Introduction
Lung cancer is the principal cause of
cancer-associated mortality worldwide, with an estimated 600,000
individual succumbing to the disease annually; the annual mortality
rate is predicted to reach 1 million by 2025 in China (1). Non-small cell lung cancer (NSCLC)
accounts for >85% of all diagnosed cases of lung cancer and
patients typically have a poor prognosis (2). At present, surgical resection remains
the only curative treatment for localized tumors confined to the
lung; however, 80–85% of patients present with un resectable
disease at diagnosis (3). Distant
metastasis leads to the majority of human cancer-associated
mortalities (4). Therefore, it is
important to identify sensitive and specific biomarkers for the
development and progression of lung cancer.
Tumor metastasis is comprised of multiple steps.
Cell invasion and migration are hallmarks of malignant
transformation. Epithelial-mesenchymal transition (EMT) refers to
the process whereby epithelial cells lose their epithelial
phenotype and obtain a mesenchymal phenotype, which is responsible
for tumor metastasis (5). During the
EMT process, epithelial cells gradually lose their epithelial
structures, including E-cadherin protein-mediated intercellular
interactions, whilst also obtaining mesenchymal characteristics,
including the upregulation of vimentin and fibronectin (6). The expression of Snail, a key
transcriptional repressor of E-cadherin that can induce EMT, is
controlled by a number of signaling pathways including the Wnt
pathway (5).
Epigenetic changes refer to functional changes to
the genomic DNA that affect gene expression but do not alter the
nucleotide sequence (7). Examples of
epigenetic alterations include histone modifications, DNA
methylation and microRNA (miRNA/miR) expression. Epigenetic
alterations have been proposed to be targets for cancer treatment
due to their vital roles in human carcinogenesis (8). DNA methylation is a type of epigenetic
modification that affects gene expression by adding a methyl group
to cytosine nucleotides across the genome at
cytosine-phosphate-guanine sites (9).
Deregulation of DNA methylation has been reported to cause abnormal
gene regulation, leading to anomalous embryonic development and
diseases (10,11). In addition, a number of regulators of
the Wnt signaling pathway that are modulated by DNA methylation
have been reported to be involved in the regulation of tumor
metastasis (11,12). DNA methylation is typically mediated
by DNA methyltransferases (DNMTs) The DNMT family consists of four
members: DNMT1, DNMT2, DNMT3A and DNMT3B (13). Among the DNMT family members, DNMT1 is
primarily responsible for maintaining DNA methylation. DNMT1 mRNA
expression has been reported to be elevated in NSCLC, and the
elevated mRNA levels of DNMT1 occur more frequently in poorly
differentiated, as compared with in well and moderately
differentiated, tumor cells. In addition, elevated mRNA levels of
DNMT1 were negatively associated with the overall survival time in
patients with NSCLC (14).
Previous studies have demonstrated that the
inhibition of DNMT1 mediates growth arrest and apoptosis in lung
cancer cells (15). DNMT1 knock down
induces Cyclin-dependent kinase inhibitor 1A and B-cell lymphoma 2
interacting killer gene expression via an underlying mechanism
independent of DNA methylation. Reduction of DNMT1 expression may
prevent epigenetically mediated gene silencing and provide a novel
clinical strategy to target tumor cells (16).
DNA methylation may be involved in the EMT process.
For example, E-cadherin, a gene involved in cell adhesion and
signaling that serves a key role in EMT, is suppressed by DNA
methylation during EMT (17). The
critical Smad signaling pathway that regulates EMT serves an
important role in maintaining the epigenetic silencing of target
genes (18). Furthermore, it has been
reported that certain therapeutic agents, including mithramycin A,
are able to repress tumor metastasis via demethylation mechanisms
by inhibiting the protein expression of DNMT1 (19). Although a number of studies have
suggested that DNMT1 is implicated in cancer progression, its role
in tumor migration, invasion and EMT remains unclear.
Thus, in the present study, the expression of DNMT1
in NSCLC cells in vitro was investigated, in addition to the
effects of small interfering (si)RNA-mediated knockdown of DNMT1
expression on tumor migration and invasion, and on the EMT of NSCLC
cells. In addition, the effects of DNMT1 siRNA on the matrix
metalloproteinase (MMP)2 and Wnt signaling pathways were
investigated in order to explore the underlying molecular
mechanisms of the effect of DNMT1 on lung tumors.
Materials and methods
Cell lines
Two invasive lung cancer cell lines, 95C (low
invasive ability) and 95D (high invasive ability), were purchased
from The Cell Bank of Type Culture Collection of Chinese Academy of
Sciences (Shanghai, China). Clonetics™ human small
airway epithelial cells (SAECs) were obtained from Lonza (Basel,
Switzerland). All the cells were cultured in RPMI-1640 medium
supplemented with 10% fetal bovine serum (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) in a humidified atmosphere at
37°C with 5% CO2.
Plasmid construction and
transfection
95D cells were seeded in 24-well plates
(1.5×105 cells/well) and incubated for 24 h in a
humidified atmosphere at 37°C with 5% CO2. Negative
control (NC) siRNA was purchased from Ambion (Thermo Fisher
Scientific, Inc.). DNMT1 siRNA was constructed as described
previously (20). NC siRNA and DNMT1
siRNA were transfected into the cells using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol.
Following incubation for at 37°C for 48 h, 95D cells transfected
with NC siRNA or DNMT1 siRNA were analyzed by reverse
transcription-polymerase chain reaction (RT-PCR) and western
blotting to validate the siRNA knockdown.
Immunofluorescence
95D cells at a concentration of 1.0×104
per coverslip were seeded onto sterile coverslips and subjected to
immunofluorescence analysis following NC or DNMT1 siRNA
transfection. Cells were fixed with 4% paraformaldehyde at room
temperature for 10 min, permeabilized using 0.3% Triton-X-100 and
then washed with PBS, followed by incubation with specific primary
antibodies (anti-fibronectin; cat. no. F3648; 1:400 dilution;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany; anti-E-cadherin;
cat. no. 24E10; 1:600 dilution; Cell Signaling Technology, Inc.,
Danvers, MA, USA; anti-vimentin; 1:200 dilution; cat. no. 5741;
Cell Signaling Technology, Inc.) at 4°C overnight Then the cells
were incubated with fluorescein isothiocyanate-conjugated secondary
antibodies (cat. no. ZF-0314; 1:100 dilution; Beijing Zhongshan
Golden Bridge Biotechnology, Co., Ltd., Beijing, China) at 37°C for
1 h. The cell nucleus was counterstained with DAPI (Invitrogen;
Thermo Fisher Scientific, Inc.). Images of the stained cells were
captured with a fluorescence microscope.
Western blot analysis for DNMT1 and
EMT markers
RIPA lysis buffer was used for cell lysis, and a BCA
Protein Assay kit (Beyotime Institute of Biotechnology, Haimen,
China) was used to determine the protein concentration. Equal
amounts of protein were loaded into each well for SDS-PAGE,
separated by electrophoresis and transferred onto polyvinylidene
fluoride membranes (EMD Millipore, Billerica, MA, USA). The
membranes were blocked with 5% fat-free milk at room temperature
for 1 h. Then the membranes were probed with anti-DNMT1 (1:1,000
dilution; cat. no. sc-271729), anti-MMP2 (1:2,000 dilution; cat.
no. sc-13594), anti-Snail (1:2,000 dilution; cat. no. sc-28199),
anti-β-catenin (1:2,000 dilution; cat. no. sc-133239), anti-GAPDH
(1:1,000 dilution; cat. no. sc-47724) (all Santa Cruz
Biotechnology, Inc., Dallas, TX, USA), anti-E-cadherin (1:2,000
dilution), anti-vimentin (1:1,000 dilution) and anti-fibronectin
(1:1,000 dilution) overnight at 4°C, followed by incubation with
horseradish peroxidase-conjugated secondary antibodies (ZB-2301;
1:3,000; ZSGB-Bio, Beijing, China) for 1 h at room temperature. The
protein bands on the membranes were visualized using an Enhanced
Chemiluminescence Detection kit (Beyotime Institute of
Biotechnology). GAPDH served as the loading control. The relative
amount of protein in the bands was quantified by densitometry using
ImageJ software (version 1.46; National Institutes of Health,
Bethesda, MD, USA).
RT-PCR
Total RNA was isolated from the 95D cells using the
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and reverse transcribed into cDNA using a PrimeScript™ RT
Reagent kit (Takara Bio, Inc., Tokyo, Japan), according to the
manufacturer's protocol. The sequences of the PCR primers employed
in the current study were as follows: DNMT1 forward,
5′-ACAGACAGCGTTTGGTTGAC-3′ and reverse, 5′-TAGGGAACAGAAGGCAAGGG-3′;
β-actin forward, 5′-CGGGAAATCGTGCGTGAC-3′ and reverse,
5′-TGGAAGGTGGACAGCGAGG-3′. PCR was conducted under the following
conditions: 95°C for 30 sec, 30 cycles of 94°C for 45 sec, 59°C
(DNMT-1) or 54°C (β-actin) for 40 sec and 72°C for 1 min, followed
by 10 min at 72°C. All the PCR reactions were run in triplicate.
β-actin served as an internal control. The intensity of each band
amplified by RT-PCR was analyzed using ImageJ software.
In vitro invasion assays
For the Matrigel® invasion assay, 8-µm
pore size Transwell migration chambers coated with Matrigel (BD
Biosciences, San Jose, CA, USA) were used to assess the cell
invasion ability. Briefly, 95D cells (5×104
cells/insert) transfected with NC or DNMT1 siRNA were seeded into
the Matrigel-coated chambers of the 24-well plates and incubated at
37°C for 48 h. After 48 h of incubation, non-invasive cells were
removed with cotton swabs. Invaded cells were fixed in 100%
methanol at room temperature for 10 min and then stained with 1%
crystal violet at room temperature for 10 min, rinsed with PBS and
then subjected to microscopic inspection. The number of invaded
cells was manually counted under an inverted microscope
(magnification, ×200, five random fields/well).
Wound healing assay
95D cells (2×105 cells/well) were seeded
into 24-well plates and grown to ~95% confluence. A scratch was
created in the cell monolayer using a 200 µl sterile pipette tip.
Serum-free RPMI-1640 medium was used to remove the floating cells
and debris. Images of the wound were captured at 0 and 48 h. The
percentage of the wound healing was calculated as (the width of
wound at 0 h-the width of wound at 48 h)/(the width of wound at 0
h), which was used to evaluate cell migration ability as previously
described (21). Wounds were
evaluated using Adobe Photoshop (version 7.0; Adobe Systems, Inc.,
San Jose, CA, USA) to measure the wound distance at 0 and 48 h.
Representative images from three independent experiments conducted
in duplicate are presented in Fig.
2A.
Gelatin zymography of MMP enzyme
activity
95D cells at 2.0×105 cells per well were
cultured in RPMI-1640 without serum at 37°C for 24 h, and
subsequently centrifuged at 1,000 × g for 10 min at 4°C. The
supernatants were collected and subjected to electrophoresis using
10% SDS-PAGE with 1 mg/ml gelatin. The electrophoresis was ended
when the bromophenol blue reached the bottom of the gel. The gel
was transferred into a beaker and washed by gentle agitation with
an eluent (2.5% Triton-X-100) for 45 min. After rinsing twice for
45 min with double distilled water, the gels were transferred into
Incubation Buffer (50 mmol/l Tris-HCl, 5 mmol/CaCl2, 1
µmol/lZnCl2, 0. 02% Brij-35, pH 76) and incubated for 42
h at 37°C. The gel was then stained with 0.5% (w/v) Coomassie
brilliant blue R-250 for 2 h at room temperature. Finally, the gel
was discolored using destaining solution A (methanol concentration,
30%; acetic acid concentration, 10%), destaining solution B
(methanol concentration, 20%; acetic acid concentration, 10%) and
destaining solution C (methanol concentration, 10%; acetic acid
concentration, 5%) for 0.5, 1 and 2 h, respectively. MMP activity
was visualized as white bands on the blue background using Quantity
One® software (version 4.6.2; Bio-Rad Laboratories,
Inc., Hercules, CA, USA). The results were quantified using ImageJ
software (version 1.47). Experiments were repeated ≥3 times.
β-catenin reporter luciferase
assay
95D cells transfected with NC or DNMT1 siRNA were
co-transfected with TOPflash or FOPflash plasmids (Upstate
Biotechnology, Inc., Lake Placid, NY, USA), along with
β-galactosidase expression plasmid pRL-SV40 (Promega Corporation,
Madison, WI, USA), using Lipofectamine® 2000 reagent.
After 48 h of incubation at 37°C, the cells were lysed and the
luciferase activity was determined as described previously
(22). The luciferase activity of
each sample was normalized against Renilla luciferase
activity to monitor transfection efficiency. 200 nM NC or DNMT1
siRNA were co-transfected with the TOPflash or FOPflash reporter.
After 48 h, luciferase activity was evaluated using the
Dual-Luciferase® Reporter Assay system (Promega
Corporation) normalized to Renilla luciferase activity.
Statistical analysis
Data are presented as the mean ± standard deviation.
Experiments were repeated ≥3 times. Comparisons between the control
and treated groups were performed using a Student's t-test. The
one-way analysis of variance test was used for multiple
comparisons. All the data were analyzed using SPSS software
(version 13.0; SPSS, Inc., Chicago, IL, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
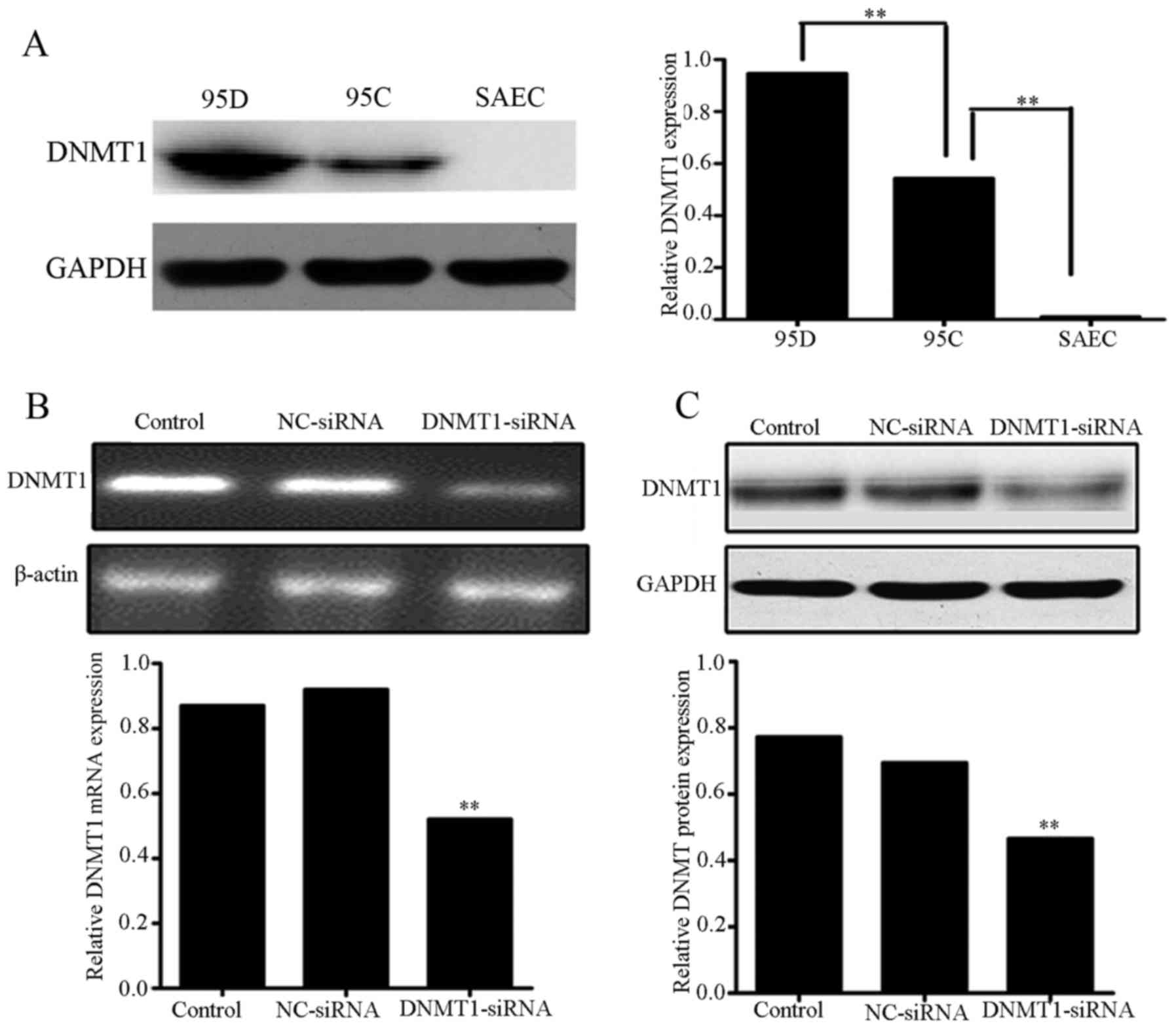
DNMT1 siRNA downregulates DNMT1
expression in 95D cells
To determine whether DNMT1 was associated with tumor
invasion and metastasis, the protein expression levels of DNMT1 in
95C (low invasive ability) and 95D (high invasive ability) cells
were compared using western blotting. The two cell lines exhibited
significantly higher expression of DNMT1 compared with the SAEC
cells (P<0.01; Fig. 1A); however,
the expression level of DNMT1 in 95D cells was significantly higher
compared with that in 95C cells (P<0.01). Thus, the result
indicated that DNMT1 expression level may be positively associated
with the in vitro invasiveness of human NSCLC cells and the
NSCLC malignancy grade. To determine whether DNMT1 may be
implicated in the invasiveness of 95C and 95D cells, DNMT1 siRNA
was transfected into 95D cells, which has higher DNMT1 expression
levels and in vitro invasiveness. After 48 h of
transfection, RT-PCR and western blot analysis demonstrated that
DNMT1 levels in the 95D cells transfected with DNMT1 siRNA were
significantly lower than those in the 95D cells transfected with NC
siRNA or the untreated control cells (P<0.01; Fig. 1B and C).
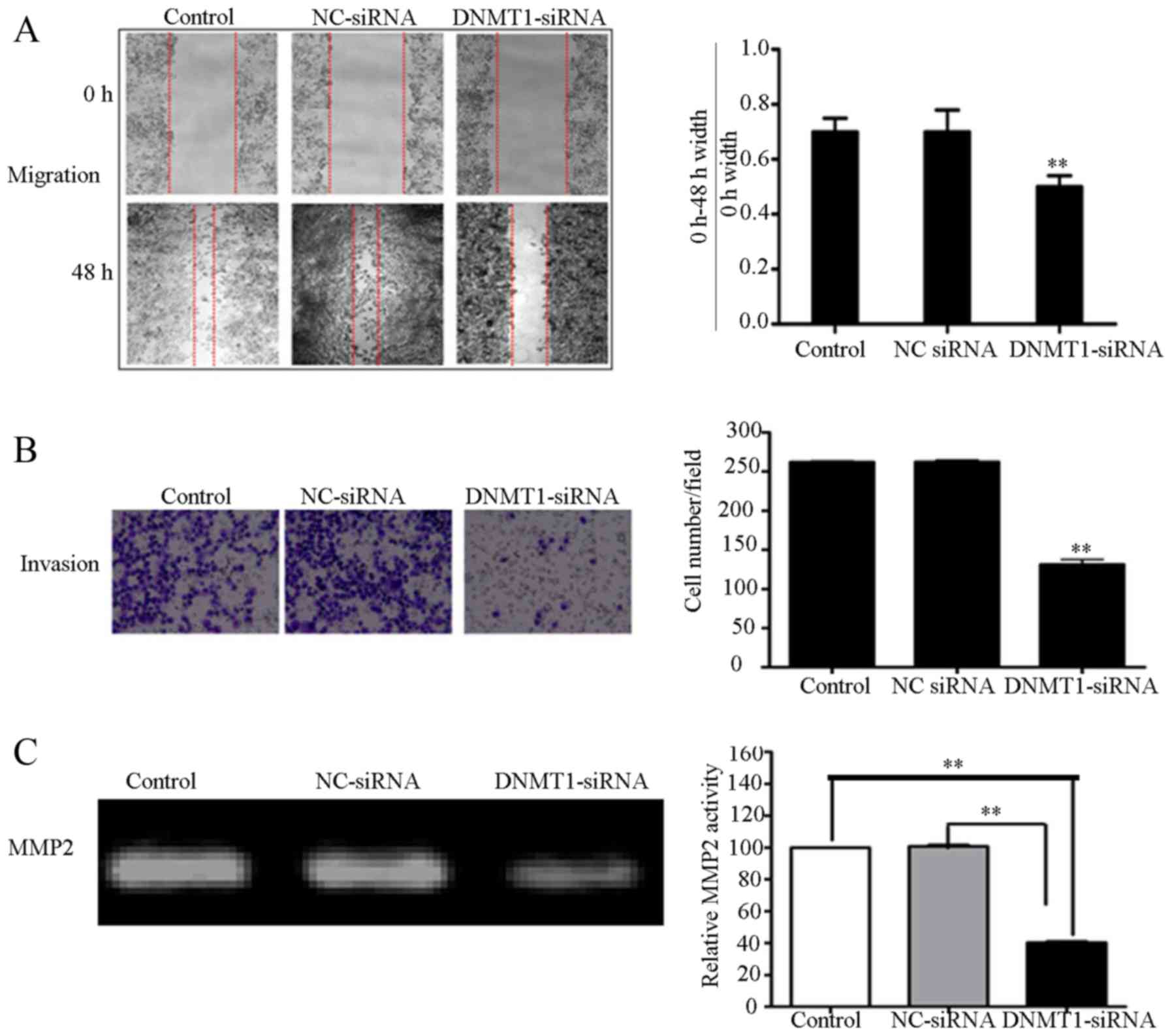
Downregulation of DNMT1 by DNMT1 siRNA
suppresses the migration and invasion of 95D cells
To determine the role of DNMT1 in the migration and
invasion of 95D cells, 95D cells transfected with NC or DNMT1 siRNA
were subjected to wound healing and Matrigel invasion assays. The
motility and invasiveness of 95D cells transfected with DNMT1 siRNA
was significantly lower compared with the control groups, which
included an untreated control group and a group transfected with NC
siRNA (P<0.01; Fig. 2A and B). To
detect MMP2 enzyme activity, gelatin zymography was performed. MMP2
activity in the serum-free medium collected from 95D cells
transfected with DNMT1 siRNA exhibited significantly lower enzyme
activity, as compared within the NC siRNA transfected and untreated
groups (P<0.01; Fig. 2C). These
results suggest that the inhibition of DNMT1 can attenuate MMP2
activity, which then inhibits tumor invasion.
Downregulation of DNMT1 reverses EMT
in 95D cells
To evaluate the role of DNMT1 on EMT, morphological
alterations in cells transfected with NC or DNMT1 siRNA were
examined by fluorescence microscope. The results demonstrated that
the cells become more compact compared with those of the control
groups (Fig. 3A). EMT-associated
markers and the principal regulator transcriptional factors Snail
and MMP2 were also detected by western blot analysis. The results
demonstrated that E-cadherin expression was significantly increased
in the DNMT1 siRNA group compared with in the NC siRNA and
untreated groups (P<0.01; Fig.
3B). However, the expression levels of vimentin, fibronectin,
MMP2 and Snail were significantly lower in the DNMT1 siRNA group
than in NC siRNA and control groups (P<0.01). To confirm these
results, immunofluorescence staining was used to detect E-cadherin,
vimentin and fibronectin expression. Concordant with the results of
the western blot analysis, E-cadherin expression was markedly
increased in the 95D cells transfected with DNMT1 siRNA, as
compared with in the untreated 95D cells and 95D cells transfected
with NC siRNA. These results suggest that the inhibition of DNMT1
can reverse EMT, and that DNMT1 promotes lung cancer metastasis by
inducing EMT.
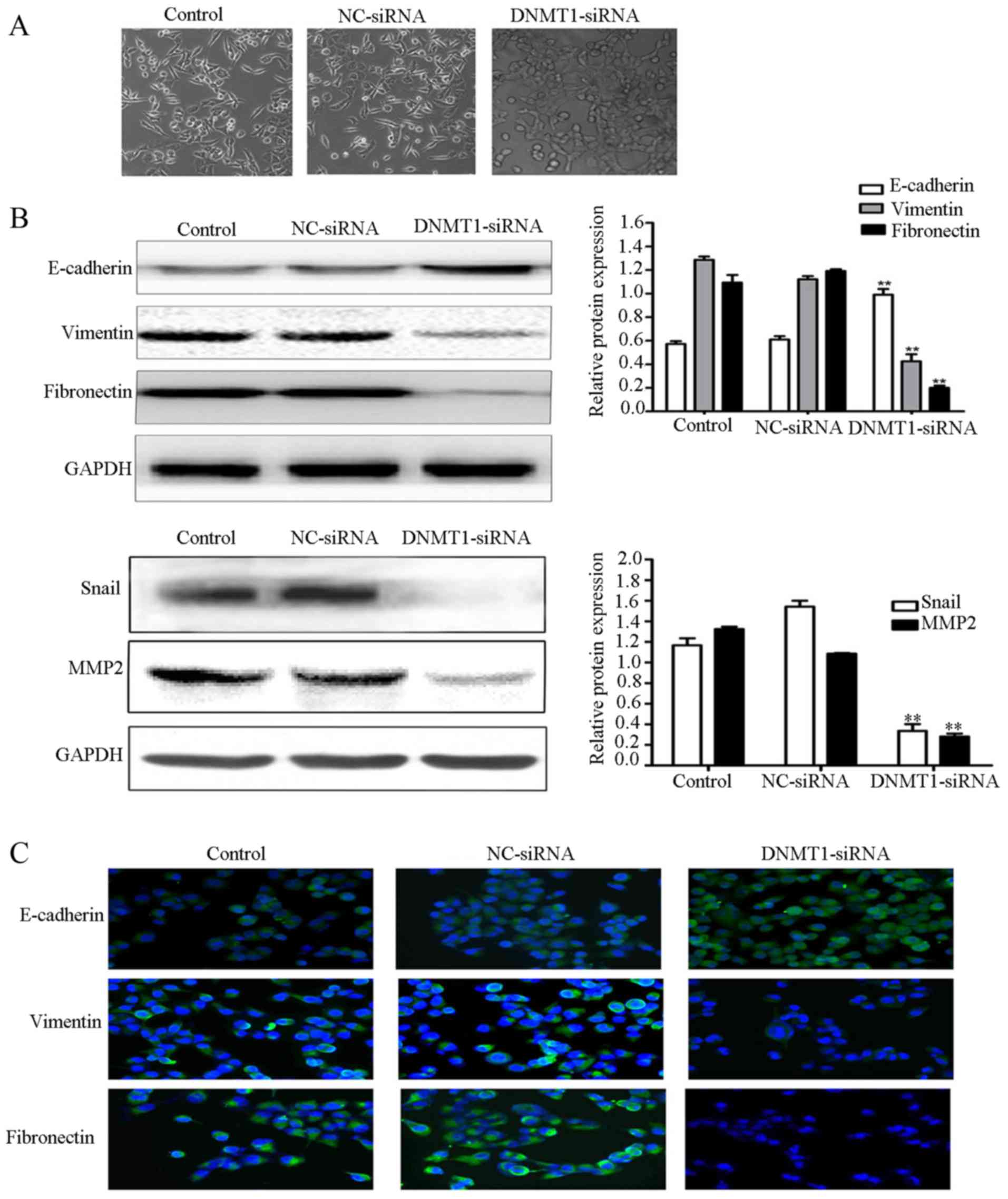 | Figure 3.Downregulation of DNMT1 inhibits the
epithelial-mesenchymal transition. (A) Light microscope images of
untreated 95D cells (control) and 95D cells transfected with NC or
DNMT1 siRNA. Magnification, ×100. (B) Western blot analysis of
E-cadherin, vimentin, fibronectin, Snail and MMP2 expression in
untreated 95D cells (control) and 95D cells transfected with NC or
DNMT1 siRNA. GAPDH served as an internal control. (C)
Immunofluorescence staining of E-cadherin, vimentin, and
fibronectin in untreated 95D cells (control) and 95D cells
transfected with NC or DNMT1 siRNA. The green signal represents
staining of the indicated proteins and the blue signal represents
nuclear DNA staining by DAPI. Magnification, ×100. The results are
representative of ≥3 independent experiments.**P<0.01, compared
with the NC siRNA group. DNMT1, DNA methyltransferase 1; siRNA,
small interfering RNA; NC, negative control. |
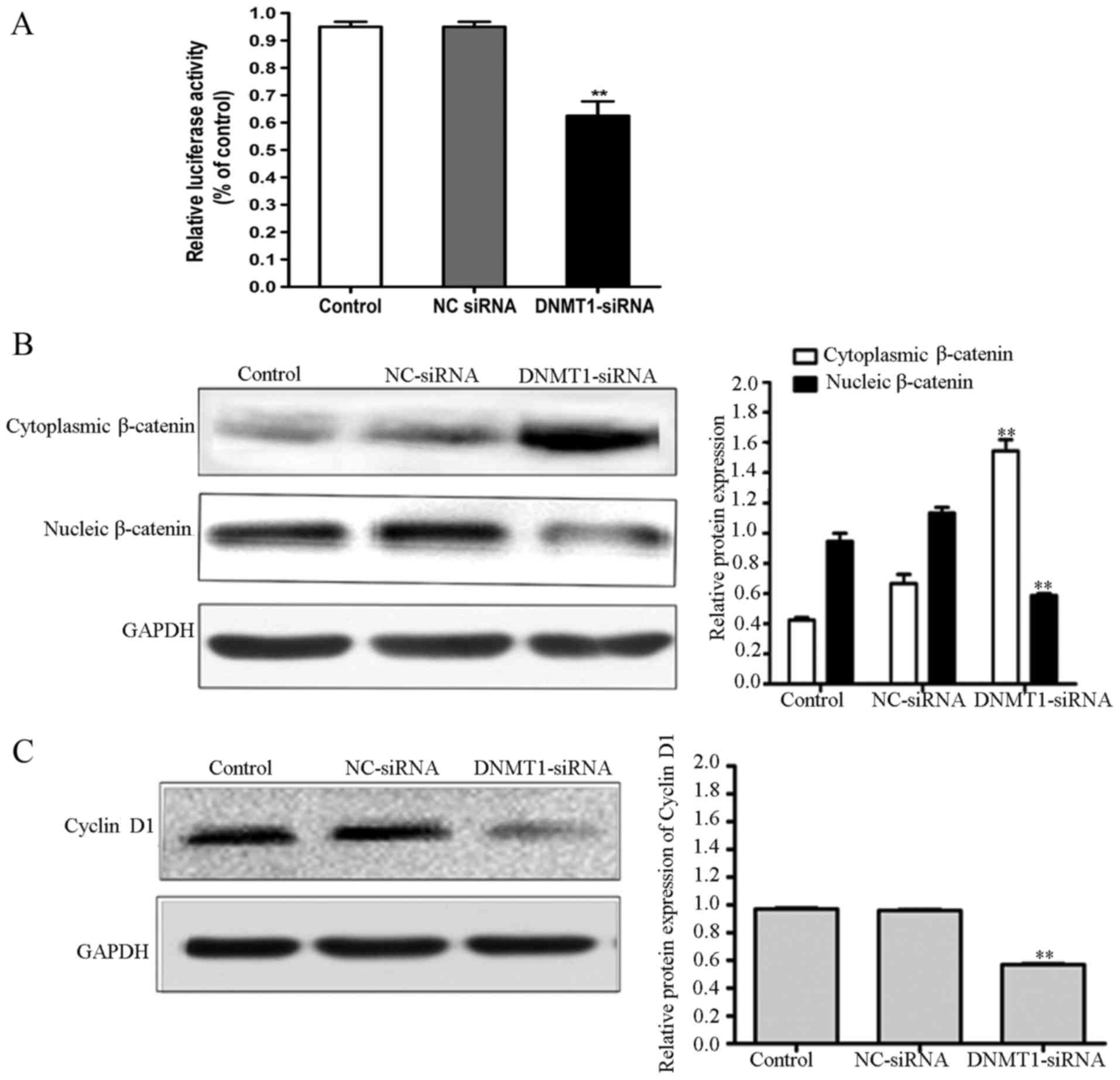
Inhibition of DNMT1 inhibits the Wnt
signaling pathway
To elucidate the involvement of Wnt signaling in the
anti-EMT effect of DNMT1 siRNA, the influence of DNMT1 siRNA on
T-cell factor/lymphoid enhancer factor (TCF/LEF)-dependent
transcriptional activity was examined using a dual-luciferase
reporter assay. DNMT1 siRNA inhibited TCF/LEF-dependent
transcriptional activity in 95D cells (Fig. 4A). Since β-catenin is an essential
mediator of the Wnt/β-catenin signaling pathway, the expression of
β-catenin in 95D cells transfected with NC or DNMT1 siRNA was
examined. The results demonstrated that cytoplasmic β-catenin
expression is significantly higher in 95D cells transfected with
DNMT1 siRNA, compared within the NC siRNA and untreated groups
(P<0.01; Fig. 4B). Nuclear
β-catenin expression in 95D cells transfected with NC siRNA was
significantly lower compared with that in 95D cells transfected
with NC siRNA or untreated groups (P<0.01). In addition,
expression of cyclin D1 was significantly lower in the 95D cells
transfected with DNMT1 siRNA group, as compared with in the control
groups (P<0.01; Fig. 4C). These
results demonstrate that the inhibition of DNMT1 can reduce tumor
cell EMT through inhibiting the Wnt/β-catenin signaling
pathway.
Discussion
Lung cancer is the most common type malignancy and
has a high incidence worldwide (23).
The majority of patients with lung cancer present with distant
metastatic lesions at the time of diagnosis; therefore, metastasis
is a major cause of cancer-associated mortality. Tumor metastasis
is a complex event involving extracellular matrix degradation and
cell migration. Cell migration and invasion is a critical step of
tumor metastasis (24). Therefore, it
is necessary to identify specific molecules and protein that may
inhibit cell invasion and migration.
MMPs serve critical roles in a number of cellular
processes, including cell apoptosis, adhesion, migration and
invasion. MMP2 is a 72-kDa type IV collagenase that degrades the
extracellular matrix, including the basement membrane (25), and is therefore associated with
angiogenesis and cell migration in lung cancer progression
(26,27). The results of the present study
demonstrated that DNMT1 siRNA significantly attenuates the motility
and invasion ability of 95D cells. MMP2 activity was decreased in
95D cells transfected with DNMT1 siRNA compared with in the NC
siRNA-treated or un-transfected 95D cells. This suggests that the
suppression of 95D cell invasion observed following DNMT1 siRNA
treatment is mediated via the inhibition of MMP2.
EMT has been reported to be an initial step in tumor
metastasis (28). During EMT,
polarized epithelial cells undergo a number of molecular changes,
leading to the acquisition of a mesenchymal cell phenotype and
subsequent increased motility and invasion. Results from the
present study demonstrated that treatment with DNMT1 siRNA leads to
the cells becoming more compact and to the upregulation of
E-cadherin. In addition, the expression of Snail, the
transcriptional suppressor of E-cadherin, and vimentin and
fibronectin was downregulated by DNMT1 siRNA. These results suggest
that DNMT1 siRNA can inhibit cell migration and invasion by
reversing the EMT process. However, the molecular mechanisms by
which DNMT1 regulates EMT remain unclear.
The Wnt signaling pathway can be divided into the
canonical β-catenin-dependent pathway and the non-canonical
β-catenin-independent pathway. Previous studies have demonstrated
that deregulation of the Wnt signal transduction pathway
contributes to EMT, cell migration, invasion and metastasis
(29,30). β-catenin, a central molecule in the
Wnt signaling pathway, is a multifunctional protein involved in
cell-cell adhesion, signal transduction, cellular differentiation
regulation and proliferation. Previous studies have indicated that
β-catenin serves a role in the onset and progression of EMT; active
β-catenin is translocated to the nucleus, where it activates
downstream target genes involved in EMT (31,32). Data
from the present study demonstrated that, in addition to the
attenuation of cell invasion and migration, treatment with DNMT1
siRNA significantly decreased the levels of activated β-catenin and
cyclin D1 while promoting the increase of inactivated β-catenin. As
demonstrated by the dual-luciferase assay, the activity of the Wnt
pathway was inhibited by DNMT1 siRNA. These results indicate that
the Wnt/β-catenin signaling pathway mediates the effects of DNMT1
siRNA on Snail-dependent EMT.
A number of molecules and proteins implicated in the
Wnt signaling pathway that are mediated by DNMT1 have previously
been investigated as targets for the diagnosis and treatment of
malignant tumors. Downregulation of Wnt inhibitory factor1 (WIF-1)
has been revealed to be due to the cooperative activity of DNMT1
and DNMT3B (33). WIF-1 is a secreted
antagonist of the Wnt signaling pathway. In addition, WIF-1 has
been demonstrated to inhibit tumor metastasis (34). Inhibition of DNMT1 restores WIF-1
expression (20); therefore, the
inhibition of DNMT1 may prevent metastasis by restoring the
expression of certain anti-metastatic genes that are the target
genes of the Wnt pathway. Snail and Slug have each been defined as
transcriptional repressors (35).
In the present study, knockdown of DNMT1 inhibited
NSCLC cell invasion and migration and downregulated β-catenin and
MMP2 expression. Downregulation of nuclear β-catenin and MMP2
inhibited the Wnt signaling pathway and decreased the expression of
its downstream target gene cyclin D1. In addition, Snail, an
important inducer of EMT, was inhibited by DNMT1 siRNA. Therefore,
the downregulation of DNMT1 leads to the downregulation of
β-catenin, MMP2 and cyclin D1, and inhibits Wnt signaling
pathway-dependent tumor cell migration and invasion.
In conclusion, the results of the present study
indicate a novel biological function for DNMT1 in the promotion of
95D cell invasion and migration. In addition, targeting DNMT1 may
represent an effective anti-invasion and anti-migration strategy
for the treatment of NSCLC, as DNMT1 may contribute directly or
indirectly to tumor metastasis. However, the underlying mechanisms
by which DNMT1 inhibits tumor invasion and migration remain
unclear. The present study indicated that depletion of DNMT1
reverses the EMT process, as E-cadherin expression increased,
whereas vimentin and fibronectin expression decreased following
DNMT1 siRNA treatment. In addition, the data from the present study
indicated that DNMT1 suppression inhibits tumor migration and
invasion by reversing the EMT process via inhibition of the Wnt
signaling pathway. Thus, a DNMT1 inhibitor may be a novel treatment
used to prevent tumor metastasis; however, in vivo studies
are required to validate the results of the present study.
Reduced miR-148a expression inhibits the metastasis
of non-small cell lung cancer tumors by depleting DNMT1 expression
(36), and miR-342 inhibits
colorectal cancer cell proliferation and invasion by directly
targeting DNMT1 (37). These findings
support further evaluation of DNMT knockdown strategies for cancer
therapy. Experiments in vitro demonstrated that DNMT1 serves
a key role in the migration and invasion of lung cancer cells;
however, alteration sin DNMT protein expression have not yet been
demonstrated in patients with cancer, to the best of our knowledge;
therefore, further studies in vivo are required to confirm
the findings from the present study and to elucidate the molecular
mechanisms by which DNTM1 mediates tumor metastasis.
Acknowledgements
The authors would like to thank Miss Qian Qiu, from
the Department of Respiratory Medicine of Southwest Hospital
(Chongqing, China) for her help.
Funding
The present study was supported by the Youth Fund of
Guizhou Provincial Center for Disease Control and Prevention, China
(grant no. 2014-E1-8).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
XCB, XYZ, JHX, HPY, designed the study. HJW and LG
performed western blotting. XCB performed immunofluorescence,
RT-PCR, wound healing and β-catenin reporter luciferase assays and
analysed the data with XYZ. JHX wrote the first draft of the
manuscript with XCB and XYZ. HPY, XDZ, HJW, LG, XCB and XYZ
contributed to the interpretation of the results and helped to
write the manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Ethics
Committee in Rizhao City Hospital of Traditional Chinese Medicine
(Shangdong, China).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
She J, Yang P, Hong Q and Bai C: Lung
cancer in China: Challenges and interventions. Chest.
143:1117–1126. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zeng L, Yu X, Yu T, Xiao J and Huang Y:
Interventions for smoking cessation in people diagnosed with lung
cancer. Cochrane Database Syst Rev: CD011751. 2015. View Article : Google Scholar
|
|
3
|
Jahangeer S, Forde P, Soden D and Hinchion
J: Review of current thermal ablation treatment for lung cancer and
the potential of electrochemotherapy as a means for treatment of
lung tumours. Cancer Treat Rev. 39:862–871. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cai C, Shi R, Gao Y, Zeng J, Wei M, Wang
H, Zheng W and Ma W: Reduced expression of sushi domain containing
2 is associated with progression of non-small cell lung cancer.
Oncol Lett. 10:3619–3624. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lee SY, Jeon HM, Ju MK, Kim CH, Yoon G,
Han SI, Park HG and Kang HS: Wnt/Snail signaling regulates
cytochrome C oxidase and glucose metabolism. Cancer Res.
72:3607–3617. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Thiery JP and Sleeman JP: Complex networks
orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell
Biol. 7:131–142. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dupont C, Armant DR and Brenner CA:
Epigenetics: Definition, mechanisms and clinical perspective. Semin
Reprod Med. 27:351–357. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Allen A: Epigenetic alterations and
cancer: New targets for therapy. IDrugs. 10:709–712.
2007.PubMed/NCBI
|
|
9
|
Bacigalupo ML, Manzi M, Espelt MV,
Gentilini LD, Compagno D, Laderach DJ, Wolfenstein-Todel C,
Rabinovich GA and Troncoso MF: Galectin-1 triggers
epithelial-mesenchymal transition in human hepatocellular carcinoma
cells. J Cell Physiol. 230:1298–1309. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li Y, Xu Z, Li B, Zhang Z, Luo H, Wang Y,
Lu Z and Wu X: Epigenetic silencing of miRNA-9 is correlated with
promoter-proximal CpG island hypermethylation in gastric cancer
in vitro and in vivo. Int J Oncol. 45:2576–2586.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Licchesi JD, Westra WH, Hooker CM, Machida
EO, Baylin SB and Herman JG: Epigenetic alteration of Wnt pathway
antagonists in progressive glandular neoplasia of the lung.
Carcinogenesis. 29:895–904. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ying J, Li H, Seng TJ, Langford C,
Srivastava G, Tsao SW, Putti T, Murray P, Chan AT and Tao Q:
Functional epigenetics identifies a protocadherin PCDH10 as a
candidate tumor suppressor for nasopharyngeal, esophageal and
multiple other carcinomas with frequent methylation. Oncogene.
25:1070–1080. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hermann A, Gowher H and Jeltsch A:
Biochemistry and biology of mammalian DNA methyltransferases. Cell
Mol Life Sci. 61:2571–2587. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim H, Kwon YM, Kim JS, Han J, Shim YM,
Park J and Kim DH: Elevated mRNA levels of DNA methyltransferase-1
as an independent prognostic factor in primary nonsmall cell lung
cancer. Cancer. 107:1042–1049. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kassis ES, Zhao M, Hong JA, Chen GA,
Nguyen DM and Schrump DS: Depletion of DNA methyltransferase 1
and/or DNA methyltransferase 3b mediates growth arrest and
apoptosis in lung and esophageal cancer and malignant pleural
mesothelioma cells. J Thorac Cardiovasc Surg. 131:298–306. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Belinsky SA, Klinge DM, Stidley CA, Issa
JP, Herman JG, March TH and Baylin SB: Inhibition of DNA
methylation and histone deacetylation prevents murine lung cancer.
Cancer Res. 63:7089–7093. 2003.PubMed/NCBI
|
|
17
|
Tan EJ, Kahata K, Idas O, Thuault S,
Heldin CH and Moustakas A: The high mobility group A2 protein
epigenetically silences the Cdh1 gene during
epithelial-to-mesenchymal transition. Nucleic Acids Res.
43:162–178. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Papageorgis P, Lambert AW, Ozturk S, Gao
F, Pan H, Manne U, Alekseyev YO, Thiagalingam A, Abdolmaleky HM,
Lenburg M and Thiagalingam S: Smad signaling is required to
maintain epigenetic silencing during breast cancer progression.
Cancer Res. 70:968–978. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lin RK, Hsu CH and Wang YC: Mithramycin A
inhibits DNA methyltransferase and metastasis potential of lung
cancer cells. Anticancer Drugs. 18:1157–1164. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu Y, Wang H, Gong L, Tang C and Yang H:
siRNAi targeted DNMT1 silence induces WIF-1 promoter
hypomethylation. Chongqing Med J. 40:1266–1268. 2011.(In
Chinese).
|
|
21
|
International symposium on lifestyle
factors and human lung cancer. Guangzhou, China, 12–16 December
1994. Proceedings and abstracts. Lung Cancer. 14 Suppl 1:S1–S245.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang Q, Sun ZX, Allgayer H and Yang HS:
Downregulation of E-cadherin is an essential event in activating
beta-catenin/Tcf-dependent transcription and expression of its
target genes in Pdcd4 knockdown cells. Oncogene. 29:128–138. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Siegel R, Ward E, Brawley O and Jemal A:
Cancer statistics, 2011: The impact of eliminating socioeconomic
and racial disparities on premature cancer deaths. CA Cancer J
Clin. 61:212–236. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hosono Y, Yamaguchi T, Mizutani E,
Yanagisawa K, Arima C, Tomida S, Shimada Y, Hiraoka M, Kato S,
Yokoi K, et al: MYBPH, a transcriptional target of TTF-1, inhibits
ROCK1, and reduces cell motility and metastasis. EMBO J.
31:481–493. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang H, Zhu Y, Zhao M, Wu C, Zhang P, Tang
L, Zhang H, Chen X, Yang Y and Liu G: miRNA-29c suppresses lung
cancer cell adhesion to extracellular matrix and metastasis by
targeting integrin β1 and matrix metalloproteinase2 (MMP2). PLoS
One. 8:e701922013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chetty C, Lakka SS, Bhoopathi P and Rao
JS: MMP-2 alters VEGF expression via alphaVbeta3 integrin-mediated
PI3K/AKT signaling in A549 lung cancer cells. Int J Cancer.
127:1081–1095. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rojiani MV, Alidina J, Esposito N and
Rojiani AM: Expression of MMP-2 correlates with increased
angiogenesis in CNS metastasis of lung carcinoma. Int J Clin Exp
Pathol. 3:775–781. 2010.PubMed/NCBI
|
|
28
|
Zhou J, Xie M, Shi Y, Luo B, Gong G, Li J,
Wang J, Zhao W, Zi Y, Wu X and Wen J: MicroRNA-153 functions as a
tumor suppressor by targeting SET7 and ZEB2 in ovarian cancer
cells. Oncol Rep. 34:111–120. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fu Y, Zheng S, An N, Athanasopoulos T,
Popplewell L, Liang A, Li K, Hu C and Zhu Y: β-catenin as a
potential key target for tumor suppression. Int J Cancer.
129:1541–1551. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Valenta T, Hausmann G and Basler K: The
many faces and functions of β-catenin. EMBO J. 31:2714–2736. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Oh SJ, Shin JH, Kim TH, Lee HS, Yoo JY,
Ahn JY, Broaddus RR, Taketo MM, Lydon JP, Leach RE, et al:
β-Catenin activation contributes to the pathogenesis of adenomyosis
through epithelial-mesenchymal transition. J Pathol. 231:210–222.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhao JH, Luo Y, Jiang YG, He DL and Wu CT:
Knockdown of β-catenin through shRNA cause a reversal of EMT and
metastatic phenotypes induced by HIF-1α. Cancer Invest. 29:377–382.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Quan H, Zhou F, Nie D, Chen Q, Cai X, Shan
X, Zhou Z, Chen K, Huang A, Li S and Tang N: Hepatitis C virus core
protein epigenetically silences SFRP1 and enhances HCC
aggressiveness by inducing epithelial-mesenchymal transition.
Oncogene. 33:2826–2835. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yee DS, Tang Y, Li X, Liu Z, Guo Y,
Ghaffar S, McQueen P, Atreya D, Xie J, Simoneau AR, et al: The Wnt
inhibitory factor 1 restoration in prostate cancer cells was
associated with reduced tumor growth, decreased capacity of cell
migration and invasion and a reversal of epithelial to mesenchymal
transition. Mol Cancer. 9:1622010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Haslehurst AM, Koti M, Dharsee M, Nuin P,
Evans K, Geraci J, Childs T, Chen J, Li J, Weberpals J, et al: EMT
transcription factors snail and slug directly contribute to
cisplatin resistance in ovarian cancer. BMC Cancer. 12:912012.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen Y, Min L, Zhang X, Hu S, Wang B, Liu
W, Wang R, Gu X, Shen W, Lv H, et al: Decreased miRNA-148a is
associated with lymph node metastasis and poor clinical outcomes
and functions as a suppressor of tumor metastasis in non-small cell
lung cancer. Oncol Rep. 30:1832–1840. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang H, Wu J, Meng X, Ying X, Zuo Y, Liu
R, Pan Z, Kang T and Huang W: MicroRNA-342 inhibits colorectal
cancer cell proliferation and invasion by directly targeting DNA
methyltransferase 1. Carcinogenesis. 32:1033–1042. 2011. View Article : Google Scholar : PubMed/NCBI
|