Introduction
Breast cancer is one of the most common forms of
cancer among Japanese women, and about 70,000 women were diagnosed
with breast cancer in 2015 (1). After
the advent of screening mammography, there was an increase in the
reported number of small invasive tumors (measuring <2 cm or
in situ carcinomas) (2).
Ductal carcinoma in situ (DCIS) is a heterogeneous disease
that currently accounts for approximately 20% of all
screening-detected breast cancers (3). This disease is characterized by a
proliferation of neoplastic ductal epithelial cells that are
confined to the basement membrane of the mammary ducts. However,
lesions initially diagnosed as DCIS in needle biopsy are
occasionally upstaged to invasive ductal carcinoma (IDC) after the
final pathology report on the completely excised specimen. It has
been reported that the IDC identification rates at final pathology
in patients initially diagnosed with DCIS by core-needle biopsy or
vacuum-assisted biopsy devices are between 8 and 38% (4,5).
Therefore, clarification of the invasive cancer in patients
diagnosed with DCIS by needle biopsy is important in deciding
treatment strategy. The ability to distinguish DCIS and IDC at an
earlier stage, would enable earlier individualized treatment
options.
MicroRNAs (miRNAs) are 18–25 nucleotides single
stranded non-coding RNAs. They regulate gene expression at the
transcriptional or post-transcriptional level, and can act as
either tumor suppressor genes or oncogenes, depending on the roles
of their target mRNAs (6). An
increasing number of studies have shown aberrant expression
profiles of miRNAs in breast cancer, and reported on the usefulness
of miRNA for diagnosis of breast cancer (7). Recently, miRNAs have been identified in
plasma and serum (plasma/serum) and are considered as minimally
invasive liquid biomarkers for diagnosis, prognosis, and
therapeutic outcome in various cancer patients, including the
breast cancer (8). Furthermore, these
circulating miRNAs have been identified in the exosome of
plasma/serum in a stable form, which is protected from endogenous
RNase activity (9). Exosomes are
50–150 nm in diameter membrane-derived vesicles that are actively
secreted from many cell types. They contain protein, lipids, mRNAs
and miRNAs, and can transfer these components to other cells
(10–12). In general, cancer cells secrete
exosomes and cancer patients show high concentrations of exosomes
in the blood. Therefore, exosomes which encapsulate intact miRNA
could be potential biomarkers of malignancy of cancer (13–15).
However, there is currently little available information regarding
the relationship between the exosomal miRNA expression profiles and
the pathological condition in patients with breast cancer. In
particular, few reports have been published on the exosome miRNA
characteristics of DICS and IDC patients with breast cancer.
In this study, we examined the usefulness of plasma
exosomal miRNAs in the selection of patients with invasive lesions
in DICS patients diagnosed using needle biopsy. Furthermore, we
aimed to clarify the relationship between this invasion-specific
exosomal miRNA and the clinicopathological factors of breast
cancer.
Patients and methods
Study design and clinical samples
In this study, 185 breast cancer patients were
included. Clinical samples were obtained from breast cancer
patients who underwent surgery between June 2014 and May 2017 at
Teikyo University School of Medicine (Tokyo, Japan). For a healthy
control, 20 healthy volunteers were included. Peripheral blood and
plasma samples were collected before the start of treatment. This
study protocol conformed to the guidelines of the ethics committee,
and was approved by the review board of the Teikyo University
(09-081-3). Written informed consent was obtained from all the
patients.
First, we selected the plasma exosomal miRNA which
can distinguish between DCIS and IDC of breast cancer by the miRNA
array using exosomes collected from DCIS patients (n=3), IDC
patients (n=3) with stage I and healthy controls (n=3). Next, we
examined the potential of selected miRNA for cell proliferation and
cell invasion using the MCF7 cell lines in vitro. Third, we
clarified the usefulness of selected plasma exosomal miRNA as
biomarker for distinguishing early IDC from another 43 patients
diagnosed with DCIS by preoperative needle biopsy. Finally, we
examined the relationship between the selected miRNA and
clinicopathological characteristics using the exosomes collected
from 179 patients with breast cancer.
Cell culture
The human breast cancer cell line MCF7 was obtained
from the Cell Resource Center for Biomedical Research Institute of
Development, Aging and Cancer, Tohoku University. The Cell line was
maintained in RPMI-1640 supplemented with 10% fetal bovine serum
(FBS) at 37°C in a 5% humidified CO2 atmosphere.
Transfection and establishment of
pre-miR-223-3p-stably transfected MCF7 cell line
The backbone plasmid pcDNA6.2-GW/EmGFP-miR was
obtained from the Block-iT Pol II miR RNAi Expression Vector kit
(Thermo Fisher Scientific, Inc., Waltham, MA, USA). The plasmids
pcDNA6.2-GW/EmGFP-pre-miR-223-3p (pCMV-pre-miR-223-3p) containing
pre-miR-223-3p, and pcDNA6.2-GW/EmGFP-miR-neg (pCMV-N) containing
an unrelated insert were constructed according to the manual for
the Block-iT Pol II miR RNAi Expression Vector kit, as described
previously (16). The sequence of
mature miR-223-3p was UGUCAGUUUGUCAAAUACCCCA (has-miR-223-3p). The
pCMV-pre-miR-223-3p and pCMV-miR-neg were transfected into the MCF7
cell line using the lipofectamine 3000 (Life Technologies, Inc,
Tokyo, Japan) according to the manufacturer's instructions. Then
stably transfected cells expressing mature miR-223-3p were selected
with G418 treatment followed by sorting with GFP by MACS. A
pCMV-miR-neg-transfected clone of the cell line was used for the
control. Expressions of miR-223-3p of pCMV-pre-miR-223-3p or
wpCMV-miR-neg-transfected MCF7 cells were measured by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
system (StepOne; Thermo Fisher Scientific, Inc.) and Digital PCR
system (Quant Studio 3D Digital PCR System; Thermo Fisher
Scientific, Inc.).
Cell proliferation assay
Cell proliferation was evaluated by performing MTT
assays using a Cell Proliferation kit 1 (Roche Applied Science,
Penzberg, Germany) according to the manufacturer's instructions.
miR-223-3p-transfected cells and mock-transfected cells were seeded
at 3,000 cells per well in triplicate 96-wells in 100 µl medium.
The coloring reaction was quantitated using a plate reader, at 570
nm with a reference filter of 650 nm. Each independent experiment
was performed in triplicate.
Cell invasion assay
Cell invasion capacities were assessed using the BD
BioCoat Tumor Invasion System, 24 Multiwell (BD Bioscience)
according to the manufacturer's instructions as described
previously (16). In brief,
miR-223-3p-transfected cells and mock-transfected cells
(105 cells/well) were placed in the upper chamber, and
the lower chamber was filled with 750 µl of RPMI-1640 with 10% FBS
and incubated in a humidified atmosphere (37°C and 5%
CO2). After 48 h incubation, the upper chamber was
transferred into a second 24-well plate, with each well containing
500 µl of 4 µg/ml calcein AM solution. The plates were incubated
for an additional 1 h (37°C and 5% CO2). Invasive cells
that advanced through the membrane were evaluated in a fluorescence
plate reader at excitation/emission wavelengths of 485/535 nm. Each
independent experiment was performed in triplicate.
Purification of exosome from
plasma
Peripheral blood was separated by centrifugation at
2,000 × g for 15 min at 4°C in order to collect plasma. Plasma of
1.0 ml was used for microarray analysis and reverse
transcription-quantitative PCR (RT-qPCR). Exosomes of plasma were
separated by ultracentrifugation at 15,000 × g for 70 min. at 4°C,
and the pellets were washed with phosphate-buffered saline (PBS)
and prepared for microarray and RT-qPCR analysis.
Confirmation of exosome by electron
microscopic image
Isolated exosomes were confirmed by transmission
electron microscopy. Isolated exosomes were dissolved in PBS
buffer, and a drop of the suspension was placed on a sheet. A
carbon-coated copper grid was floated on the drop for 10s. Then the
grid was removed and excess liquid drained by filter paper. The
grid was put in contact with a drop of 2% uranyl acetate of
phosphotungstic acid for 5s. After remove of the excess liquid, the
grid was allowed to dry for several min and was then observed using
electron microscope (HITACHI H-7600; Hitachi, Ltd., Tokyo,
Japan).
Total RNA isolation from exosomes and
tissues
Total RNAs (including the miRNA) of exosomes were
isolated using the miRNeasy serum/plasma kit (Qiagen, Inc.,
Valencia, CA, USA), and total RNAs (including the miRNA) of tissues
were extracted using the miRNeasy Mini kit (Qiagen, Inc.).
Subsequent procedure was performed according to the manufacturer's
protocol described previously (17).
Using an Agilent 2100 Bioanalyzer, quality of extracted RNA was
examined (Agilent Technologies, Inc., Santa Clara, CA, USA).
miRNA microarray analysis
Exosomal miRNA expression profiles were examined
using a 3D-Gene Human miRNA Oligo chips ver.21 (Toray Industries
Inc., Tokyo, Japan), according to the manufacturer's protocol.
Fluorescence signals were scanned and analyzed using the 3D-Gene
Scanner (Toray). The number of genes mounted in this chip was
2,565. The raw data for each spot were normalized to the mean
intensity of background signals determined by all blank signal
intensities at a 95% confidence intervals. Valid measurements were
considered those in which the signal intensity of both duplicate
spots was >2SD of the background signal intensity.
RT-qPCR for miRNA of exosomes and
tumor tissues
miRNA expressions of plasma exosomes and tissues
were assayed using RT-qPCR as described previously (18). All primers, reagents and assay kits
for TaqMan RT-qPCR assays were purchased from ThermoFisher
Scientific, Inc. Complementary DNA (cDNA) of exosomes or tissues
samples was synthesized from total RNA using TaqMan MicroRNA
primers specific for miR-223-3p (002295) and TaqMan MicroRNA RT
kit. miR-16a (000391) was used as an internal control for exosomal
samples, and RNU-6B (001093) was used as an internal control for
tissue samples. RT-qPCR was performed using TaqMan Universal PCR
Master Mix and StepOne, following the manufacturer's protocol. Each
sample was analyzed in triplicate. Relative quantification of miRNA
expression was calculated using the 2−ΔΔCq method as
described previously (18).
RT-qPCR for EPB41L3 mRNA of tumor
tissues
Total RNAs were extracted by miRNeasy Mini kit
(Qiagen, Inc.) and cDNA synthesis was performed using random
hexamer primers and SuperScript II reverse transcriptase according
to the manufacturer's protocol (Thermo Fisher Scientific. Inc.).
The amplifications of EPB41L3 mRNA (Hs00202360) and GAPDH mRNA
(Hs03929097) were performed using the TaqMan primer and probe set
(Thermo Fisher Scientific, Inc.) and the RT-qPCR of these mRNAs was
measured using StepOne. GAPDH mRNA was used as an internal control
for this assay. All the experiments were performed in triplicate.
The expression levels of EPB41L3 mRNA were normalized to GAPDH mRNA
expression levels.
Statistical analysis
The data were expressed as mean ± standard
deviation. The relationships between microRNA expression and
clinicopathological factors were analyzed using the Student's
t-test, the chi-squire test and one-way analysis of variance
(ANOVA). Tukey-HSD was used as post hoc test after one-way ANOVA.
Univariate analysis was examined for each factor, and multivariate
analysis was performed for factors that showed significance in
univariate analysis. All P-values are two-sided, and P<0.05 was
considered to indicate a statistically significant difference.
Statistical analyses were performed using the JMP 9.0 software (SAS
Institute, Inc., Cary, NC, USA).
Results
Identification of exosome in
plasma
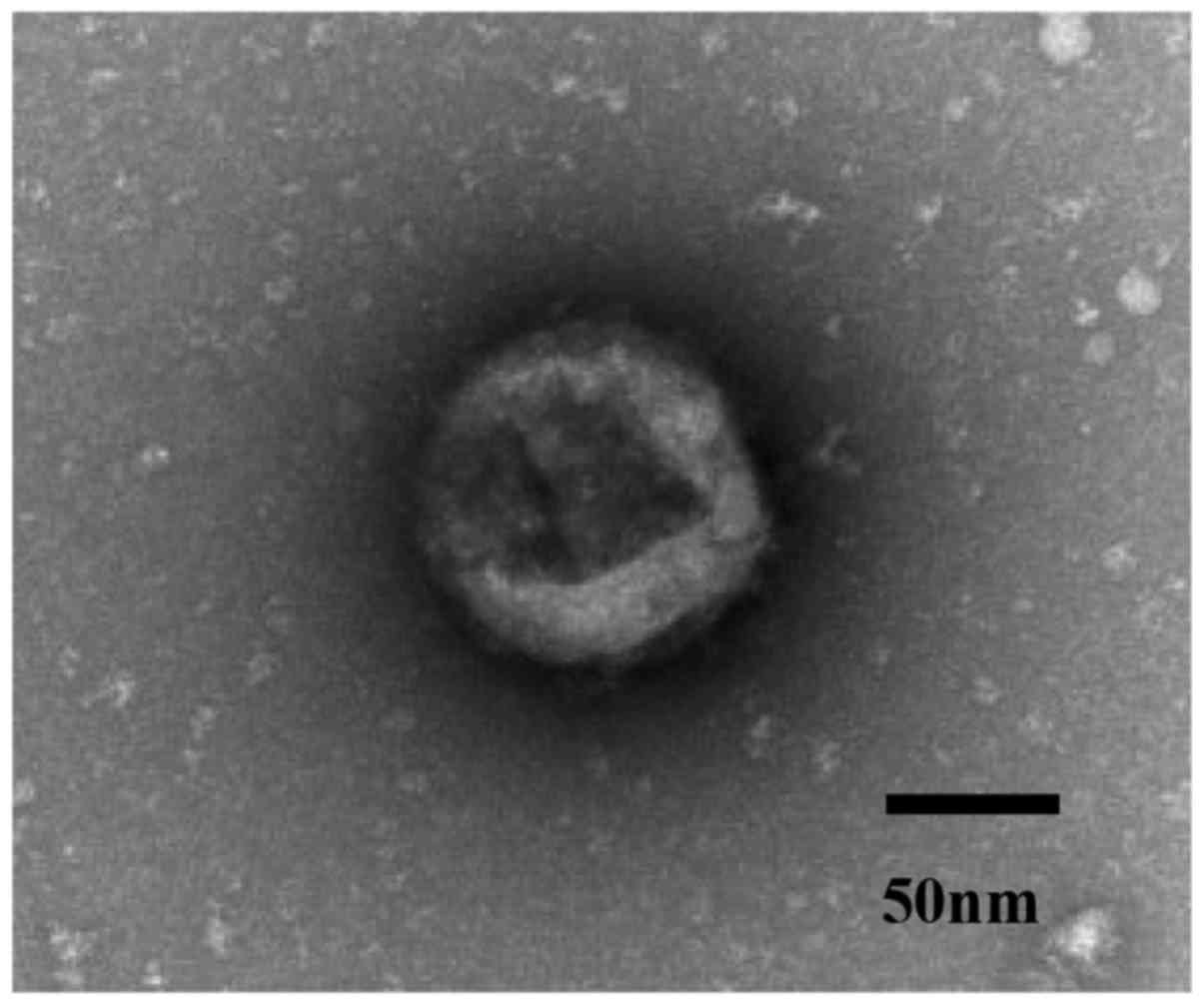
To confirm the isolation of exosome, we examined the
ultracentrifugation samples from the plasma of breast cancer
patients using transmission electron microscopy. We captured images
of micro vesicles with diameters of about 100 nm (Fig. 1).
Exosomal miRNA profile of DCIS and IDC
breast cancer patients
To clarify the exosomal miRNA useful for
discriminating between the DCIS and IDC of the breast cancer
patients, we examined the miRNA microarray analyses of samples from
3 DCIS patients, 3 IDC patients with stage I and 3 healthy
controls. The clinical characteristics of these patients are shown
in Table I. The average age of DCIS
and IDC patients was 66 years (range, 49–78) and 59 years (range,
38–75), respectively. The degree of vascular invasion, lymph node
metastasis, nuclear grade (NG), estrogen receptor (ER) and
progesterone receptor (PgR) were the same in DCIS and IDC patients.
The average value of Ki67 labeling index (Ki67) of IDC patients is
higher than that of DCIS patients; however it was not a significant
difference. Table II shows the 5
most highly upregulated exosomal miRNAs among 2565 miRNAs in these
breast cancer patients. In the exosomal samples, the miR-223-3p
(MIMAT0000280) of IDC patients showed the highest fold-change (3.45
times) as compared with that of the healthy controls. And the
miR-223-3p of IDC patients also showed a high fold-change (2.85
times) as compared with that of the DCIS patients. We selected
miR-223-3p as a potential marker for diagnosis of early invasion in
breast cancer patients.
 | Table I.Characteristics of patients used for
miRNA microarray analyses. |
Table I.
Characteristics of patients used for
miRNA microarray analyses.
| Variable | DCIS (n=3) | IDC (n=3) |
|---|
| Age, year
(range) | 66 (49–78) | 59 (38–75) |
| Tumor size, cm
(range) | 0 | 1.8 (0.3–2.0) |
| Pathological
stage |
|
|
| 0 | 3 | 0 |
| 1 | 0 | 3 |
| Vascular
invasion |
|
|
|
(−) | 3 | 3 |
|
(+) | 0 | 0 |
| Lymph node
metastasis |
|
|
|
(−) | 3 | 3 |
|
(+) | 0 | 0 |
| NG |
|
|
| 1 | 2 | 2 |
| 2 | 1 | 1 |
| 3 | 0 | 0 |
| ER |
|
|
|
(−) | 1 | 1 |
|
(+) | 2 | 2 |
| PgR |
|
|
|
(−) | 1 | 1 |
|
(+) | 2 | 2 |
| HER2 |
| 3 |
|
(−) | −a | 3 |
|
(+) | −a | 0 |
| Ki67, %
(range) | 9 (5–15) | 14 (7–20) |
 | Table II.Five most highly upregulated miRNAs
based on the miRNA microarray. |
Table II.
Five most highly upregulated miRNAs
based on the miRNA microarray.
|
|
|
| Fold-changes |
|---|
|
|
|
|
|
|---|
| Rank | miR healthy | MirBase no. | IDC vs.
controls | IDC vs. DCIS |
|---|
| 1 | miR-223-3p | MIMAT 0000280 | 3.45 | 2.85 |
| 2 | miR-130a-3p | MIMAT 0000278 | 3.11 | 2.03 |
| 3 | miR-191-5p | MIMAT 0000440 | 3.25 | 2.14 |
| 4 | miR-146a | MIMAT 0000092 | 2.93 | 2.08 |
| 5 | miR-221-3p | MIMAT 0000278 | 2.73 | 2.24 |
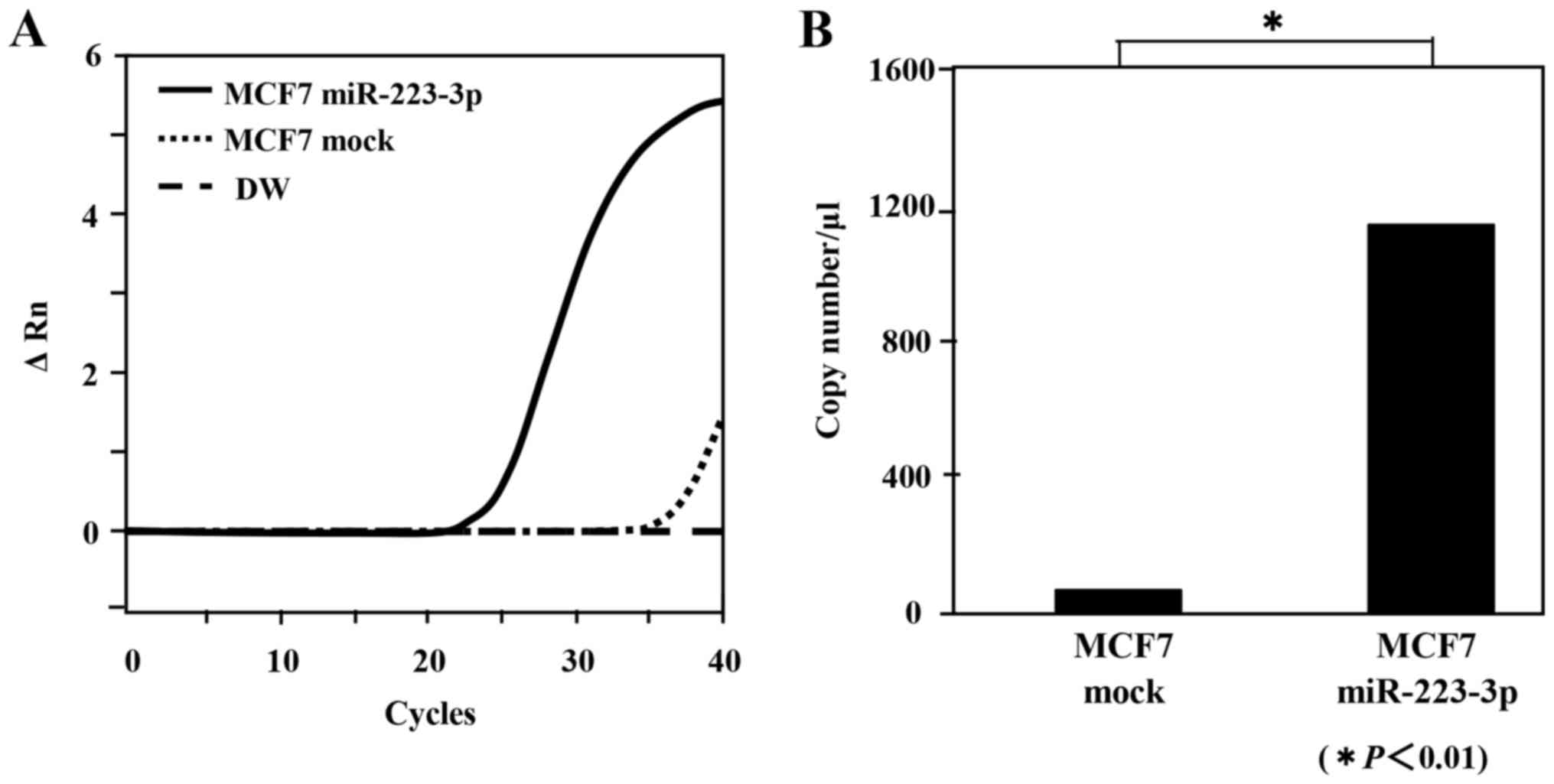
Expression of miR-223-3p in
miR-223-3p-transfected MCF7 cells
The miR-223-3p gene expressions of
pCMV-pre-miR-223-3p transfected (miR-223-3p-transfected) MCF7 cells
and pCMV-N transfected (mock-transfected) MCF7 cells were measured
by RT-qPCR and Digital PCR. In the RT-qPCR assay, a significant
amplification of miR-223-3p was shown in the miR-223-3p-trasnfected
MCF7 cells as compared with that of the mock-transfected cells
(Fig. 2A). In the digital PCR assay,
the copy number of miR-223-3p of the miR-223-3p-transfected MCF7
cells was significantly higher than that of the mock-transfected
MCF7 cells (Fig. 2B).
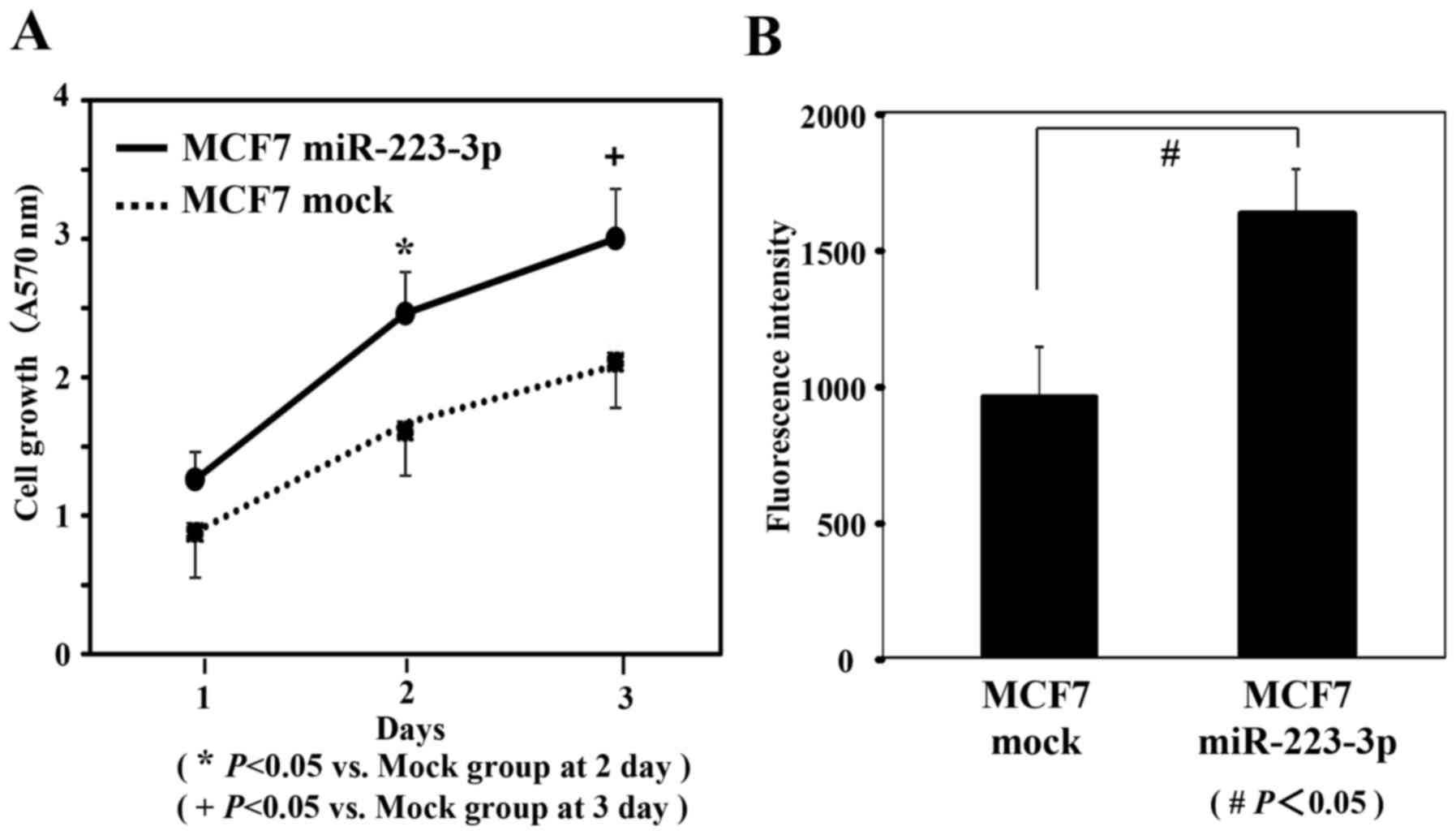
Cell proliferation and cell invasion
potentials
To examine the proliferation potential of the MCF7
cells transfected with miR-223-3p, MTT assays were performed. As
shown in Fig. 3A, MCF7 cells
transfected with miR-223-3p showed a significant increase of
proliferation compared to that of mock-transfected MCF7 cells
(P<0.05). To evaluate the cell invasion potential of the MCF7
cells transfected with miR-223-3p, invasion assays were performed.
As presented in Fig. 3B, the invasive
ability of MCF7 cells transfected with miR-223-3p was significantly
increased compared to that of mock-transfected MCF7 cells
(P<0.05).
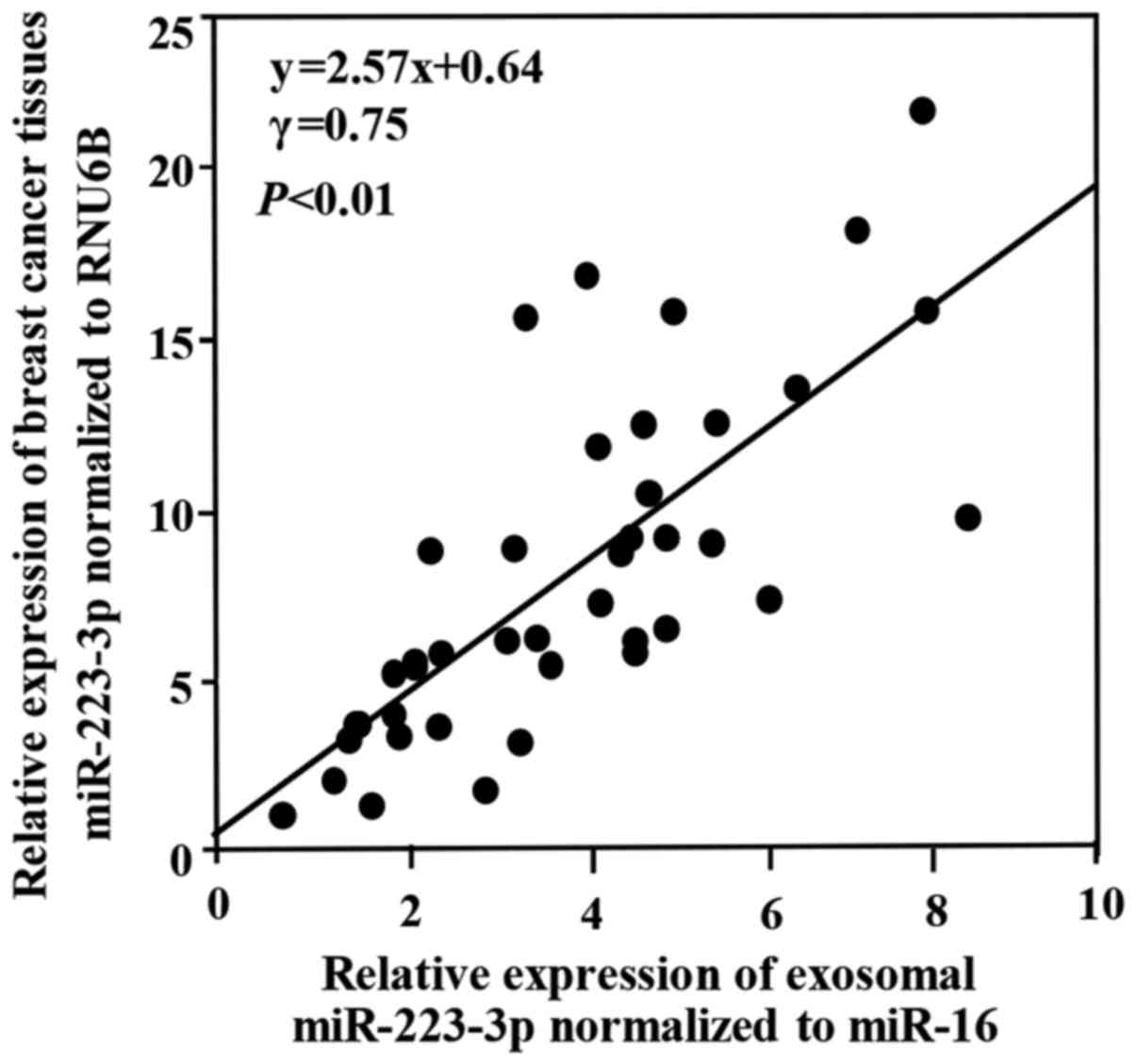
Expression of miR-223-3p in the plasma
exosomes and breast cancer tissues
We examined the correlation between the exosomal
miR-223-3p levels and the miR-223-3p expressions in primary tumor
tissues collected from same breast cancer patients. This study
consisted of 40 breast cancer patients, including DCIS patients
(n=5) and stage I patients (n=15), stage II patients (n=15) and
stage III patients (n=5). As shown in Fig. 4, a positive significant correlation
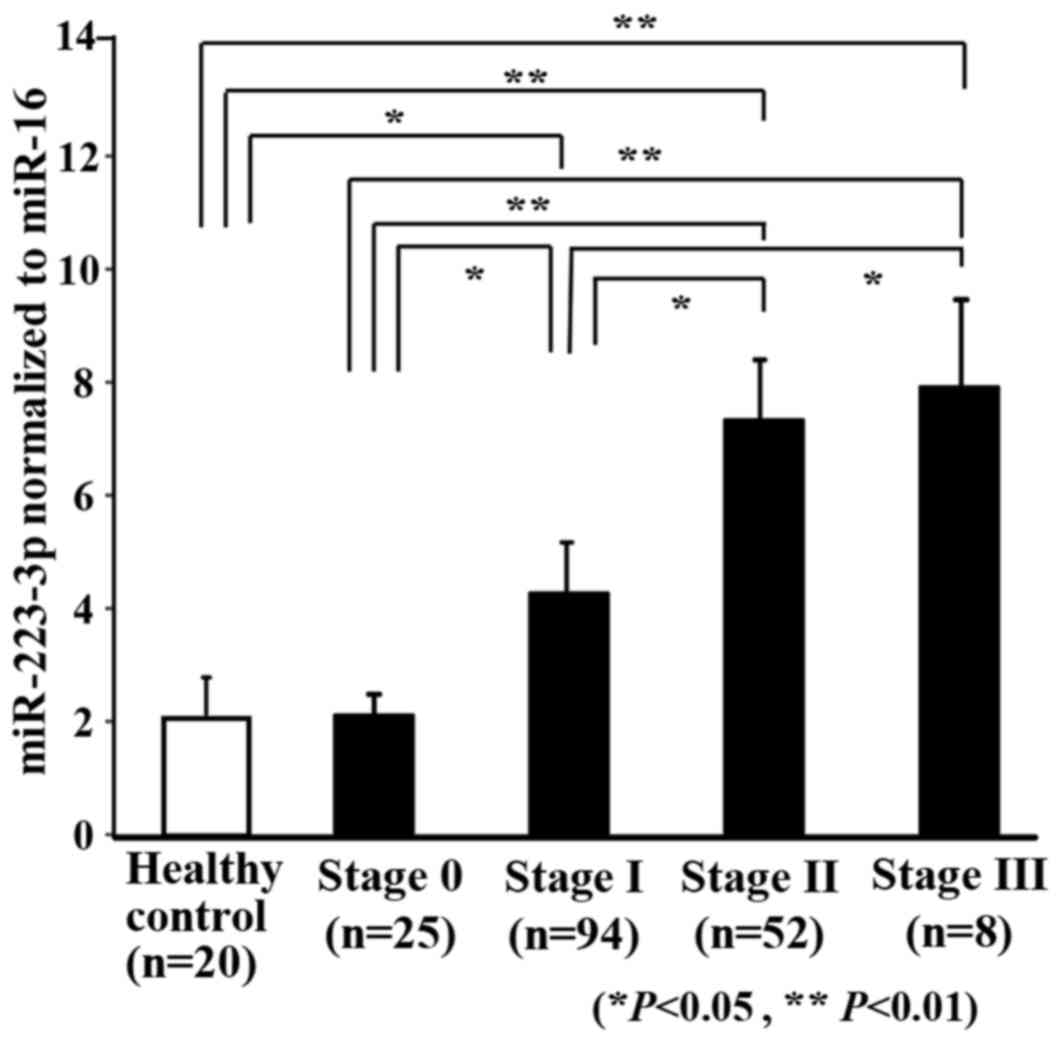
was demonstrated between them (P<0.01). Fig. 5 shows a comparison of exosomal
miR-223-3p levels between the breast cancer patients and the
healthy controls. The exosomal miR-223-3p levels of the breast
cancer patients were significantly higher than those of the healthy
controls (P<0.01). Furthermore, exosomal miR-223-3p levels
increased according to the advance in stage (stage 0 vs. stage II
and stage III: P<0.01; stage I vs. stage 0, stage II and stage
III: P<0.05).
Relationship between
clinicopathological factors and exosomal miR-223-3p levels
To evaluate the correlation between the miR-223-3p
expression levels and the clinicopathological characteristics, 179
breast cancer patients were divided into two groups (high and low
expression). The cut-off level of miR-223-3p was determined as the
median of the relative quantity (median=2.80). The average age was
58 years (range, 33–88). All patients were female. As shown in
Table III, a statistically
significant association was observed between high miR-223-3p
expression and histological type, pT stage, pN stage, pathological
stage, lymphatic invasion and NG (P<0.05).
 | Table III.Association between
clinicopathological factors and exosomal miR-223-3p levels. |
Table III.
Association between
clinicopathological factors and exosomal miR-223-3p levels.
| Variable | No. of patients
n=179 | High n=84 (%) | Low n=95 (%) | P-value |
|---|
| Histological
type |
|
|
|
<0.001 |
|
DCIS | 25 | 3 (3.6) | 22 (23.2) |
|
|
IDC | 154 | 81 (96.4) | 73 (76.8) |
|
| pT Stage |
|
|
|
<0.001 |
| 0 | 27 | 5 (6.0) | 22 (23.2) |
|
| 1 | 119 | 56 (66.6) | 63 (66.3) |
|
| 2 | 33 | 23 (27.4) | 10 (10.5) |
|
| pN Stage |
|
|
| 0.020 |
| 0 | 140 | 58 (69.1) | 82 (86.3) |
|
| 1 | 31 | 21 (25.0) | 10 (10.5) |
|
| 2 | 8 | 5 (5.9) | 3 (3.2) |
|
| Pathological
stage |
|
|
|
<0.001 |
| 0 | 25 | 3 (3.6) | 22 (23.2) |
|
| I | 94 | 41 (48.8) | 53 (55.8) |
|
| II | 52 | 34 (40.5) | 18 (18.9) |
|
|
III | 8 | 6
(7.1) | 2 (2.1) |
|
| Lymphatic
invasion |
|
|
| 0.048 |
|
(−) | 107 | 44 (52.4) | 63 (66.3) |
|
|
(+) | 72 | 40 (47.6) | 32 (33.7) |
|
| Vascular
invasion |
|
|
| 0.559 |
|
(−) | 119 | 54 (64.3) | 65 (68.4) |
|
|
(+) | 60 | 30 (35.7) | 30 (31.6) |
|
| NG |
|
|
| 0.020 |
| 1 | 122 | 61 (72.6) | 61 (64.2) |
|
| 2 | 24 | 5 (6.0) | 19 (20.0) |
|
| 3 | 33 | 18 (21.4) | 15 (15.8) |
|
| ER |
|
|
| 0.510 |
|
(−) | 20 | 8
(9.5) | 12 (12.6) |
|
|
(+) | 159 | 76 (90.5) | 83 (87.4) |
|
| PgR |
|
|
| 0.736 |
|
(−) | 49 | 24 (28.6) | 25 (26.3) |
|
|
(+) | 130 | 60 (71.4) | 70 (73.7) |
|
| HER2a |
|
|
| 0.072 |
|
(−) | 143 | 76 (97.4) | 67 (90.5) |
|
|
(+) | 9 | 2 (2.6) | 7 (9.5) |
|
| Ki67 |
|
|
| 0.800 |
|
≥15% | 39 | 19 (22.6) | 20 (21.1) |
|
|
<15% | 140 | 65 (77.4) | 75 (78.9) |
|
| Intrinsic
subtype |
|
|
| 0.228 |
| Luminal
A | 108 | 61 (78.2) | 47 (63.5) |
|
| Luminal
B | 28 | 11 (14.1) | 17 (23.0) |
|
| HER2
enricheda | 4 | 1 (1.3) | 3 (4.0) |
|
| Triple
negative | 12 | 5 (6.4) | 7 (9.5) |
|
Comparison of exosomal miR-223-3p
levels of DCIS, upstaged IDC and IDC patients
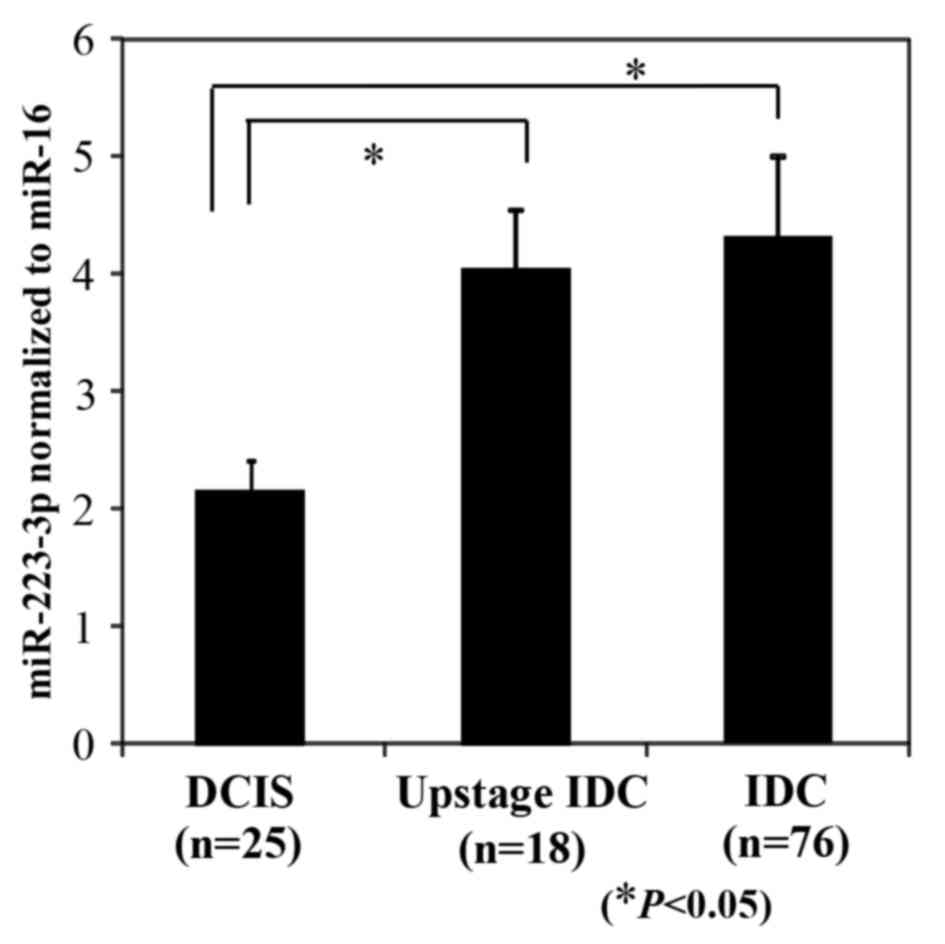
At the time of diagnosis by biopsy before surgery,
there were 43 DCIS patients and 76 IDC patients with stage I. Of
the 43 DCIS patients diagnosed by biopsy, 18 patients (41.9%)
upstaged to IDC patients with stage I in the final pathological
diagnosis by completely excised specimen. Fig. 6 shows a comparison of the plasma
exosomal miR-223-3p levels of DCIS patients diagnosed by completely
excised specimen (n=25), upstaged IDC patients with stage I (n=18),
and IDC patients with stage I (n=76). In our study, miR-223-3p
levels of IDC patients and upstaged IDC patients were significantly
higher than those of DCIS patients (P<0.05).
Correlation between miR-223-3p levels
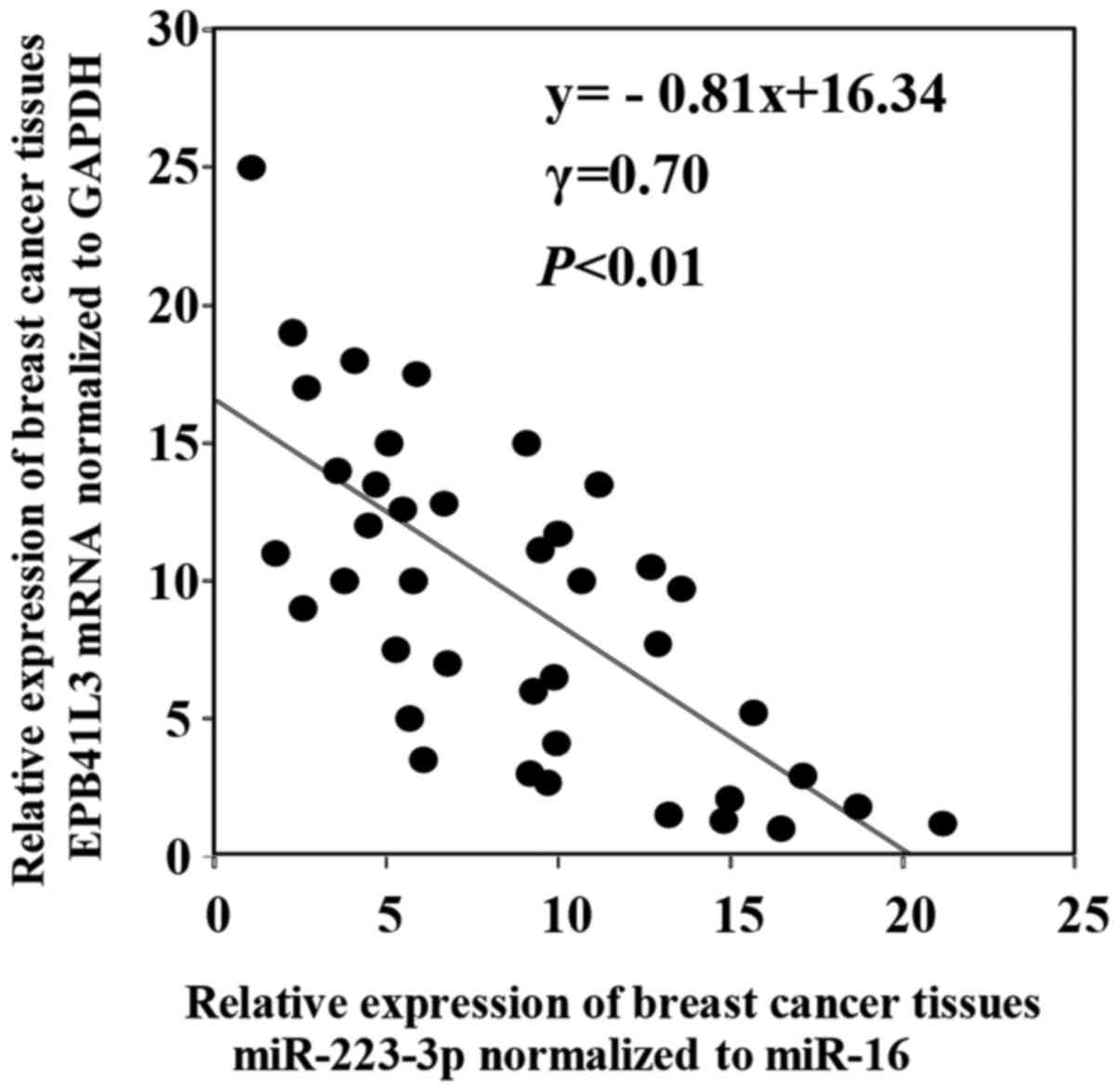
and target gene
Using the target gene prediction program TargetScan
and PicTar, we selected EPB41L3 as one of the direct target genes
of miR-223-3p (data not shown). This study consisted of 40 breast
cancer patients, including DCIS patients (n=5) and stage I patients
(n=15), stage II patients (n=15) and stage III patients (n=5). The
correlation between the miR-223-3p and the EPB41L3 mRNA levels in
the breast cancer tissues was examined. As shown in Fig. 7, a significant inverse correlation was
shown between miR-223-3p and the EPB41L3 mRNA expressions in the
breast cancer tissues (γ=0.70, P<0.01).
Discussion
In the present study, we demonstrated that
miR-223-3p has the potential to enhance cell proliferation and
invasion of breast cancer cells. In breast cancer patients,
exosomal miR-223-3p levels showed a significant relationship with
histological type, pT stage, pN stage, pathological stage,
lymphatic invasion and NG. Furthermore, we clarified that the
plasma exosomal miR-223-3p levels of patients upstaged from DCIS to
IDC were significantly higher than those of the DCIS patients.
A number of recent studies have supported a model of
breast cancer development in which atypical ductal hyperplasia
evolves into DCIS. It is known that disruption of the myoepithelial
layer and the basement membrane in DCIS progresses to IDC (19). DCIS is considered to be a direct
precursor to IDC. Some studies have reported that lesions initially
diagnosed as DCIS by core-needle biopsy or vacuum-assisted biopsy
are occasionally upstaged to IDC after the final diagnosis of the
completely excised specimen. One study (n=336) has reported that
this underestimation rate was 33.6% and another meta-analysis study
(n=7350) showed an underestimation rate was 25.9% (5,20). In our
study, 41.9% (18/43) of DCIS patients upstage to IDC after the
final diagnosis of the completely excised specimen. By accurate
prediction of DCIS and IDC before surgery, more appropriate
individualized treatment for these patients can be planned at an
earlier time. Early diagnosis of IDC may lead to more effective
treatment. This underestimation is mostly occurring due to inherent
limitations in biopsy sampling techniques, whereby a small invasive
lesion may escape detection within the large area of the
intraductal lesion. Previous studies have focused on
clinico-histological or clinico-radiological predictors for
underestimation of invasion in biopsy samples (21,22).
Meta-analysis has indicated that the preoperative variables most
significantly associated with underestimation include the biopsy
device and guidance method, size, grade, mammographic features, and
palpability (5). However, the
relative importance of these various predictors of invasive breast
cancer and their impact on patient outcome has not been clearly
established. In our study, these clinical factors did not show a
relationship with the invasion of breast cancer (23). Therefore, a novel biomarker to
distinguish the invasion lesion hidden in the DCIS patients is
necessary.
Recently, some researchers have reported on the
potential of diagnostic biomarkers of circulating plasma/serum
miRNA in breast cancer patients using various kinds of miRNA
(8,24). In particular, exosome of plasma/serum
is attractive because the miRNAs are preserved in a stable form in
the exosome. However, until now, there are few reports that have
compared IDC and DCIS with miRNA microarrays using exosome samples.
First, we compared the profile of miRNA in DCIS patients, IDC
patients and healthy controls, using the miRNA microarray. In our
study, exosomal miR-223-3p in IDC patients revealed the highest
fold-change compared with the DCIS patients and healthy controls.
Therefore we selected miR-223-3p as on invasion specific biomarker
for breast cancer patients.
Next, we examined the potential of miR-223-3p
against proliferation and invasion of breast cancer cells. Our
results demonstrated that breast cancer MCF7 cells transfected with
miR-223-3p significantly increased proliferation and invasion. As
for the function of miR-223, Huang et al have revealed that
miR-223 increase proliferation and promote invasion of lung cancer
A549 cells via activation of the NF-κB signaling pathway (25). Also of interest is a reported by Li
et al that miR-223 is overexpressed in metastatic gastric
cancer cells and promotes gastric cancer invasion and metastasis
(26). These manuscripts support our
results. In contrast, Pinatel et al reported that a
suppressive role of miR-223 in the migration of breast cancer
(27). The exact reasons for the
discrepancy in these results remain unknown and wait further study.
Since this manuscript does not separately describe miR-223,
miR-223-3p and miR-223-5p, further analysis in which patients are
divided into miR-223-3p and miR-223-5p may be necessary.
The clinical characteristics of exosomal miR-223-3p
in breast cancer patients have not been fully reported. In this
study, we clarified that exosomal miR-223-3p levels of breast
cancer were significantly higher than those found in healthy
controls. In addition, we found a significant association between
exosomal miR-223-3p levels and miR-223-3p expression in primary
breast cancer tissues collected from the same patients. These
results suggest the possibility that tumor tissues may be the
source of plasma exosomal miR-223-3p. Next, we investigated the
relationship between exosome miR-223-3p levels and
clinicopathological factors. We found that exosome miR-223-3p has
relevance to the histological type, pT stage, pN stage,
pathological stage, lymphatic invasion and NG. Furthermore, as a
special point to note, we have revealed that exosomal miR-223-3p
levels of IDC patients upstaged from DCIS by final diagnosis were
significantly higher than those of the DCIS patients. To the best
of our knowledge, this is the first study to show the potential of
exosomal miR-223-3p in the detection of invasive lesions in DCIS
patients diagnosed by biopsy. These results suggest the possibility
that exosomal miR-223-3p may be a useful, less invasive biomarkers
for the selection of IDC patients hidden among DCIS patients
diagnosed by biopsy.
It is known that miRNAs downregulate gene expression
by either inducing degradation of target mRNA or impairing their
translation. By the data base analysis using the target gene
prediction program TargetScan and PicTar, we selected EPB41L3 as a
candidate target gene for miR223-3p. We found a significant inverse
correlation between the miR-223-3p levels and the EPB41L3 mRNA
levels. These results suggest that the expression of EPB41L3 mRNA
may be negatively regulated by miR-223-3p. Li et al also
reported that EPB41L3, which is a tumor suppressor gene, is a
direct target gene for miR-223, and overexpression EPB41L3 can
strongly inhibit migration and invasion of gastric cancer cells
(26). As another target of miR-223,
HAX-1 and STAT5A genes are reported (27,28).
One of the limitations of our study is the small
number of patients. Further research, conducted on a larger number
of cases is required in order to validate the usefulness of
miR-223-3p in the accurate detection of invasive lesions in DCIS
patients diagnosed by biopsy. Furthermore, on inhibition test using
the RNAi of miR-223-3p is also required in order to clarify the
function of miR-223-3p in breast cancer cells. We are planning to
investigate these points in our next study.
In summary, we have shown that miR-223-3p promotes
the invasion of breast cancer cells, and exosomal miR-223-3p may be
useful as a minimally invasive biomarker for the selection of
patients with invasion from DSIC patients diagnosed by biopsy. We
hope to be able to report on future developments in subsequent
larger scale studies.
Acknowledgements
The authors would like to thank Miss J. Tamura for
her technical assistance.
Funding
This study was supported by JSPS KAKENHI (grant no.
JP17K10608).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Author's contributions
HI designed the study. MY, HI and HJ wrote the
manuscript. MY and HI performed the experiments. MY, AM, YU and TY
collected the clinical data. MY, HI and HJ performed the
statistical analysis. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The study protocol conformed to the guidelines of
the Teikyo University Ethics Committee and was approved by the
review board of Teikyo University (approval no. 09-081-3). Written
informed consent was obtained from all patients.
Consent for publication
Written informed consent was obtained from all
patients for the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
DCIS
|
ductal carcinoma in situ
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
IDC
|
invasive ductal carcinoma
|
|
HER2
|
human epidermal growth factor receptor
type 2
|
|
NG
|
nuclear grade
|
|
ER
|
estrogen receptor
|
|
PgR
|
progesterone receptor
|
References
|
1
|
The Editorial Board of the Cancer
Statistics in Japan: Cancer Statistics in Japan 2015Foundation for
Promotion of Cancer Research. National Cancer Center; Tokyo: 2016,
https://ganjoho.jp/data/reg_stat/statistics/brochure/2015/cancer_statistics_2015.pdfMarch
30–2016
|
|
2
|
Welch HG, Prorok PC, O'Malley AJ and
Kramer BS: Breast-cancer tumor size, overdiagnosis and mammography
screening effectiveness. N Engl J Med. 375:1438–1447. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fentiman IS: The dilemma of in situ
carcinoma of the breast. Int J Clin Pract. 55:680–683.
2001.PubMed/NCBI
|
|
4
|
Cox CE, Nguyen K, Gray RJ, Salud C, Ku NN,
Dupont E, Hutson L, Peltz E, Whitehead G, Reintgen D and Cantor A:
Importance of lymphatic mapping in ductal carcinoma in situ (DCIS):
Why map DCIS? Am Surg. 67:513–519. 2001.PubMed/NCBI
|
|
5
|
Brennan ME, Turner RM, Ciatto S,
Marinovich ML, French JR, Macaskill P and Houssami N: Ductal
carcinoma in situ at core needle biopsy: Meta-analysis of
underestimation and predictors of invasive breast cancer.
Radiology. 260:119–128. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Grady WM and Tewai M: The next thing in
prognostic molecular markers: microRNA signatures of cancer. Gut.
59:706–708. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Graveel CR, Calderone HM, Westerhuis JJ,
Winn ME and Sempere LF: Critical analysis of the potential for
microRNA biomarkers in breast cancer management. Breast Cancer
(Dove Med Press). 7:59–79. 2015.PubMed/NCBI
|
|
8
|
Shimomura A, Shiino S, Kawauchi J,
Takizawa S, Sakamoto H, Matsuzaki J, Ono M, Takeshita F, Niida S,
Shiizu C, et al: Novel combination of serum microRNA for detedting
brest cnacer in the early stage. Cancer Sci. 107:326–334. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ge Q, Zhou Y, Lu J, Bai Y, Xie X and Lu Z:
miRNA in plasma exosome is stable under different storage
conditions. Molecules. 19:1567–1575. 2014. View Article : Google Scholar
|
|
10
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Simons M and Raposo G: Exosomes-vesicular
carriers for intercelluar communication. Curr Opin Cell Biol.
21:575–581. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hannafon BN and Ding WQ: Intercelluar
communication by exosome-derived microRNAs in cancer. Int J Mol
Sci. 14:14240–14269. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ogata-Kawata H, Izumiya M, Kurioka D,
Honma Y, Yamada Y, Furuta K, Gunji T, Ohta H, Okamoto H, Sonoda H,
et al: Circulating exosomal microRNAs as biomarkers of colon
cancer. PLoS One. 9:e929212014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Joyce DP, Kerin MJ and Dwyer RM:
Exosome-encapsulated microRNAs as circulating biomarkers for breast
cancer. Int J Cancer. 139:1443–1448. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hannafon BN, Trigoso YD, Calloway CL, Zhao
YD, Lum DH, Welm AL, Zhao ZJ, Blick KE, Dooley WC and Ding WQ:
Plasma exosome microRNAs are indicative of breast cancer. Breast
Cancer Res. 18:902016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kurashige J, Hasegawa T, Niida A,
Sugimachi K, Deng N, Mima K, Uchi R, Sawada G, Takahashi Y, Eguchi
H, et al: Integrated molecular profiling of human gastric cancer
identifies DDR2 as a potential regulator of peritoneal
dissemination. Sci Rep. 6:223712016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dejima H, Iinuma H, Kanaoka R, Matsutani N
and Kawamura K: Exosomal microRNA in plasma as a non-invasive
biomarker for the recurrence of non-small cell lung cancer. Oncol
Lett. 13:1256–1263. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tsukamoto M, Iinuma H, Yagi T, Matsuda K
and Hashiguchi Y: Circulating exosomal MicroRNA-21 as a biomarker
in each tumor stage of colorectal cancer. Oncology. 92:360–370.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bombonati A and Sgroi DC: The molecular
pathology of breast cancer progression. J Pathol. 223:307–317.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Osako T, Iwase T, Ushijima M, Horii R,
Fukami Y, Kimura K, Matsuura M and Akiyama F: Incidence and
prediction of invasive disease and nodal metastasis in
preoperatively diagnosed ductal carcinoma in situ. Cancer Sci.
105:576–582. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chan MY and Lim S: Predictors of invasive
breast cancer in ductal carcinoma in situ initially diagnosed by
core biopsy. Asian J Surg. 33:76–82. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim J, Han W, Lee JW, You JM, Shin HC, Ahn
SK, Moon DG, Cho N, Moon WK, Park IA and Noh DY: Factors associated
with upstaging from ductal carcinoma in situ following core needle
biopsy to invasive cancer in subsequent surgical excision. Breast.
21:641–645. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yoshikawa M: Sentinel node biopsy in
patients diagnosed with ductal carcinoma in situ preoperatively.
Japanese Soc Sentinel Node Navigation Surg. 18:452016.
|
|
24
|
Kodahl AR, Lyng MB, Binder H, Cold S,
Gravgaard K, Knoop AS and Ditzel HJ: Novel circulating microRNA
signature as a potential non-invasive multi-marker test in
ER-positive early-stage breast cancer: A case control study. Mol
Oncol. 8:874–883. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang L, Li F, Deng P and Hu C:
MicroRNA-223 promotes tumor progression in lung cancer A549 cells
via activation of the NF-κB signaling pathway. Oncol Res.
24:405–413. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li X, Zhang Y, Zhang H, Liu X, Gong T, Li
M, Sun L, Ji G, Shi Y, Han Z, et al: miRNA-223 promotes gastric
cancer invasion and metastasis by targeting tumor suppressor
EPB41L3. Mol Cancer Res. 9:824–833. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pinatel EM, Orso F, Penna E, Cimino D,
Elia AR, Circosta P, Dentelli P, Brizzi MF, Provero P and Tavern D:
miR-223 is a coordinator of breast cancer progression as reveraled
by bioinformatics predictions. PLoS One. 9:e848592014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sun X, Li Y, Zheng M, Zuo W and Zheng W:
MicroRNA-223 increases the sensitivity of triple-negative breast
cancer stem cells to TRAIL-induced apoptosis by targeting HAX-1.
PLoS One. 11:e01627542016. View Article : Google Scholar : PubMed/NCBI
|