Introduction
Colorectal cancer (CRC) is currently the third most
common malignancy and the fourth leading cause of cancer-related
death worldwide, with an estimated 1.2 million new cases and over 6
hundred thousand deaths each year (1). CRC develops as a result of the
pathological transformation of normal colonic epithelium to
adenomatous polyp, which ultimately leads to invasive cancer
(2). Though the five-year survival
rate of patients with resectable CRC has recently improved,
exceeding 90%, that of CRC patients with unresectable metastases
remains discouraging at less than 10% (3). Notably, 50–60% of CRC patients can
develop distant metastases and the five-year survival rate will
decrease to only 5% in patients with distant metastases (4). Therefore, early diagnosis and therapy
holds a significant prognostic value, and it is an urgent necessity
to identify new biomarkers as well as specific therapeutic targets
for CRC (5,6). Tumor metastasis consists of multiple
sequential biological processes, including the invasion of cancer
cells into surrounding tissues, intravasation, survival in
circulation, arrest at distant organ sites, extravasation, and
finally growth in distant organs (7).
However, despite considerable advances have been made concerning
the molecular mechanisms underlying tumor metastasis, it is still
far from well understood.
MicroRNAs (miRNAs) belong to a class of small
endogenous RNAs that do not encode for proteins and yet influence
many physiological processes through binding to the 3′-untranslated
region (3′-UTR) of target messenger RNAs (mRNAs), mediating either
mRNA degradation or translational repression (8). The role of miRNAs in human malignancies
has been intensively studied in recent years. Dysregulated
expressions of miRNAs are associated with tumor initiation,
promotion and progression by acting on various oncogenes or tumor
suppressors (9–11). Moreover, miRNAs have been demonstrated
to be closely correlated with clinical stage, metastasis and
survival in numerous human cancers, including CRC (12). microRNA-888 (miR-888), a newly
identified miRNA, was reported to be implicated in the tumorigenic
process of endometrial cancer, prostate cancer, and breast cancer
(13–16). Lewis et al found that
overexpression of miR-888 significantly increases the proliferation
and migration of prostate cancer cells (14), and Huang et al showed that
miR-888 is a repressor of the adherens junction pathway in breast
cancer (15). What's more, evidences
have even pointed to the involvement of miR-888 in maintaining
cancer stem cell-related properties and regulating the
epithelial-mesenchymal transition (EMT) and cancer metastasis
(17). Nevertheless, little is known
about the roles of miR-888 in the clinical pathological
correlations and biological functions in CRC tumorigenesis. Thus,
in the present study, we sought to determine the biological and
clinicopathological implications of miR-888 in the development and
metastasis of CRC.
Materials and methods
Human samples and cell lines
A total of 126 CRC patients that received surgery
resection in Subei People's Hospital of Jiangsu Province between
2009 and 2014 were enrolled in this study. None of them had
undergone chemotherapy, radiotherapy or immunotherapy before.
Pathological analyses were used to confirm the diagnosis, and the
patients were staged according to the tumor-node-metastasis (TNM)
staging system of American Joint Committee on Cancer (AJCC).
Detailed clinical informations of the patients are shown in
Tables I and II. Tumor tissues and the paired normal
tissues were obtained after surgical resection and immediately
placed in liquid nitrogen or 10% formalin for further analyses. The
present study was approved by the Ethics Committee of Subei
People's Hospital of Jiangsu Province and written informed consent
was obtained from all of the patients. The overall survival (OS)
time was defined as the date of surgery to the date of death from
any cause, or to the last follow-up date; while the disease-free
survival (DFS) was computed from the operation date to the date of
local or distant recurrence or death from any cause.
 | Table I.Correlation between
clinicopathological parameters and miR-888 expression in patients
with colorectal cancer. |
Table I.
Correlation between
clinicopathological parameters and miR-888 expression in patients
with colorectal cancer.
|
|
| miR-888
expression |
|
|---|
|
|
|
|
|
|---|
|
Characteristics | No. (%) | High n=93 (%) | Low n=33 (%) | Pa |
|---|
| Age |
|
|
| 0.453 |
|
<60 | 60 (47.6) | 44 (47.3) | 16 (48.5) |
|
|
≥60 | 66 (52.4) | 49 (52.7) | 17 (51.5) |
|
| Sex |
|
|
| 0.251 |
|
Male | 71 (56.3) | 53 (57.0) | 18 (54.5) |
|
|
Female | 55 (43.7) | 40 (43.0) | 15 (45.5) |
|
| Tumor location |
|
|
| 0.235 |
|
Left | 79 (62.7) | 59 (63.4) | 20 (60.6) |
|
|
Transverse | 13 (10.3) | 9 (9.7) | 4 (12.1) |
|
|
Right | 34 (27.0) | 25 (26.9) | 9 (27.3) |
|
| pT status |
|
|
| 0.009a |
|
T1/T2 | 28 (22.2) | 14 (15.1) | 14 (42.4) |
|
|
T3/T4 | 98 (77.8) | 79 (84.9) | 19 (57.6) |
|
| pN status |
|
|
| 0.031a |
|
Absent | 72 (57.1) | 48 (51.6) | 24 (72.7) |
|
|
Present | 54 (42.9) | 45 (48.4) | 9 (27.3) |
|
| pMstatus |
|
|
| 0.018a |
|
Absent | 116 (92.1) | 84 (90.3) | 32 (97.0) |
|
|
Present | 10 (7.9) | 9 (9.7) | 1 (3.0) |
|
| AJCC stage |
|
|
| 0.042a |
|
I/II | 71 (56.3) | 48 (51.6) | 23 (69.7) |
|
|
III/IV | 55 (43.7) | 45 (48.4) | 10 (30.3) |
|
|
Differentiation |
|
|
| 0.005a |
|
Well | 27 (21.4) | 13 (14.0) | 14 (42.4) |
|
|
Moderate | 78 (61.9) | 60 (64.5) | 18 (54.5) |
|
|
Poor | 21 (16.7) | 20 (21.5) | 1 (3.0) |
|
 | Table II.Univariate and multivariate analyses
of the overall survival (OS) and disease-free survival (DFS) in
patients with colorectal cancer. |
Table II.
Univariate and multivariate analyses
of the overall survival (OS) and disease-free survival (DFS) in
patients with colorectal cancer.
|
| Overall survival
univariate | OS
multivariate | Disease-free
survival univariate | DFS
multivariate |
|
|
|
|
|
|
|
| HR (95%CI) | pa | HR (95%CI) | pa | HR (95%CI) | pa | HR (95%CI) | Pa |
|---|
| miR-888 |
|
|
|
|
|
|
|
|
|
Low | – |
| – |
| – |
| – |
|
|
High | 5.42 (3.69,
11.55) | 0.005a | 4.89 (2.44,
9.08) | 0.008a | 6.01 (2.85,
11.75) | 0.018a | 5.37 (2.96,
10.03) | 0.011a |
| Age |
|
|
|
|
|
|
|
|
|
<60 | – |
|
|
| – |
| – |
|
|
≥60 | 1.72 (1.11,
2.93) | 0.290 |
|
| 1.56 (1.07,
2.67) | 0.345 |
|
|
| Sex |
|
|
|
|
|
|
|
|
|
Female | – |
|
|
| – |
|
|
|
|
Male | 1.42 (1.02,
1.99) | 0.284 |
|
| 1.28 (0.96,
1.85) | 0.368 |
|
|
| Tumor location |
|
|
|
|
|
|
|
|
|
Transverse | – |
|
|
| – |
|
|
|
|
Right | 0.85 (0.55,
1.34) | 0.523 |
|
| 0.81 (0.49,
1.28) | 0.673 |
|
|
|
Left | 1.16 (0.91,
1.66) | 0.301 |
|
| 1.07 (0.85,
1.50) | 0.455 |
|
|
| pT status |
|
|
|
|
|
|
|
|
|
T1/T2 | – |
| – |
| – |
| – |
|
|
T3/T4 | 2.60 (1.17,
4.36) | 0.025a | 2.11 (1.01,
4.22) | 0.036a | 2.28 (1.25,
4.64) | 0.017a | 1.98 (1.12,
3.87) | 0.024a |
| pN status |
|
|
|
|
|
|
|
|
|
Absent | – |
| – |
| – |
| – |
|
|
Present | 3.18
(2.03,7.67) | 0.003a | 2.88
(1.21,4.70) |
<0.001a | 3.03 (2.43,
7.07) | 0.004a | 2.78 (1.27,
5.45) | 0.011a |
| pM status |
|
|
|
|
|
|
|
|
|
Absent | – |
| – |
| – |
| – |
|
|
Present | 5.21
(2.42,10.61) | 0.023a | 4.98
(2.33,9.75) | 0.037a | 5.75
(2.21,10.87) | 0.028a | 5.18
(2.08,9.15) | 0.033a |
| AJCC stage |
|
|
|
|
|
|
|
|
|
I/II | – |
| – |
| – |
| – |
|
|
III/IV | 4.12
(2.37,8.86) | 0.009a | 3.82
(1.55,6.71) | 0.011a | 4.92
(2.49,9.19) | 0.024a | 3.37 (1.43,
6.77) | 0.031a |
|
Differentiation |
|
|
|
|
|
|
|
|
|
Well | – |
| – |
| – |
| – |
|
|
Moderate | 3.59 (1.02,
6.12) | 0.023a | 2.54
(1.35,4.71) | 0.017a | 3.03 (1.25,
5.66) | 0.008a | 2.69 (1.09,
4.96) | 0.013a |
|
Poor | 5.45
(2.22,12.34) |
<0.001a | 4.76
(2.29,11.23) | 0.004a | 5.75
(2.41,11.63) | 0.019a | 5.08 (2.83,
10.21) | 0.005a |
Human HEK 293 cell and colorectal cancer cell line
SW620 were obtained from the American Type Culture Collection
(ATCC). Cells were maintained in a humidified 5% CO2
atmosphere at 37°C and were grown in complete growth medium
(Invitrogen, Carlsbad, CA) as recommended by the manufacturer.
Cells were regularly authenticated by checking their morphology and
confirming the absence of mycoplasma contamination (MycoAlert,
Lonza, Rockland, ME).
In situ hybridization and staining
assessment
In situ hybridization (ISH) was performed using
double-digoxigenin (DIG) labeled 2′O-methyl locked nucleic acid
(LNA)-ZEN probes (Integrated DNA Technologies) complimentary to
miR-888 (5′-DIG-U/ZEN/GACUGACAGCUTUUUGAGU/ZEN/A-DIG-3′) along with
a scrambled negative control probe
(5′-DIG-C/ZEN/GUAUUAUAGCCGAUUAACG/ZEN/A-DIG-3′), where LNA
modifications are underlined. Hybridization, washing, and scanning
were carried out according to previous reports (18).
The staining index (SI), a semiquantitative
evaluation system incorporating the intensity and percentage of
positive cells, was conducted to assess the miR-888 staining
results (19). The staining intensity
was classified into four grades: 0, no staining; 1, weak; 2,
moderate; 3, strong. The percentage of cells stained was graded as
follows: 0, no staining; 1, <10%; 2, 10–50%; and 3, >50%
tumor cells. SI was calculated by multiplying the grade for
percentage staining by the grade for intensity. Each sample was
evaluated by two experienced pathologists in a blind manner. Scores
of 4 or greater were defined as positive staining and high
expression.
RNA isolation and quantitative
real-time RT-PCR
RNA samples were isolated using Trizol reagent
(Invitrogen) according to the manufacturer's instructions. miR-888
expression was quantified by two-step quantitative RT-PCR,
beginning with first-strand cDNA synthesis using the One-step
primeScript miRNA cDNA Synthesis kit, followed by quantitative
real-time PCR amplification using the miRscript SYBR-Green PCR kit
(Takara); while the quantitative SYBR-Green PCR kit (Qiagen,
Hilden, Germany) was used to quantify the mRNA level of genes in
Smad4 pathway. All primers used are shown in Table III. miRNA quantity was normalized
using the small nuclear RNA U6 while the other genes were
normalized to GAPDH. Fold change of miRNA/mRNA expression was
calculated using the 2-∆∆cq method.
 | Table III.Primers used in this study |
Table III.
Primers used in this study
| Oligo | Sequence |
|---|
| Smad4 |
5′-CGGACATTACTGGCCTGTTC-3′ |
| (Foward) |
|
| Smad4 |
5′-TAGGGCAGCTTGAAGGAAACC-3′ |
| (Reverse) |
|
| E-cadherin |
5′-GAAGTGTCCGAGGACTTTGG-3′ |
| (Forward) |
|
| E-cadherin |
5′-CAGTGTCTCTCCAAATCCGATA-3′ |
| (Reverse) |
|
| TGF-β1 |
5′-GAGGCGGTGCTCGCTTTGTA-3′ |
| (Forward) |
|
| TGF-β1 |
5′-GCACTGCTTCCCGAATGTCTG-3′ |
| (Reverse) |
|
| Snail |
5′-GTCCTTGCTCCACAAACACCA-3′ |
| (Forward) |
|
| Snail |
5′-CTGCCTTCCATCAGCCATCT-3′ |
| (Reverse) |
|
| Twist |
5′-GTCCGCAGTCTTACGAGGAG-3′ |
| (Forward) |
|
| Twist |
5′-CCAGCTTGAGGGTCTGAATC-3′ |
| (Reverse) |
|
| N-cadherin |
5′-TTGGTTTGGGGAGGGAGA-3′ |
| (Forward) |
|
| N-cadherin |
5′-CTGGGGTCAGAGGTGTATCATTT-3′ |
| (Reverse) |
|
| GAPDH |
5′-ACAGTCAGCCGCATCTTCTT-3′ |
| (Forward) |
|
| GAPDH |
5′-ACGACCAAATCCGTTGACTC-3′ |
| (Reverse) |
|
| miR-888 |
5′-ATGTGGCAGATCCCACAGGAGTTT-3′ |
| (Forward) |
|
| miR-888 |
5′-ACTGGGTTTGACTTCGTAGCCCTT-3′ |
| (Reverse) |
|
| U6 (Forward) |
5′-CTGCTTCGGCAGCACA-3′ |
| U6 (Reverse) |
5′-AACGCTTCACGAATTTGCGT-3′ |
|
Western blotting
Western blot analysis was performed as described
previously (20), using the Smad4
antibodies (1:5000 dilution; Abcam, UK) and β-actin antibodies
(1:5000 dilution; Abcam).
Vector construction and cell
transfection
The miR-888 expression plasmid was generated by
cloning the genomic pre-miR-888 (5′-UACUCAAAAAGCUGUCAGUCA-3′) into
OriGene's pCMV6-Mir Vector (Promega, Madison, WI, USA) to generate
the plasmid pCMV-miR-888. The empty vector was used as a negative
control (NC).
Vectors were transfected into cells with
Lipofectamine 2000 (Invitrogen). The day before transfection, the
cells were seeded in 6-well plates and reached a confluence of
approximately 80%. For each transfection experiment, 2 µg of DNA
and 6 µl of Lipofectamine 2000 were added to 100 µl of Opti-MEM
medium. Medium was replaced 6 h later. After a 72 h incubation,
cells were harvested for further experiments.
Luciferase reporter assay
Luciferase constructs were generated by ligating
oligonucleotides containing the wild-type or mutant putative target
site of the Smad4 3′-UTR into the Psi-CHECK2 vector (Promega)
downstream of the luciferase gene. HEK 293 or human CRC cells were
transfected with miR-888 or the NC in combination with each
individual psiCHECK2 luciferase vector. Cells were collected at 24
h post-transfection, and Firefly and Renilla luciferase activities
were measured using the Dual Luciferase Assay System (Promega)
according to the manufacturer's protocol.
Cell proliferation assay
The cells were plated in 96-well plates at
5×103 cells/well and allowed to grow for 24–72 h.
Subsequently, the cells were incubated with 10 µl CCK-8 (Beyotime,
Shanghai, China) at 37°C for 4 h. Absorbance was measured at 450 nm
using a microplate reader.
Wound healing assay
Cells were seeded in 6-well plates and incubated
under permissive conditions until approximately 90% confluence.
After serum starvation for 24 h, wounds were created in the
confluent cells using a pipette tip. Wound healing within the
scrape line was then observed and photographed at indicated time
points. Each experiment was repeated at least three times.
Transwell invasion assay
A Boyden chamber with 8 µm-pore filter membrane was
used. Briefly, cells (1×105) in culture medium
containing free FBS were seeded in the upper chamber, and the
culture medium with 20% FBS was added in the lower chamber as a
chemoattractant. The upper side of the filter was coated with 0.2%
Matrigel (BD Biosciences, San Jose, CA, USA). After incubation for
48 h, the cells on the upper side of the filter were removed with
cotton swabs, while the cells that migrated to the lower side were
fixed in 4% paraformaldehyde and stained with crystal violet. The
cells invaded to the lower surface of the filter were counted at 10
random fields.
Statistical analysis
All statistical analyses were carried out using the
SPSS v20.0 program. Data were expressed as mean ± standard
deviation (SD). The two-tailed paired Student's t-test was
conducted for analyzing two groups. Group differences were
statistically analyzed using the χ2 test. Survival curves were
described by the Kaplan-Meier method and compared with the log-rank
test. Prognostic factors and survival data were evaluated using
univariate and multivariate Cox regression analyses. P<0.05 was
considered to indicate a statistically significant difference.
Results
High level of miR-888 expression is
correlated with aggressive CRC
To determine the clinical and pathological relevance
of miR-888 in human CRC, we explored the expression of miR-888 in
126 cases of primary CRC biopsies by qRT-PCR and found that the
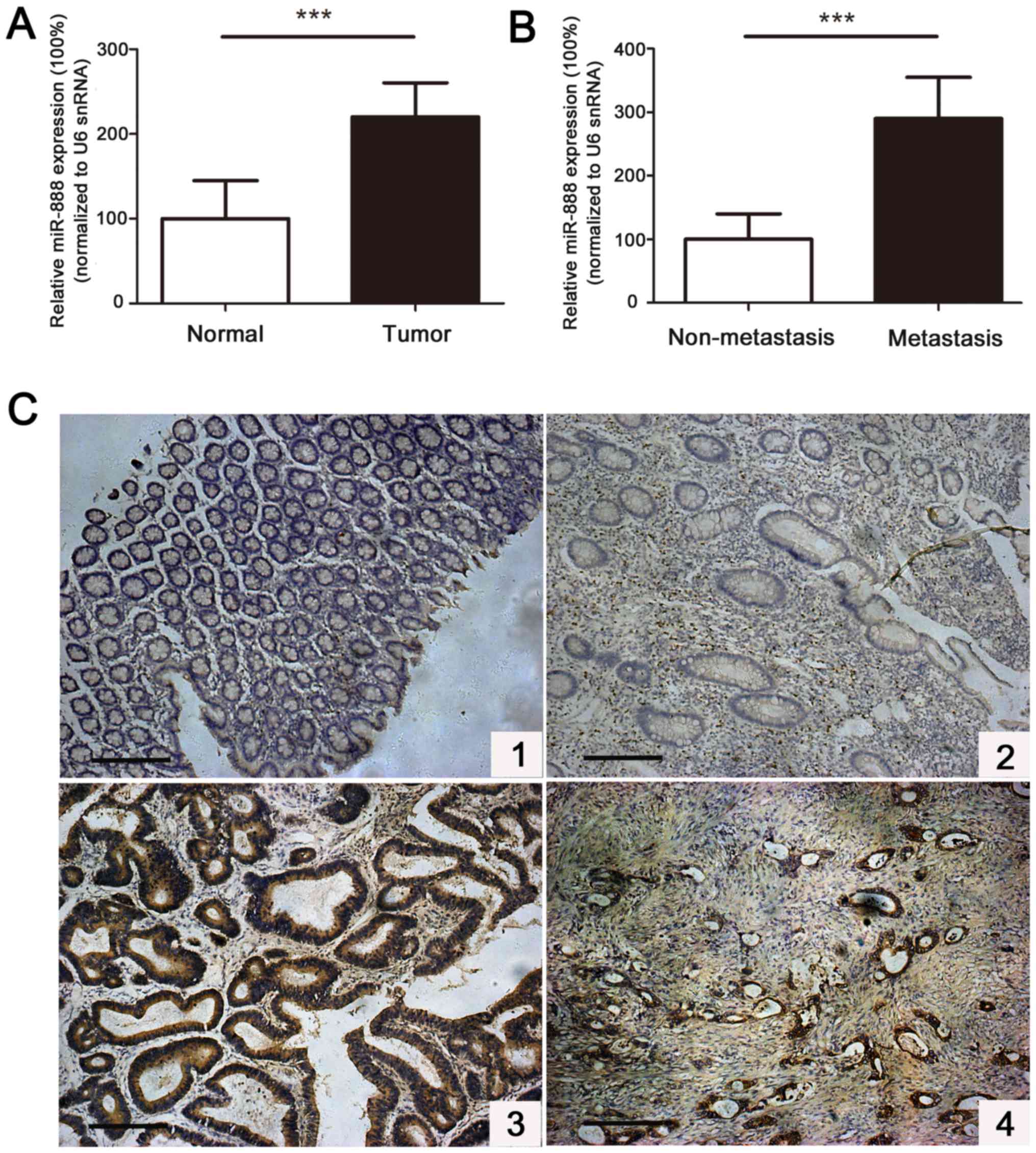
miR-888 expression level was significantly upregulated in tumor
tissues compared with the paired adjacent normal tissues (Fig. 1A). Furthermore, patients with distant
metastatic tumors showed a remarkably higher miR-888 level than
those without tumor metastasis. The level of miR-888 in metastatic
tumors was 2.9-fold higher than that in non-metastatic tumors
(Fig. 1B). Further in situ
hybridization (ISH) analysis demonstrated that high miR-888
expression was detected in 93/126 (73.8%) cases of CRC patients,
whereas low miR-888 expression was observed in 33/126 (26.2%) of
the patients (Fig. 1C).
Clinicopathological investigations confirmed that increased miR-888
expression was positively associated with pT status (P=0.009), pN
status (P=0.031), pM status (P=0.018), AJCC stage (P=0.042), and
histological differentiation (P=0.005) of tumors, but not with
patients' age (P=0.453), sex (P=0.251), or tumor location (P=0.235)
(Table I). These results indicate
that miR-888 plays an important role in the progression and
metastasis of CRC and represents a potential predictive marker.
Increased miR-888 expression predicts
poor prognosis in patients with CRC
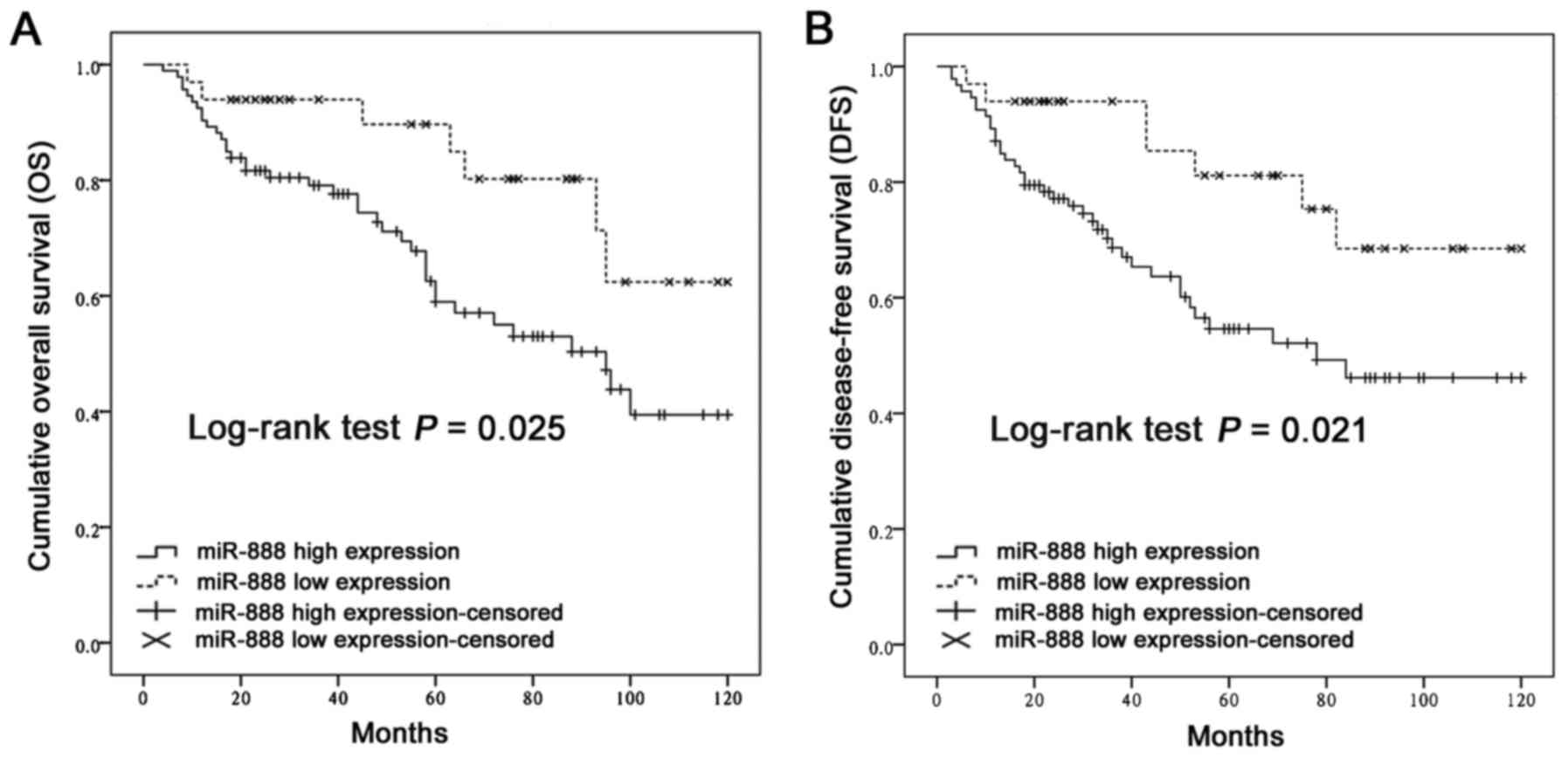
Kaplan-Meier analysis and the log-rank test were
used to determine the prognostic significance of miR-888 in CRC. It
was observed that patients with high miR-888 expression presented
significantly shorter OS (P=0.025, Fig.
2A) and DFS (P=0.021, Fig. 2B)
times than those with low miR-888 expression. Subsequently,
univariate and multivariate analyses were performed to identify the
risk factors correlated with the prognosis of CRC patients. The
univariate Cox proportional hazard analysis showed that high
miR-888 expression, advanced pT, N, and M status, and poor
histological differentiation contributed significantly to poor OS
and DFS rates in patients with CRC (Table II, P<0.05 for all). Furthermore,
after adjusting for all clinicopathological factors, multivariate
analysis confirmed that miR-888 expression, the TNM stage, and
histological grade were independent prognostic factors for survival
(Table II, P<0.05 for all). These
above findings strongly suggest that miR-888 may serve as a
predictor of poor survival among CRC patients.
miR-888 contributes to proliferation,
invasion, and metastasis of CRC cells
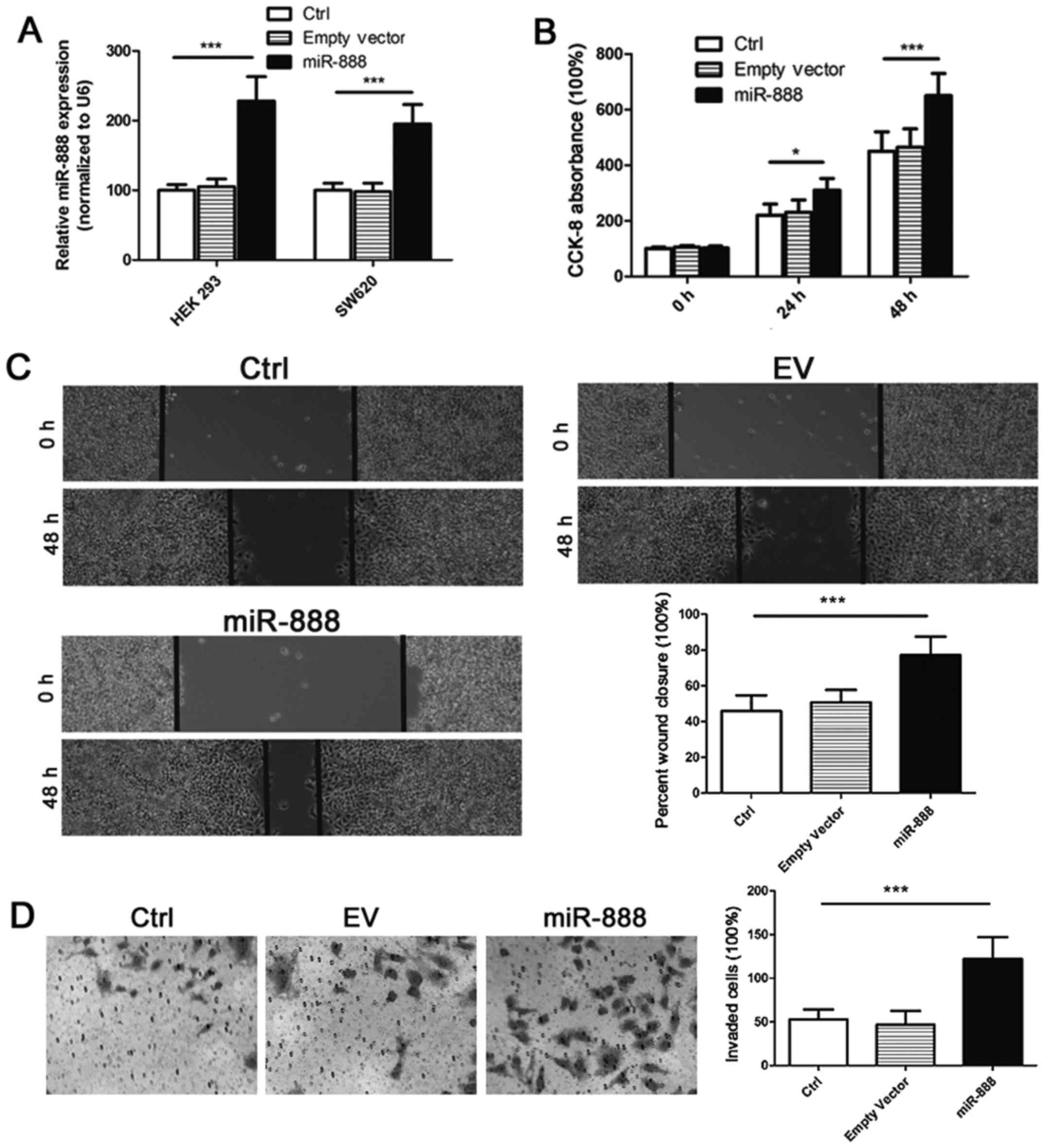
To explore the biological role of miR-888 in CRC,
miR-888 expression vectors and the empty control vectors were
constructed and transfected into targeted cells. RT-PCR analysis
validated that the miR-888 level was increased by 2.3-fold in HEK
293 cells and approximately 2-fold in SW620 cells after
transfection of expression vectors (Fig.
3A). We then investigated the effects of miR-888 on CRC cell
proliferation by CCK-8 assay. Overexpressing miR-888 in SW620 cells
markedly upregulated the proliferation rate of cancer cells in
vitro, whereas transfection of empty controls showed no
significant effects on cell proliferation (Fig. 3B). We next examined the influence of
miR-888 on cell migration and invasion by the wound healing assay
and the transwell assay, respectively. It was found that CRC cells
transfected with miR-888 expression vectors showed significantly
enhanced capability of migration and invasion than those
transfected with empty controls (Fig. 3C
and D). Taken together, these results suggest that miR-888 may
function as an oncogenic miRNA in CRC tumorigenesis.
miR-888 inhibits Smad4 expression in
CRC cells by directly binding to its 3′-UTR
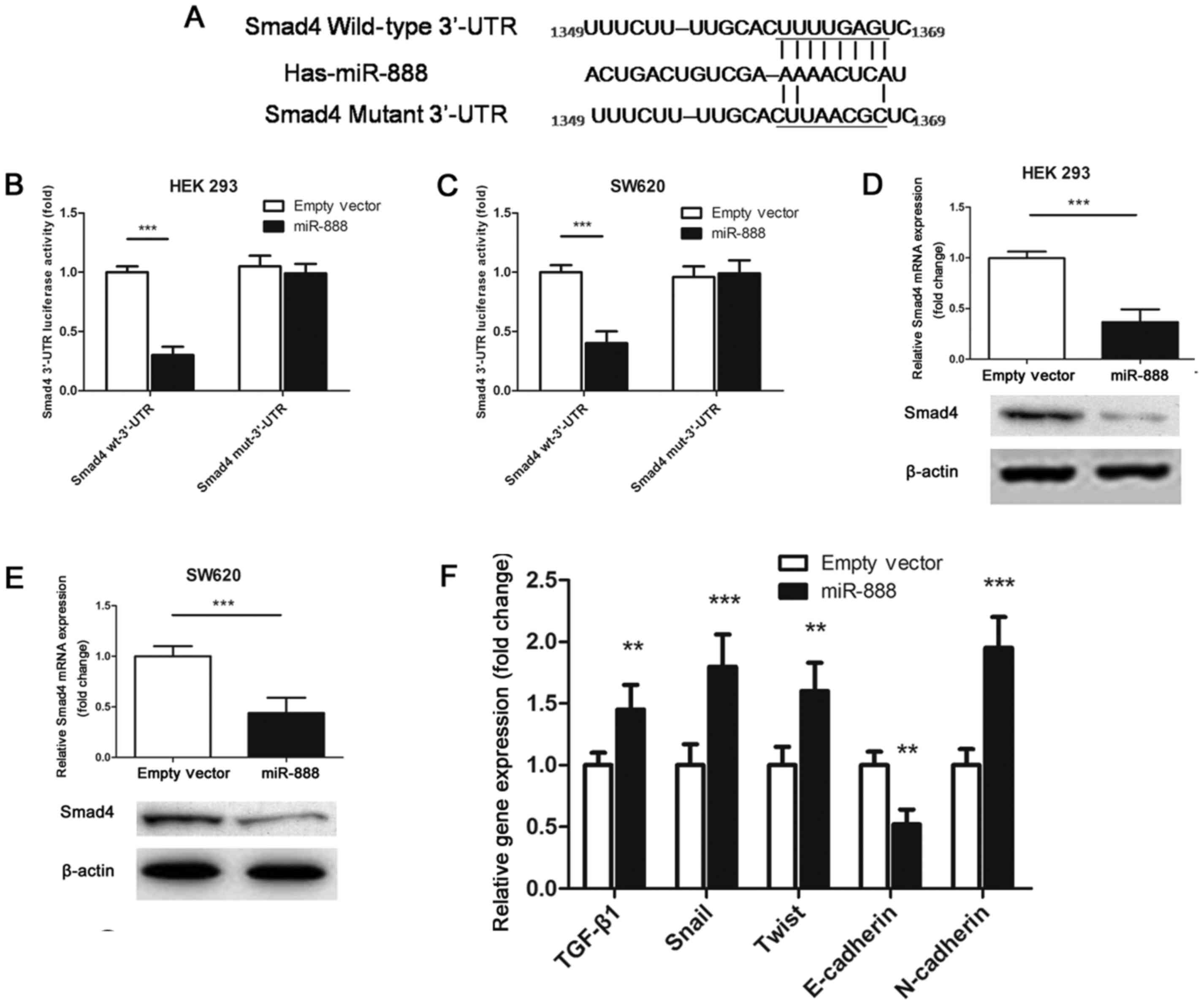
After reviewing the published literature and
searching the miRNA prediction databases, we identified a putative
miR-888 binding site in the 3′-UTR of Smad4, a well-established
tumor suppressor whose mutation or loss of function causes an
acceleration of tumor progression in a variety of human
malignancies (21,22). We thus cloned the wild-type or mutant
Smad4 3′-UTR, and inserted it into a luciferase reporter vector
(Fig. 4A). Our experiments showed
that overexpressing miR-888 remarkably reduced the luciferase
activity of the wild-type 3′-UTR of Smad4, but not the mutant
reporter constructs, in both HEK 293 cells and SW620 cells
(Fig. 4B and C). Further PCR and
western blot verifications confirmed the direct inhibition of
miR-888 on the gene and protein levels of Smad4 expression
(Fig. 4D and E).
Thereafter, we investigated the expression of key
factors involved in the molecular signaling of Smad4-regulated
epithelial-mesenchymal transition (EMT) and tumor metastasis. PCR
analysis showed that by upregulation of miR-888, the expression
levels of TGF-β1, Snail, Twist and N-cadherin in SW620 cells were
significantly increased, compared with that in cells transfected
with empty vectors; but in contrast, the expression level of
E-cadherin was decreased (Fig. 4F).
These results demonstrate the importance of miR-888-mediated Smad4
inhibition in the regulation of CRC invasion and metastasis.
Discussion
CRC remains one of the major lethal causes of
patients with cancer. Early diagnosis is an effective means to
reduce the mortality of CRC patients, and the detection of
biomarkers is an important component of diagnosis, as well as
treatment (23,24). An ideal biomarker for diagnosis should
be highly specific, sensitive, and noninvasive (25). Evidences have revealed that miRNAs
exhibit unique expression profiles in different tumor types and are
important in the initiation and progression of human cancers
(26). The crucial functions of
miRNAs in carcinogenesis have promoted intensive research into
miRNA-based diagnostic and therapeutic strategies for the treatment
of cancer. In this study, for the first time, we characterized
miR-888 as a novel colorectal cancer-associated miRNA that
correlates with disease status and shares functional features of an
oncogenic factor in human CRC. By systematic clinicopathological
investigations, we found that miR-888 expressed significantly
higher in CRC tissues than in normal colon tissues. Increased
miR-888 expression was positively associated with advanced TNM
stage and poor histological differentiation of tumors. We also
showed that CRC patients with low miR-888 expression experienced
better overall survival as well as disease-free survival outcomes;
while patients with high miR-888 expression demonstrated
significantly reduced survival rates. Further univariate and
multivariate analyses identified miR-888 upregulation as an
independent prognostic factor for survival in patients with CRC.
All these findings provide strong evidence for the translational
potential of miR-888 in the prediction and diagnosis of human
CRC.
Recent studies have highlighted the pivotal role of
miRNAs in a broad range of developmental processes associated with
tumor progression and metastasis, which are controlled by complex
and multistep genetic and/or epigenetic changes (27). The TGF-β signaling has been reported
to play a bilateral role in tumor development depending on the
condition of the Smad family proteins (28–30).
Smad4, a key signal transducer of the TGF-β/Smad pathway, generally
acts as a tumor suppressor in CRC (31). It has been documented that Smad4
mutations are observed in approximately 10% of sporadic CRC
patients. Loss of the functional Smad4 protein occurs frequently in
the adenoma-to-carcinoma sequence, and contributes to distant
metastasis, non-response to chemotherapy, and undesirable prognosis
(32). In our present study, we
identified miR-888 as a novel miRNA that directly binds to the
3′-UTR of Smad4 in CRC cells and causes its downregulation. By
targeting the Smad4 signaling, miR-888 significantly promoted the
proliferation, migration, invasion and metastasis of CRC cells.
Thus, we first characterized miR-888 as an important oncomiR in
human CRC. However, given that the off-target effects, which
derived from non-specific binding of miRNAs to target sequences
sharing high homologies, are frequently existed when performing
miRNA-related gene-silencing experiments and could possibly cause
confusion on the mechanism by which the target gene is regulated
(33,34), microarray analysis should be used to
further validate the specificity of miR-888-mediated Smad4
silencing in the future.
EMT has been validated an important step involved in
cancer invasion and metastasis. During the EMT process, cells lose
the epithelial characteristics, gaining instead an invasive and
migratory mesenchymal phenotype, which permits these cells to leave
the tissue parenchyma and enter the systemic circulations (35,36).
TGF-β1 is a well-established metastatic inducer by interacting with
Smad2/3/4 and promoting EMT in late-stage tumor progression
(37–39). Here we demonstrated that miR-888 was
capable of inducing the upregulation of the mesenchyme cell marker
N-cadherin and the intracellular transcription factors Snail and
Twist; whereas the downregulation of the epithelial marker
E-cadherin. Our findings confirmed that miR-888 is important for
TGF-β1-mediated induction of EMT and that targeting miR-888 might
be an effective strategy to inhibit or reverse CRC metastasis.
In conclusion, the current study showed that miR-888
acts as an oncogenic miRNA in CRC by targeting Smad4 and represents
a promising predictive and prognostic factor for CRC patients.
Thus, interrupting or inhibiting miR-888 expression may provide a
therapeutic approach for CRC treatment.
Glossary
Abbreveations
Abbreviations:
|
miRNA
|
microRNA
|
|
CRC
|
colorectal cancer
|
|
OS
|
overall survival
|
|
DFS
|
disease-free survival
|
|
3′-UTR
|
3′-untranslated region
|
|
EMT
|
epithelial-mesenchymal transition
|
|
AJCC
|
American Joint Committee on Cancer
|
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Verma AM, Patel M, Aslam MI, Jameson J,
Pringle JH, Wurm P and Singh B: Circulating plasma microRNAs as a
screening method for detection of colorectal adenomas. Lancet. 385
Suppl 1:S1002015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lan YT, Yang SH, Chang SC, Liang WY, Li
AF, Wang HS, Jiang JK, Chen WS, Lin TC and Lin JK: Analysis of the
seventh edition of American Joint Committee on colon cancer
staging. Int J Colorectal Dis. 27:657–663. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Van Cutsem E, Cervantes A, Nordlinger B
and Arnold D: ESMO Guidelines Working Group: Metastatic colorectal
cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment
and follow-up. Ann Oncol. 25 Suppl 3:iii1–9. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
De Rosa M, Rega D, Costabile V, Duraturo
F, Niglio A, Izzo P, Pace U and Delrio P: The biological complexity
of colorectal cancer: Insights into biomarkers for early detection
and personalized care. Therap Adv Gastroenterol. 9:861–886. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Aghagolzadeh P and Radpour R: New trends
in molecular and cellular biomarker discovery for colorectal
cancer. World J Gastroenterol. 22:5678–5693. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guan X: Cancer metastases: Challenges and
opportunities. Acta Pharm Sin B. 5:402–418. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Svoronos AA, Engelman DM and Slack FJ:
OncomiR or tumor suppressor? The duplicity of microRNAs in cancer.
Cancer Res. 76:3666–3670. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shah MY, Ferrajoli A, Sood AK,
Lopez-Berestein G and Calin GA: microRNA therapeutics in cancer-an
emerging concept. EBioMedicine. 12:34–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Behbahani GD, Ghahhari NM, Javidi MA,
Molan AF, Feizi N and Babashah S: MicroRNA-mediated
post-transcriptional regulation of epithelial to mesenchymal
transition in cancer. Pathol Oncol Res. 23:1–12. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mohammadi A, Mansoori B and Baradaran B:
The role of microRNAs in colorectal cancer. Biomed Pharmacother.
84:705–713. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hovey AM, Devor EJ, Breheny PJ, Mott SL,
Dai D, Thiel KW and Leslie KK: miR-888: A Novel cancer-testis
antigen that targets the progesterone receptor in endometrial
cancer. Transl Oncol. 8:85–96. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lewis H, Lance R, Troyer D, Beydoun H,
Hadley M, Orians J, Benzine T, Madric K, Semmes OJ, Drake R and
Esquela-Kerscher A: miR-888 is an expressed prostatic
secretions-derived microRNA that promotes prostate cell growth and
migration. Cell Cycle. 13:227–239. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang S and Chen L: MiR-888 regulates side
population properties and cancer metastasis in breast cancer cells.
Biochem Biophys Res Commun. 450:1234–1240. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bader AG: miR-888: Hit it when you see it!
Cell Cycle. 13:3512014. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang S, Cai M, Zheng Y, Zhou L, Wang Q
and Chen L: miR-888 in MCF-7 side population sphere cells directly
targets E-cadherin. J Genet Genomics. 41:35–42. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen Z, Liu S, Tian L, Wu M, Ai F, Tang W,
Zhao L, Ding J, Zhang L and Tang A: miR-124 and miR-506 inhibit
colorectal cancer progression by targeting DNMT3B and DNMT1.
Oncotarget. 6:38139–38150. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cao ZG, Li JJ, Yao L, Huang YN, Liu YR, Hu
X, Song CG and Shao ZM: High expression of microRNA-454 is
associated with poor prognosis in triple-negative breast cancer.
Oncotarget. 7:64900–64909. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shen J, Song G, An M, Li X, Wu N, Ruan K,
Hu J and Hu R: The use of hollow mesoporous silica nanospheres to
encapsulate bortezomib and improve efficacy for non-small cell lung
cancer therapy. Biomaterials. 35:316–326. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Duff EK and Clarke AR: Smad4 (DPC4)-a
potent tumour suppressor? Br J Cancer. 78:1615–1619. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Demagny H and De Robertis EM: Smad4/DPC4:
A barrier against tumor progression driven by RTK/Ras/Erk and
Wnt/GSK3 signaling. Mol Cell Oncol. 3:e9891332016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Weng M, Wu D, Yang C, Peng H, Wang G, Wang
T and Li X: Noncoding RNAs in the development, diagnosis, and
prognosis of colorectal cancer. Transl Res. 181:108–120. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sinicrope FA, Okamoto K, Kasi PM and
Kawakami H: Molecular biomarkers in the personalized treatment of
colorectal cancer. Clin Gastroenterol Hepatol. 14:651–658. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Marrero JA and Lok AS: Newer markers for
hepatocellular carcinoma. Gastroenterology. 127 5 Suppl
1:S113–S119. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Heneghan HM, Miller N and Kerin MJ: MiRNAs
as biomarkers and therapeutic targets in cancer. Curr Opin
Pharmacol. 10:543–550. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang J, Du Y, Liu X, Cho WC and Yang Y:
MicroRNAs as regulator of signaling networks in metastatic colon
cancer. Biomed Res Int. 2015:8236202015.PubMed/NCBI
|
|
28
|
Zhang Q, Yu N and Lee C: Vicious cycle of
TGF-β signaling in tumor progression and metastasis. Am J Clin Exp
Urol. 2:149–155. 2014.PubMed/NCBI
|
|
29
|
Drabsch Y and ten Dijke P: TGF-β
signalling and its role in cancer progression and metastasis.
Cancer Metastasis Rev. 31:553–568. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Matsuzaki K, Seki T and Okazaki K: TGF-β
signal shifting between tumor suppression and fibro-carcinogenesis
in human chronic liver diseases. J Gastroenterol. 49:971–981. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Inamoto S, Itatani Y, Yamamoto T,
Minamiguchi S, Hirai H, Iwamoto M, Hasegawa S, Taketo MM, Sakai Y
and Kawada K: Loss of SMAD4 promotes colorectal cancer progression
by accumulation of myeloid-derived suppressor cells through the
CCL15-CCR1 Chemokine Axis. Clin Cancer Res. 22:492–501. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fleming NI, Jorissen RN, Mouradov D,
Christie M, Sakthianandeswaren A, Palmieri M, Day F, Li S, Tsui C,
Lipton L, et al: SMAD2, SMAD3 and SMAD4 mutations in colorectal
cancer. Cancer Res. 73:725–735. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Singh S, Narang AS and Mahato RI:
Subcellular fate and off-target effects of siRNA, shRNA and miRNA.
Pharm Res. 28:2996–3015. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jackson AL and Linsley PS: Recognizing and
avoiding siRNA off-target effects for target identification and
therapeutic application. Nat Rev Drug Discov. 9:57–67. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Serrano-Gomez SJ, Maziveyi M and Alahari
SK: Regulation of epithelial-mesenchymal transition through
epigenetic and post-translational modifications. Mol Cancer.
15:182016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Steinestel K, Eder S, Schrader AJ and
Steinestel J: Clinical significance of epithelial-mesenchymal
transition. Clin Transl Med. 3:172014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Saitoh M: Epithelial-mesenchymal
transition is regulated at post-transcriptional levels by
transforming growth factor-β signaling during tumor progression.
Cancer Sci. 106:481–488. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Katsuno Y, Lamouille S and Derynck R:
TGF-β signaling and epithelial-mesenchymal transition in cancer
progression. Curr Opin Oncol. 25:76–84. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shah PP and Kakar SS: Pituitary tumor
transforming gene induces epithelial to mesenchymal transition by
regulation of Twist, Snail, Slug, and E-cadherin. Cancer Lett.
311:66–76. 2011. View Article : Google Scholar : PubMed/NCBI
|