to disease, 7 (8.97%) were
alive with disease and 53 (67.95%) were alive without disease. The
present study was approved by the Ethics Committee of the Fourth
Hospital of Hebei Medical University (Hebei, China) and written
informed consent was obtained from all individuals.
Western blot analysis
Total protein was extracted from tissues using a
lysis buffer containing 1% NP-40, 150 mM NaCl, 50 mM Tris (pH 7.5),
0.5% sodium deoxycholate and 1 mM phenylmethanesulfonyl fluoride.
Total protein was quantified using the Lowry method. Equal amount
of protein (30 µg) was separated by SDS-PAGE (10% gels) and
transferred onto polyvinylidene fluoride membranes. The membranes
were blocked with 5% non-fat milk in Tris-buffered saline with
Tween-20 (TBST) at room temperature for 2 h, and then incubated
with rabbit polyclonal antibodies against SM22α (dilution, 1:1,000;
Abcam, Cambridge, UK) and GAPDH (dilution, 1:800; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) at 4°C overnight. Membranes
were washed four times with TBST. Following primary incubation,
were incubated with Anti-Rabbit IgG (H&L) (Goat) Antibody
IRDye® 800CW conjugated (dilution, 1:20,000; cat. no.
611-131-122; Rockland Immunochemicals, Inc., Gilbertsville, PA,
USA) for 1 h at room temperature. The immunoreactive bands were
visualized using an Odyssey infrared imaging system (LI-COR
Biosciences, Lincoln, NE, USA). For quantification, the bands were
analyzed using ImageQuant software (version 3.0; LI-COR
Biosciences), and the signal densities of the SM22α bands were
normalized to those of the GAPDH bands. SM22α expression was
quantified as a ratio to GAPDH expression (SM22α/GAPDH ratio). The
experiment was repeated three times.
DNA extraction and bisulfite
modification
Total DNA was extracted from tissues using the
TIANamp Genomic DNA kit (Tiangen Biotech Co., Ltd., Beijing,
China). The purity and concentration of the DNA was evaluated using
a UV spectrophotometer. DNA (500 ng) was modified by sodium
bisulfite using the EZ DNA Methylation-Gold kit (Zymo Research
Corp., Irvine, CA, USA), according to the manufacturer's
protocol.
Methylation-specific polymerase chain
reaction (PCR) (MSP)
Methylation-specific PCR primers were designed using
MethPrimer software (Sangon Biotech Co., Ltd., Shanghai, China).
The primers for methylated SM22α were as follows:
5′-AATAGTGAAGTAGGAGTAGTCGTAAGTTC-3′ (forward) and
5′-AATCTACCGAAACTACCGAAAC-3′ (reverse). The primers for
unmethylated SM22α were as follows:
5′-GAATAGTGAAGTAGGAGTAGTTGTAAGTTT-3′ (forward) and
5′-CAATCTACCAAAACTACCAAAAC-3′ (reverse). The PCR reaction contained
Platinum SYBR-Green qPCR SuperMix-UDG (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) (12.5 µl), template DNA (1 µl),
primers (each 0.5 µl) and diethylpyrocarbonate H2O (10.5
µl). The thermocycling conditions were as follows: Initial
denaturation at 95°C for 5 min, followed by 40 cycles at 95°C for
30 sec, annealing at 64°C for 30 sec, elongation at 72°C for 45 sec
and extension at 72°C for 10 min. The PCR product for methylated
and unmethylated SM22α was 149 and 151 bp, respectively.
Amplification products were analyzed by 2% agarose gel
electrophoresis. Peripheral blood DNA that was treated with SssI
methyltransferase (New England BioLabs, Inc., Ipswich, MA, USA) was
used as a positive control and deionized water was used as a
negative control. The gel was visualized under UV illumination.
Statistical analysis
Data were analyzed using SPSS software (version
21.0; IBM Corp., Armonk, NY, USA). Quantitative values of protein
expressions in CRC tissues and adjacent normal tissues were
analyzed with paired Student's t-test and expressed as mean ±
standard deviation (SD). The methylation rate of SM22α between CRC
tissues and adjacent normal tissues were analyzed using the
χ2 test. The Association of the protein expression and
methylation status of SM22α with clinical parameters of patients
with CRC was compared using the χ2 test. The log-rank
test and Kaplan-Meier survival curve method were used for survival
analysis. P<0.05 was considered to indicate a statistically
significant difference.
Results
Protein expression and methylation
level of SM22α in CRC tissues and adjacent normal tissues
The relative protein expression of SM22α was
analyzed in 78 pairs of CRC tissues and their adjacent normal
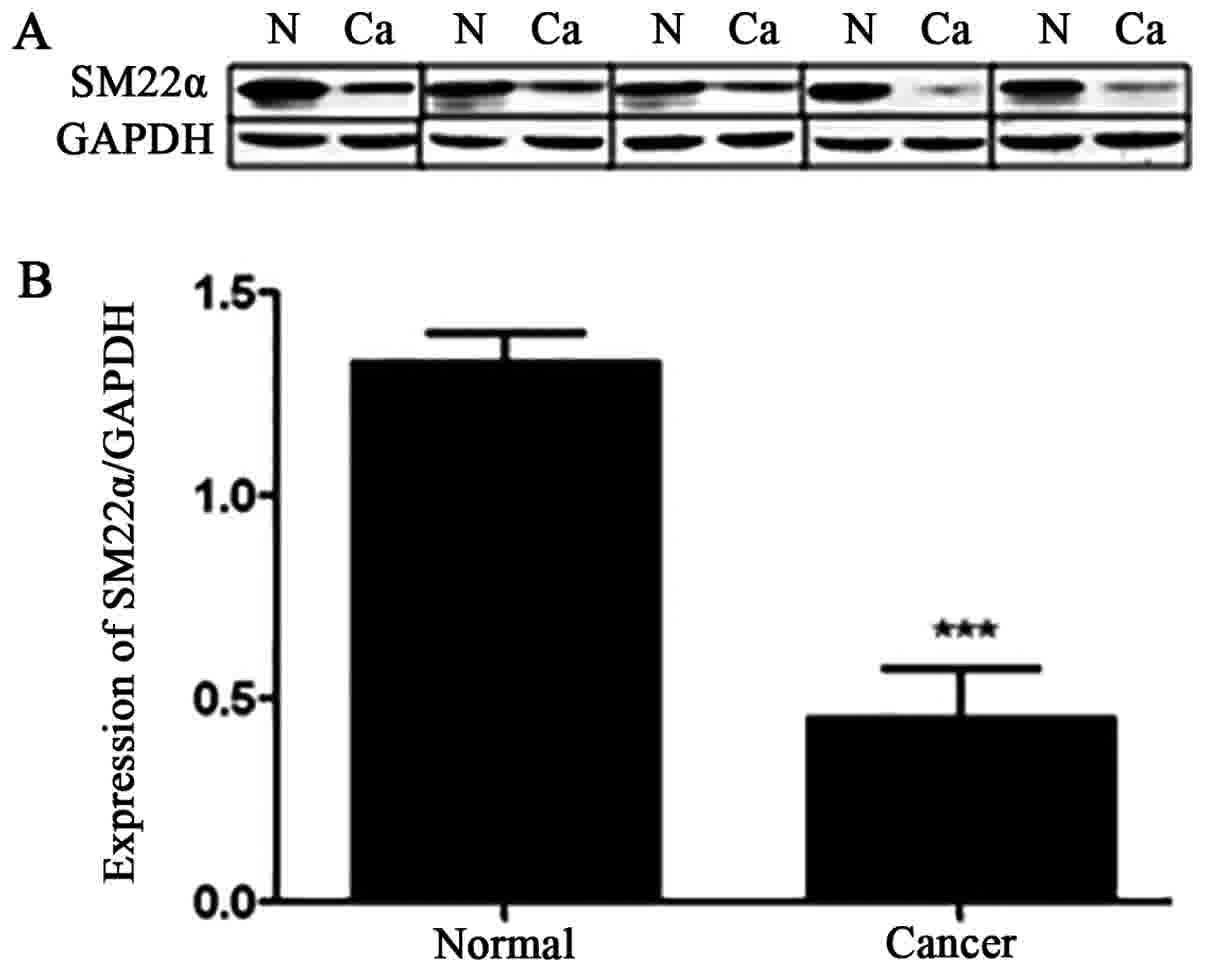
tissues using western blot analysis (Fig.
1). The results demonstrated a significantly decreased
expression of SM22α in 50 (68.5%) cases of CRC. Additionally, 28
(31.5%) cases of CRC exhibited an unchanged or upregulated
expression of SM22α. Fig. 1A shows
representative western blot analysis of the expression of SM22α
from five cases of CRC. SM22α was decreased by 2-fold in CRC
tissues compared with that in adjacent normal tissues (mean ± SD,
0.7280±0.1412 vs. 1.4458±0.3433; paired Student's t-test,
P<0.001; Fig. 1B).
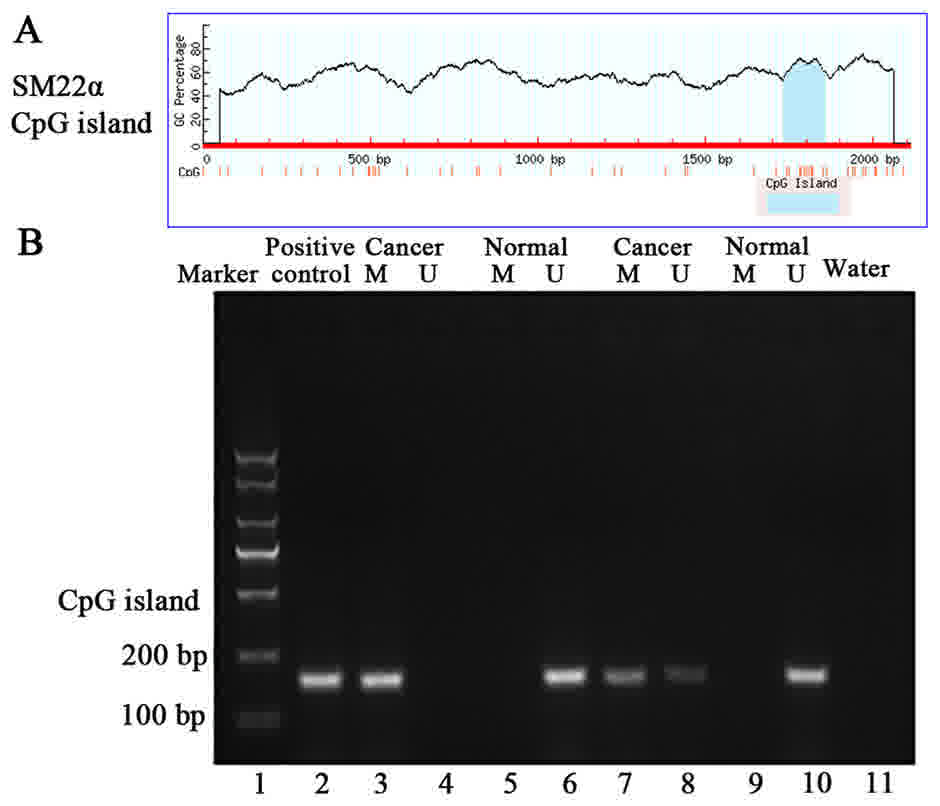
To investigate the molecular mechanisms of SM22α
gene regulation in CRC, DNA methylation levels within promoter CpG
islands, were evaluated. The results demonstrated that methylation
of SM22α CpG island in CRC tissues and adjacent normal tissues was
59.0% (43/78) and 21.9% (19/78), respectively. The difference was
statistically significant (χ2=15.418, P<0.001;
Fig. 2) (Data not shown). Fig. 2B shows data from two cases of CRC.
Association of the protein expression
and methylation status of SM22α with clinical parameters of
patients with CRC
Statistical analysis of SM22α protein expression,
SM22α gene methylation and clinical variables was performed using
the χ2 test. The expression of SM22α protein and
methylation levels of SM22α gene were not associated with age, sex,
tumor differentiation, tumor stage, lymph node metastasis, tumor
infiltration depth or tumor location in patients with colorectal
cancer (Table I).
 | Table I.Association between protein expression
and methylation level of SM22α and clinicopathological
characteristics. |
Table I.
Association between protein expression
and methylation level of SM22α and clinicopathological
characteristics.
|
|
| Decrease of SM22α
protein expression | SM22α
methylation |
|---|
|
|
|
|
|
|---|
| Clinical
parameters | Number (n) | Cases (n) | P-valuea | M (n) | P-valuea |
|---|
| Age (years) |
|
|
|
|
|
|
<60 | 31 | 22 | 0.305 | 19 | 0.374 |
| ≥60 | 47 | 28 |
| 24 |
|
| Sex |
|
|
|
|
|
|
Male | 45 | 26 | 0.174 | 23 | 0.405 |
|
Female | 33 | 24 |
| 20 |
|
|
Differentiation |
|
|
|
|
|
| Level
I–II | 62 | 41 | 0.463 | 35 | 0.644 |
| Level
III | 16 | 9 |
| 8 |
|
| TNM stage |
|
|
|
|
|
|
I–II | 49 | 29 | 0.239 | 25 | 0.343 |
|
III | 29 | 21 |
| 18 |
|
| Lymphatic
metastasis |
|
|
|
|
|
| No | 51 | 30 | 0.182 | 28 | 0.956 |
|
Yes | 27 | 20 |
| 15 |
|
| Infiltration
depth |
|
|
|
|
|
|
T1-T2 | 21 | 13 | 0.806 | 9 | 0.186 |
|
T3-T4 | 57 | 37 |
| 34 |
|
| Tumor location |
|
|
|
|
|
|
Colon | 29 | 19 | 0.841 | 16 | 0.995 |
|
Rectal | 49 | 31 |
| 27 |
|
Association of the protein expression
and methylation status of SM22α in CRC tissues
Patients with CRC were divided into SM22α high- and
low-expression groups on the basis of western blot analysis. The
association between the protein expression and the methylation
status of SM22α was determined in CRC tissues. SM22α was methylated
in 40 of 43 cases of CRC with low protein expression of SM22α.
SM22α was unmethylated in 25 of 35 cases of CRC with increased
protein expression of SM22α. Therefore, there was a negative
association between the protein expression and methylation levels
of SM22α in CRC (P<0.001; Table
II).
 | Table II.Association between protein
expression and methylation level of SM22α in colorectal cancer
tissues. |
Table II.
Association between protein
expression and methylation level of SM22α in colorectal cancer
tissues.
|
| SM22α protein
expression |
|
|---|
|
|
|
|
|---|
| SM22α promoter
methylation | Higha | Lowb | χ2 | P-value |
|---|
| Methylated | 3 | 40 |
|
|
| Unmethylated | 25 | 10 | 34.832 | <0.001 |
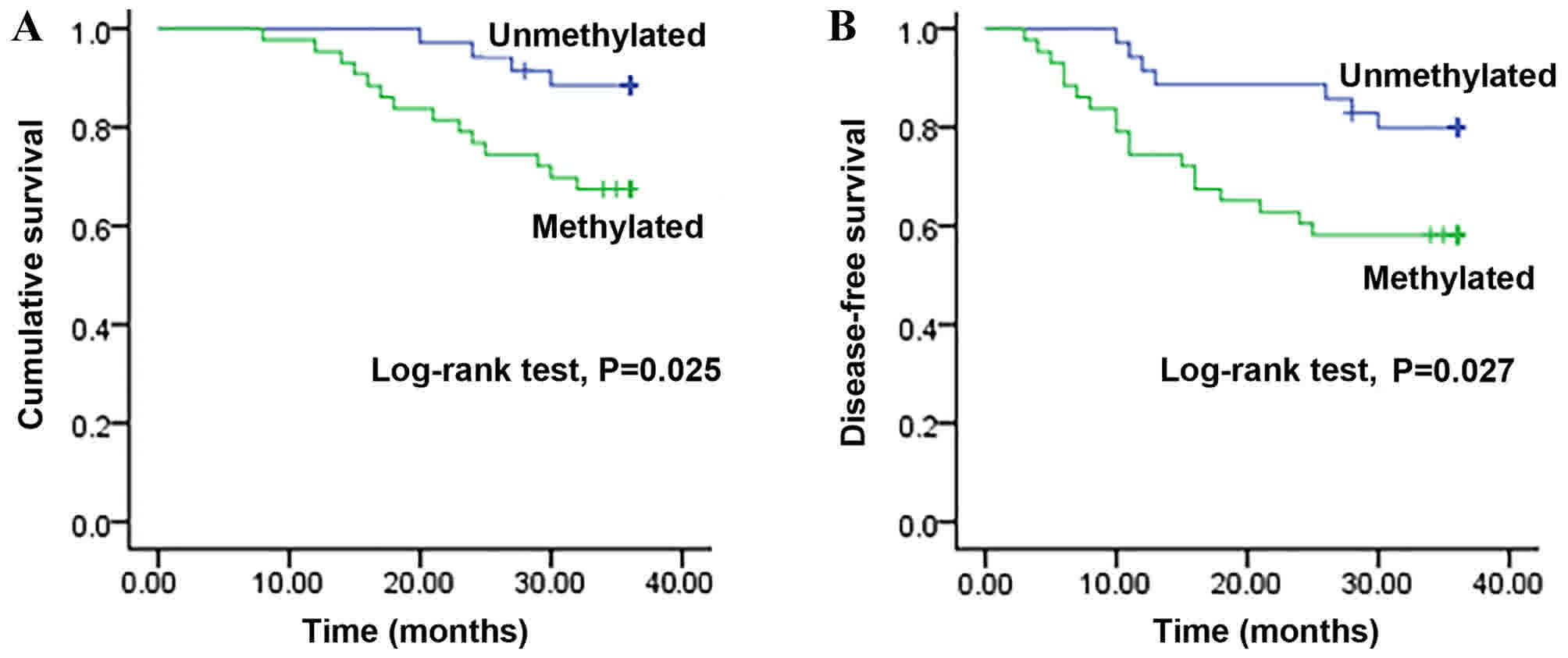
Methylation status of SM22α and
survival time in patients with CRC
Kaplan-Meier survival curves revealed that patients
with CRC with unmethylated promoter of SM22α presented a
significantly longer overall survival time (34.8±0.6 months)
compared with that in patients with a methylated SM22α promoter
status (30.9±1.3 months; log-rank test, P=0.025; Fig. 3A). Additionally, patients with CRC
with unmethylated promoter of SM22α exhibited a longer disease-free
survival time (32.5±1.3 months) compared with that in patients with
a methylated SM22α promoter status (26.0±1.9 months; log-rank test,
P=0.027; Fig. 3B).
Discussion
SM22α is an early differentiation marker of smooth
muscle cells, which is expressed in fibroblasts and epithelial
cells (13,14). SM22α serves an important function in
stabilizing the cellular structure and maintaining the
differentiated phenotype of smooth muscle cells via association
with actin (24,25). Previous studies have demonstrated that
SM22α may be involved in the development and progression of
malignant tumors. Decreased expression of SM22α has been reported
in lung, prostate, renal and breast cancer (17–20). Zhang
et al (21) reported that
SM22α may induce apoptosis via interacting with p53 in prostate
cancer cells. Nair et al (22)
demonstrated that SM22α repressed the expression of MMP-9 via
reducing the transactivation of activating protein 1-dependent and
compromising the activation of extracellular signal-regulated
kinase. Li et al (26)
demonstrated that SM22α may decrease proliferation and invasion,
and increase apoptosis in colorectal carcinoma cells. Xu et
al (27) reported that SM22α may
prevent the metastasis of CRC. These findings indicate that SM22α
may act as a tumor suppressor. However, the pathological function
of SM22α may depend on the type of cancer. For example,
upregulation of SM22α has been reported in gastric cancer and
esophageal squamous cell carcinoma (28,29).
Therefore, further research on the expression and function of SM22α
in various tumor types is required.
The results of the present study demonstrated a
decreased expression of SM22α in CRC tissues compared with that in
adjacent normal tissues. However, the protein level of SM22α was
not associated with age, sex, tumor differentiation, tumor stage,
lymph node metastasis, tumor infiltration depth or tumor location.
This implies that SM22α may be used as a biomarker of colorectal
carcinogenesis and may serve as a tumor suppressor. These results
are consistent with those of previous studies (23,30,31).
However, Zhou et al (32) and
Lin et al (33) reported that
elevated levels of SM22α increased invasiveness and lymph node
metastasis in CRC. The discrepancy between these results and the
present findings may arise from the limited sample size.
The expression of SM22α is regulated at the
transcriptional level (34). Yamamura
et al (35) demonstrated that
the transcriptional activity of SM22α was regulated by the
methylation of the promoter region in smooth muscle cells. Zhao
et al (30) revealed that
treatment with 5-aza-2-deoxycytidine may restore the mRNA and
protein levels of SM22α in the human intestinal epithelial cell
line HT29. However, the association between the protein expression
and methylation status of SM22α in CRC tissues and adjacent normal
tissues remains unclear. The results of the present study revealed
an increased methylation level of SM22α in CRC tissues compared
with that in adjacent normal tissues. Additionally, there was a
negative association between the methylation level and protein
expression of SM22α in CRC tissues. The results indicated that
hypermethylation of SM22α may be important in the regulation of
SM22α transcription.
DNA hypermethylation results in gene silencing, and
silencing of tumor suppressor genes is involved in carcinogenesis
(8,36). Aberrantly methylated tumor suppressor
genes may act as potential biomarkers for the early detection of
tumors. The results of the present study demonstrated that the
methylation level of SM22α promoter was increased in CRC tissues
compared with that in adjacent normal tissues. However, methylation
of SM22α promoter was not associated with age, sex, tumor
differentiation, tumor stage, lymph node metastasis, tumor
infiltration depth or tumor location. These results suggested that
hypermethylation of SM22α gene may occur at early stages of
colorectal carcinogenesis and therefore may be a biomarker for the
early diagnosis of CRC. Additionally, patients with an unmethylated
promoter of SM22α gene exhibited a longer survival time compared
with that in patients with methylated promoter of SM22α gene,
indicating that the methylation status of SM22α promoter region may
be associated with the prognosis of patients with CRC.
The present study has several limitations. First,
the sample size was small and further large-scale studies are
required to confirm the results of the present study. Secondly,
methylation status and protein expression of SM22α was not
evaluated in healthy individuals. Finally, further studies are
required to evaluate the methylation levels of SM22α in the plasma
or serum in patients with CRC.
In conclusion, the results of the present study
demonstrated that the protein expression of SM22α was significantly
decreased in patients with CRC. Additionally, the methylation level
in the SM22α gene promoter region was increased in CRC tissues
compared with that in adjacent normal tissues. There was a negative
association between the protein expression and methylation levels
of SM22α. Kaplan-Meier survival analysis revealed that patients
with CRC with an unmethylated promoter of SM22α gene exhibited an
improved survival time compared with that in patients with
methylated promoter of SM22α gene. Therefore, the methylation level
of SM22α promoter may be a useful biomarker for early detection,
prognosis and prediction of CRC.
Acknowledgements
The authors would like to acknowledge the doctors in
the Department of General Surgery of the Fourth Hospital of Hebei
Medical University, China, for their assistance in recruiting study
participants.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81372150) and the
Health and Family Planning Commission of Hebei Province (Hebei,
China; grant no. 20170732).
Authors' contributions
BL, YL and EW designed the study and applied for
approval from the Research Ethics Board. YL and DK recruited the
patients and collected the data. BL, YL and JZ analyzed the data
and prepared draft figures and tables. YL prepared the manuscript
draft with important intellectual input from BL, EW and JZ.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Fourth Hospital of Hebei Medical University
(Hebei, China) and written informed consent was obtained from all
individuals.
Consent for publication
All study participants provided written informed
consent for the data to be published.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shaukat A, Mongin SJ, Geisser MS, Lederle
FA, Bond JH, Mandel JS and Church TR: Long-term mortality after
screening for colorectal cancer. N Engl J Med. 369:1106–1114. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Migliore L, Igheli F, Spisni R and Coppede
F: Genetics, Cytogenetics, and epigenetics of colorectal cancer. J
Biomed Biotech. 2011:7923622011. View Article : Google Scholar
|
|
5
|
Grady WM and Ulrich CM: DNA alkylation and
DNA methylation: Cooperating mechanisms driving the formation of
colorectal adenomas and adenocarcinomas? Gut. 56:318–320. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Inbar-Feigenberg M, Choufani S, Butcher
DT, Roifman M and Weksberg R: Basic concepts of epigenetics. Fertil
Steril. 99:607–615. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Esteller M: Epigenetic gene silencing in
cancer: The DNA hypermethylome. Hum Mol Genet 16 Spec No.
1:R50–R59. 2007. View Article : Google Scholar
|
|
8
|
Fakhr Ghavifekr M, Hagh Farshdousti M,
Shanehbandi D and Baradaran B: DNA methylation pattern as important
epigenetic criterion in cancer. Genet Res Int.
2013:3175692013.PubMed/NCBI
|
|
9
|
Jaenisch R and Bird A: Epigenetic
regulation of gene expression: How the genome integrates intrinsic
and environmental signals. Nat Genet. 33 Suppl:S245–S254. 2003.
View Article : Google Scholar
|
|
10
|
Carmona FJ, Azuara D, Berenguer-Llergo A,
Fernández AF, Biondo S, de Oca J, Rodriguez-Moranta F, Salazar R,
Villanueva A, Fraga MF, et al: DNA methylation biomarkers for
noninvasive diagnosis of colorectal cancer. Cancer Prev Res
(Phila). 6:656–665. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ahn JB, Chung WB, Maeda O, Shin SJ, Kim
HS, Chung HC, Kim NK and Issa JP: DNA methylation predicts
recurrence from resected stage III proximal colon cancer. Cancer.
117:1847–1854. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang JC, Kim S, Helmke BP, Yu WW, Du KL,
Lu MM, Strobeck M, Yu Q and Parmacek MS: Analysis of
SM22alpha-deficient mice reveals unanticipated insights into smooth
muscle cell differentiation and function. Mol Cell Biol.
21:1336–1344. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Assinder SJ, Stanton JA and Prasad PD:
Transgelin: An actin-binding protein and tumour suppressor. Int J
Biochem Cell Biol. 41:482–486. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lawson D, Harrison M and Shapland C:
Fibroblast transgelin and smooth muscle SM22alpha are the same
protein, the expression of which is down-regulated in many cell
lines. Cell Motil Cytoskeleton. 38:250–257. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shields JM, Rogers-Graham K and Der CJ:
Loss of transgelin in breast and colon tumors and in RIE-1 cells by
Ras deregulation of gene expression through Raf-independent
pathways. J Biol Chem. 277:9790–9799. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Albiges-Rizo C, Destaing O, Fourcade B,
Planus E and Block MR: Actin machinery and mechanosensitivity in
invadopodia, podosomes and focal adhesions. J Cell Sci.
122:3037–3049. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li LS and Kim H, Rhee H, Kim SH, Shin DH,
Chung KY, Park KS, Paik YK, Chang J and Kim H: Proteomic analysis
distinguishes basaloid carcinoma as a distinct subtype of nonsmall
cell lung carcinoma. Proteomics. 4:3394–3400. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Prasad PD, Stanton JA and Assinder SJ:
Expression of the actin-associated protein transgelin (SM22) is
decreased in prostate cancer. Cell Tissue Res. 339:337–347. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Klade CS, Voss T, Krystek E, Ahorn H,
Zatloukal K, Pummer K and Adolf GR: Identification of tumor
antigens in renal cell carcinoma by serological proteome analysis.
Proteomics. 1:890–898. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sayar N, Karahan G, Konu O, Bozkurt B,
Bozdogan O and Yulug IG: Transgelin gene is frequently
downregulated by promoter DNA hypermethylation in breast cancer.
Clin Epigenetics. 7:1042015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang ZW, Yang ZM, Zheng YC and Chen ZD:
Transgelin induces apoptosis of human prostate LNCaP cells through
its interaction with p53. Asian J Androl. 12:186–195. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nair RR, Solway J and Boyd DD: Expression
cloning identifies transgelin (SM22) as a novel repressor of 92-kDa
type IV collagenase (MMP-9) expression. J Biol Chem.
281:26424–26436. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xie XL, Liu YB, Liu YP, Du BL, Li Y, Han M
and Li BH: Reduced expression of SM22 is correlated with low
autophagy activity in human colorectal cancer. Pathol Res Pract.
209:237–243. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gimona M, Kaverina I, Resch GP, Vignal E
and Burgstaller G: Calponin repeats regulate actin filament
stability and formation of podosomes in smooth muscle cells. Mol
Biol Cell. 14:2482–2491. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Han M, Dong LH, Zheng B, Shi JH, Wen JK
and Cheng Y: Smooth muscle 22 alpha maintains the differentiated
phenotype of vascular smooth muscle cells by inducing filamentous
actin bundling. Life Sci. 84:394–401. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li Q, Shi R, Wang Y and Niu X: TAGLN
suppresses proliferation and invasion, and induces apoptosis of
colorectal carcinoma cells. Tumour Biol. 34:505–513. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu L, Gao Y, Chen Y, Xiao Y, He Q, Qiu H
and Ge W: Quantitative proteomics reveals that distant
recurrence-associated protein R-Ras and Transgelin predict
post-surgical survival in patients with Stage III colorectal
cancer. Oncotarget. 7:43868–43893. 2016.PubMed/NCBI
|
|
28
|
Yu B, Chen X, Li J, Qu Y, Su L, Peng Y,
Huang J, Yan J, Yu Y, Gu Q, et al: Stromal fibroblasts in the
microenvironment of gastric carcinomas promote tumor metastasis via
upregulating TAGLN expression. BMC Cell Biol. 14:172013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen JY, Xu L, Fang WM, Han JY, Wang K and
Zhu KS: Identification of PA28β as a potential novel biomarker in
human esophageal squamous cell carcinoma. Tumour Biol.
39:10104283177197802017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhao L, Wang H, Deng YJ, Wang S, Liu C,
Jin H and Ding YQ: Transgelin as a suppressor is associated with
poor prognosis in colorectal carcinoma patients. Mod Pathol.
22:786–796. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yeo M, Park HJ, Kim DK, Kim YB, Cheong JY,
Lee KJ and Cho SW: Loss of SM22 is a characteristic signature of
colon carcinogenesis and its restoration suppresses colon
tumorigenicity in vivo and in vitro. Cancer. 116:2581–2589.
2010.PubMed/NCBI
|
|
32
|
Zhou HM, Fang YY, Weinberger PM, Ding LL,
Cowell JK, Hudson FZ, Ren M, Lee JR, Chen QK, Su H, et al:
Transgelin increases metastatic potential of colorectal cancer
cells in vivo and alters expression of genes involved in cell
Motility. BMC Cancer. 16:552016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lin Y, Buckhaults PJ, Lee JR, Xiong H,
Farrell C, Podolsky RH, Schade RR and Dynan WS: Association of the
actin-binding protein transgelin with lymph node metastasis in
human colorectal cancer. Neoplasia. 11:864–873. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Prinjha RK, Shapland CE, Hsuan JJ, Totty
NF, Mason IJ and Lawson D: Cloning and sequencing of cDNAs encoding
the actin cross-linking protein transgelin defines a new family of
actin-associated proteins. Cell Motil Cytoskeleton. 28:243–255.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yamamura H, Masuda H, Ikeda W, Tokuyama T,
Takagi M, Shibata N, Tatsuta M and Takahashi K: Structure and
expression of the human SM22alpha gene, assignment of the gene to
chromosome 11, and repression of the promoter activity by cytosine
DNA methylation. J Biochem. 122:157–167. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mohn F, Weber M, Rebhan M, Roloff TC,
Richter J, Stadler MB, Bibel M and Schübeler D: Lineage-specific
polycomb targets and de novo DNA methylation define restriction and
potential of neuronal progenitors. Mol Cell. 30:755–766. 2008.
View Article : Google Scholar : PubMed/NCBI
|