Introduction
Kidney cancer is the ninth most frequent type of
cancer in men and fourteenth in women worldwide; the incidence of
kidney cancer is increasing throughout the world amongst all age
groups and races (1). Kidney cancer
was the cause of ~143,000 mortalities in 2012 worldwide, among
which, clear cell renal cell carcinoma (ccRCC) accounted for ~70%
(2). Numerous studies have been
performed to explore the pathogenesis of ccRCC, and many genes were
identified as having an association with the tumorigenesis of ccRCC
(3–5).
For example, a previous study reported that Von Hippel-Lindau (VHL)
loss induced gene expression changes that were independent on
hypoxia inducible factor (HIF) and were responsible for the
development of renal cancer (6). It
was demonstrated that drugs targeting the pVHL-HIF-vascular
endothelial growth factor (VEGF) pathway had been applied in the
clinic and proven to have superiority over cytokine therapies
(7). However, the VHL mutant alone
was inadequate for ccRCC development (8).
In recent decades, high-throughput technology,
including microarray analysis and RNA sequencing, have provided
researchers with large expression data sets. Bioinformatics and
computational techniques have been well applied in the studies of
various tumors, and confirmed to be efficient and reliable in
identifying novel tumor markers for cancer diagnosis and targeted
treatments (9). In the present study,
the microarray data (GSE15641), containing 32 ccRCC samples and 23
normal kidney samples, was selected and analyzed by a series of
bioinformatics analyses. Furthermore, the results were verified by
OncoLnc, Gene Expression Profiling Interactive Analysis (GEPIA) and
the Human Protein Atlas database data. The purpose of the present
study was to identify potential target genes that may serve a role
in ccRCC development.
Materials and methods
Microarray data
Gene-Cloud of Biotechnology Information (GCBI;
Shanghai, China) is a powerful platform that provides services,
including molecular medical information solutions, platform data
services and cloud genetic analysis (www.gcbi.com.cn). It contains 120 million copies of
genomic samples, ~90,000 tumor samples and as much as 17 million
copies of genetic information. Additionally, the GCBI tool can be
used to retrieve and analyze data from The National Center for
Biotechnology Information (10,11). The
gene expression profiles of GSE15641, which were obtained based on
the platform of GPL96, Affymetrix Human Genome U133A Array, were
screened on GCBI to conduct the subsequent analysis. This dataset
contained 92 samples, including 32 ccRCC tumors, 11 papillary RCC
kidney tumors, 6 chromophobe RCC kidney tumors, 20 non-RCC renal
tumors and 23 normal kidney samples. The 32 ccRCC samples and 23
normal kidney controls were selected to perform the analysis in the
current study.
Identification of differentially
expressed genes (DEGs)
The GCBI online laboratory provides seven function
modules, including sample grouping, differential expression
analysis, gene ontology (GO) function analysis, Kyoto Encyclopedia
of Genes and Genomes (KEGG) pathway enrichment analysis, and
network analysis (pathway relation network, for example). Sample
data were uploaded to GCBI online laboratory to conduct the
subsequent analysis and DEGs with a fold change ≥2 and a false
discovery rate (FDR) <0.05 were selected. The hierarchical
clustering were implemented using two methods: Unweighted
pair-group method with arithmetic averages (12) and Pearson correlation.
Function and pathway enrichment
analysis
The function and signaling pathway analysis of the
selected DEGs were performed on the GCBI online laboratory using
its GO function and KEGG pathway enrichment analysis modules.
P<0.05 was set in the aforementioned analyses and the results
were visualized using two maps via the ggplot2 R package
(https://CRAN.R-project.org/package=ggplot2) (13). GO/pathway analysis was performed using
Fisher's exact test and represented in a 2×2 contingency table
(Table I). To correct errors
following multiple comparison analysis, the Benjamini-Hochberg
step-up method was used to control the FDR.
 | Table I.2×2 contingency table of Fisher's
exact test in the GO/pathway analysis. |
Table I.
2×2 contingency table of Fisher's
exact test in the GO/pathway analysis.
| Genes | DEGs | Non-DEGs | Total |
|---|
| Genes in the | nf | nf-nf | n |
| GO/pathway |
|
|
|
| Genes out of
the | Nf-nf |
N-Nf-(nf-nf) | N-n |
| GO/pathway |
|
|
|
| Total | Nf | N-Nf | N |
Protein-protein interaction (PPI)
network and pathway association network
Search Tool for the Retrieval of Interacting Genes
(STRING, www.string-db.org/) is a database of
known and predicted protein interactions, and provides a platform
for users to evaluate the PPI information freely (14). To evaluate the interactive
associations among DEGs, these were mapped to STRING, and only the
interactions with a combined score >0.4 were selected.
Subsequently, PPI networks were constructed using Cytoscape v3.4.0
software (cytoscape.org/) (15). The pathway association network was
performed on GCBI using its network analysis module.
Hub gene selection and validation
The most significant module of PPI network was
discriminated using the Molecular Complex Detection (MCODE) plug-in
with the MCODE score >3 and number of nodes >4 (16). Genes in this module were considered as
hub genes. Subsequently, transcriptional level analysis on the
GEPIA database (gepia.cancer-pku.cn/index.html) (17), translational level analysis on the
Human protein atlas database (www.proteinatlas.org/) (18) and survival analysis on the OncoLnc
database (www.oncolnc.org/) (19) of hub genes were performed to verify
the present results.
Results
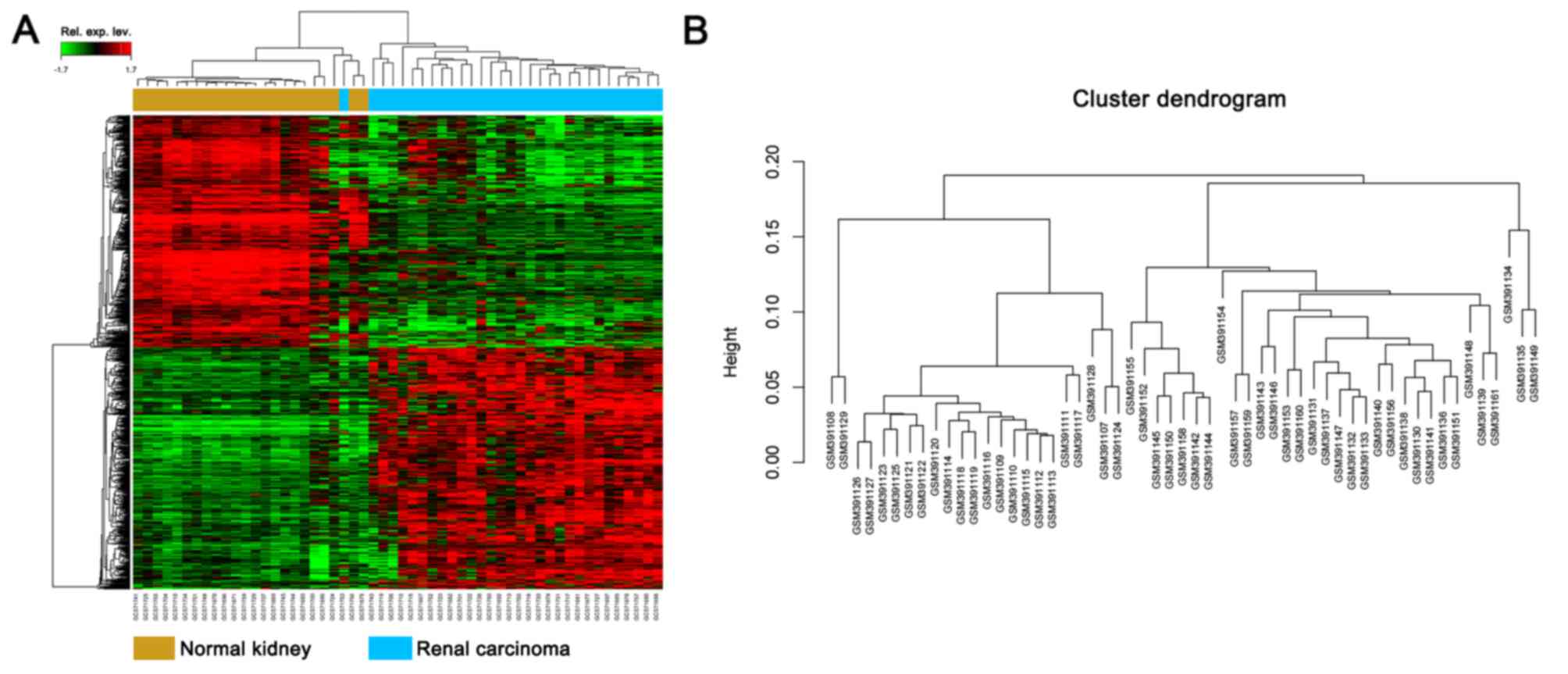
Identification of DEGs
Under the threshold of FDR <0.05 and fold change
≥2, a total of 805 genes were identified, including 403 up- and 402
downregulated genes. DEG expression heat map (top 50 up- and
downregulated genes) and sample cluster analysis are presented in
Fig. 1.
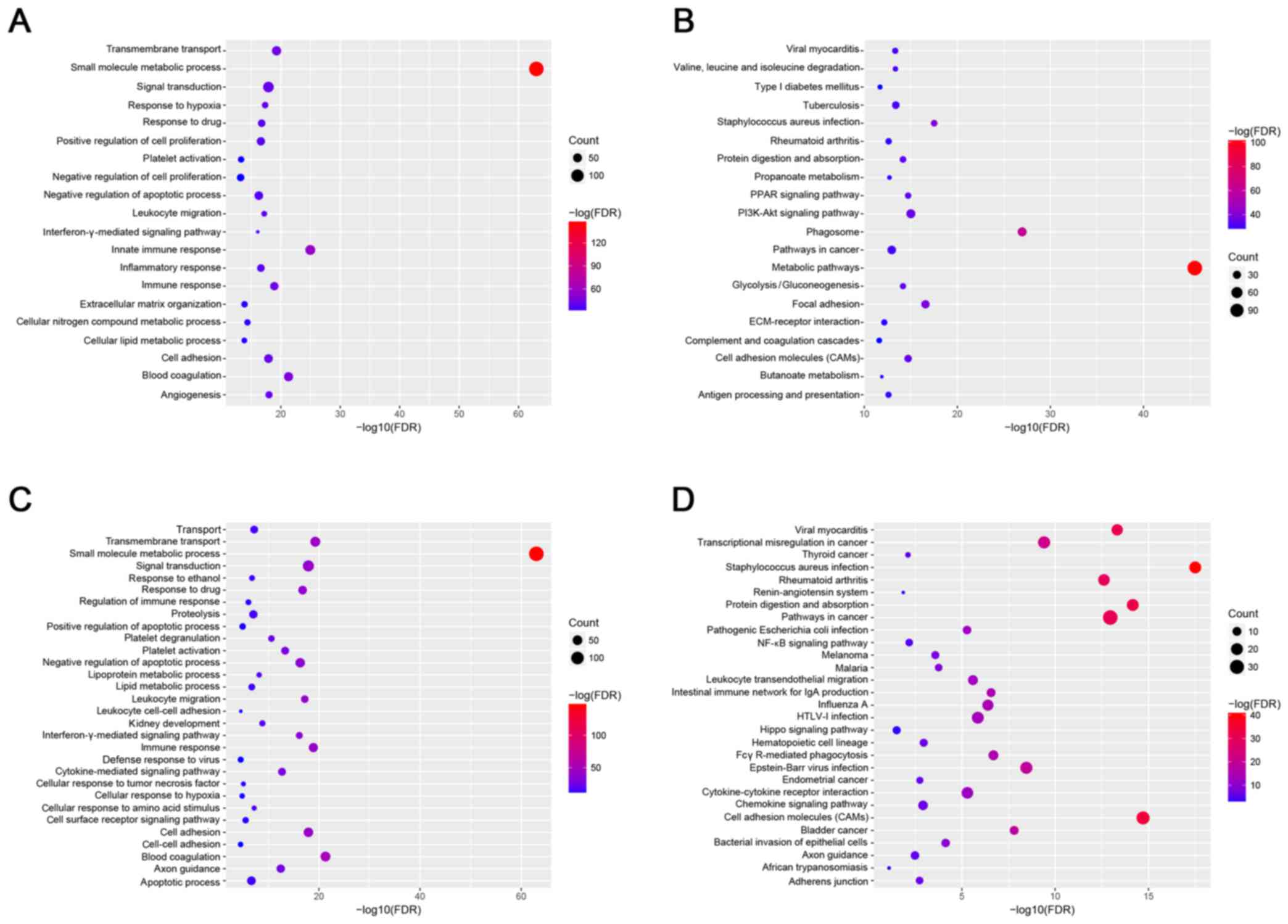
Functional and pathway enrichment
analysis
The top 20 GO and KEGG terms are listed in Fig. 2. As illustrated, the small molecule
metabolic process and the metabolic pathway were significantly
enriched. In addition, analysis of the association between the hub
genes, and GO and KEGG terms was also performed. GO function
analysis demonstrated small molecule metabolic processes, as well
as pathways involved in viral myocarditis, staphylococcus
infection, rheumatoid arthritis, protein digestion absorption,
‘pathways in cancer’ (KEGG map no. 05200, referring to signaling
pathways associated with tumorigenesis) and cell adhesion molecules
were also significantly enriched. Functional and pathway enrichment
analysis models of GCBI were performed based on all DEGs.
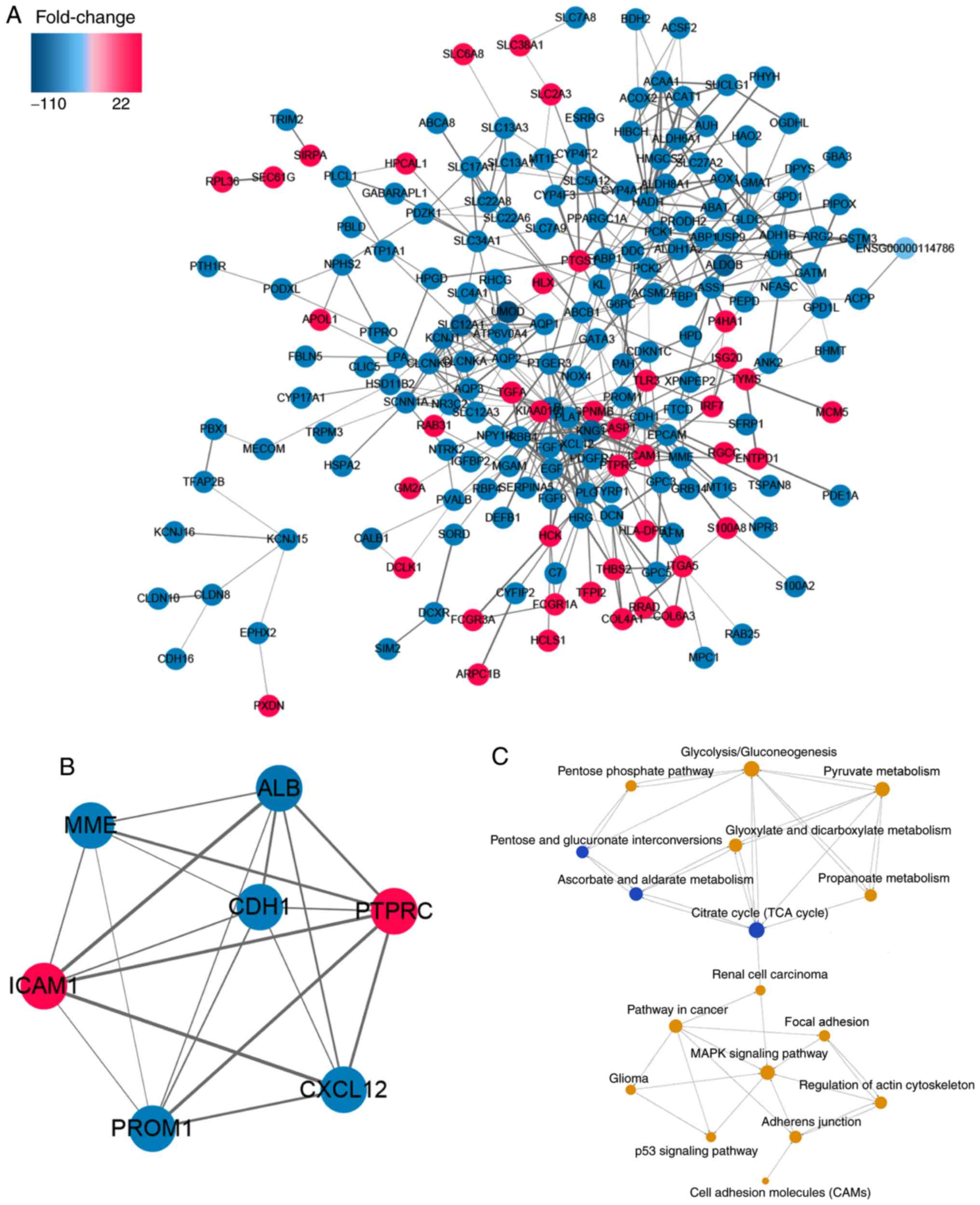
PPI network and pathway relation
network
Based on the STRING database, the PPI network was
constructed using Cytoscape software (Fig. 3A). The most significant module with a
MCODE score of 6.667 is illustrated in Fig. 3B. Pathway relation network (Fig. 3C) demonstrated that the
mitogen-activated protein kinase (MAPK) signaling pathway and KEGG
map no. 05200, ‘pathways in cancer’ were associated with RCC and
that energy metabolism (centered on the citrate cycle) served a
crucial role in RCC development.
Hub gene and validation
The most significant module with a MCODE score of
6.667 is illustrated in Fig. 3B.
There were 5 downregulated hub genes, including membrane
metallo-endopeptidase (MME), albumin (ALB), cadherin
1 (CDH1), prominin 1 (ROM1), chemokine (C-X-C
motif) ligand 12 (CXCL12), and 2 upregulated hub genes
including protein tyrosine phosphatase receptor type C
(PTPRC), intercellular adhesion molecule 1 (ICAM1) in
this module. The mRNA and protein expression levels of the 5
downregulated hub genes are demonstrated in Fig. 4, and that of the 2 upregulated hub
genes are shown in Fig. 5. As
suggested by Figs. 4 and 5, MME, CXCL12, CDH1, ALB and
PROM1 expression levels were significantly lower in ccRCC
tissue compared with normal controls, whereas ICAM1 and
PTPRC expression levels were significantly higher
(calculated by one-way ANOVA test). Survival analysis of the hub
genes is presented in Fig. 6,
indicating that altered MME, CDH1 and ICAM1 were
associated with the prognosis of ccRCC. However, the prognosis
value of CXCL12, PROM1, ALB and PTPRC had no
statistical significance.
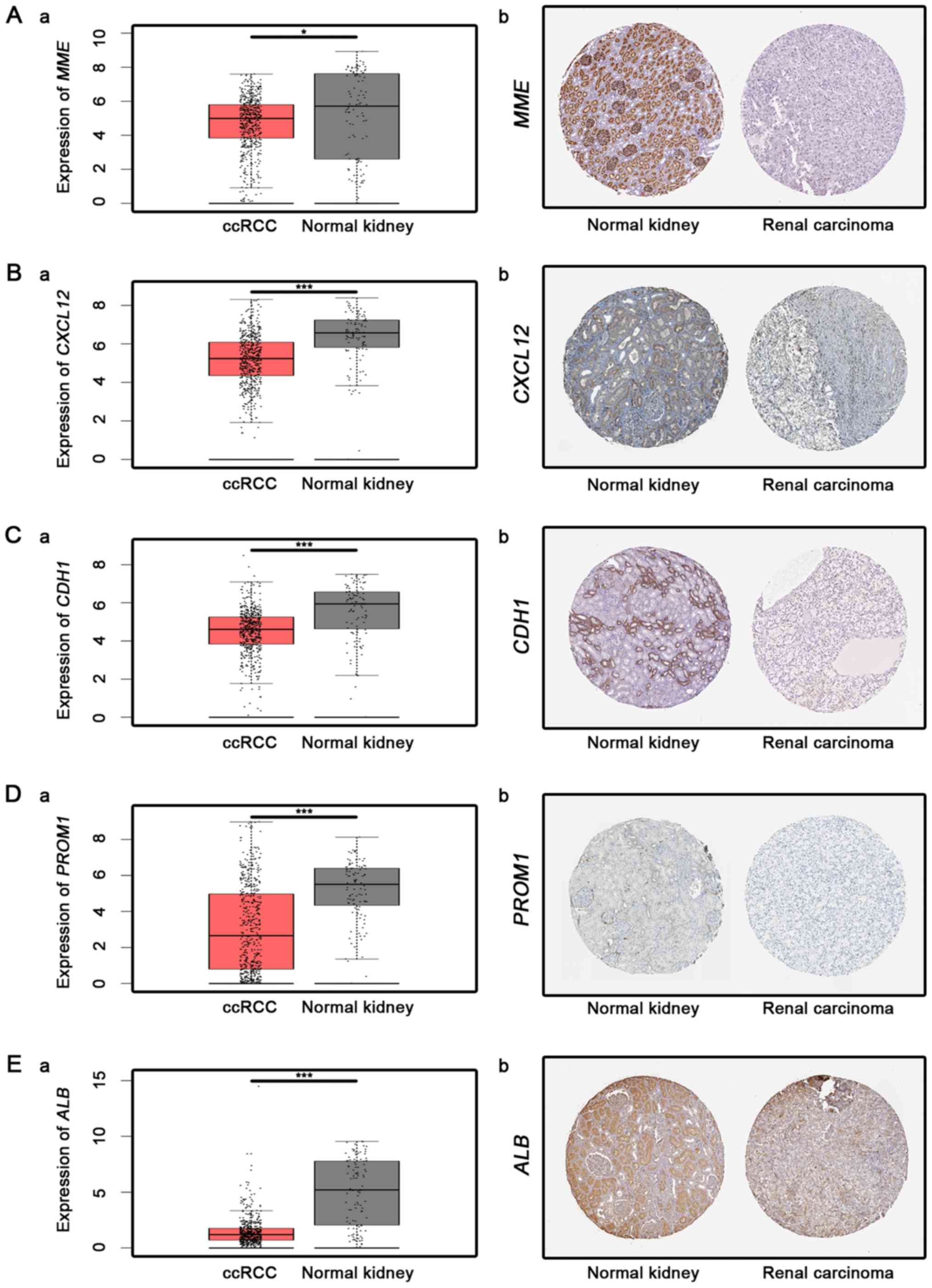 | Figure 4.Analysis of downregulated hub genes.
Analysis of 5 downregulated hub genes in ccRCC tissues and normal
kidney tissues as follows: (A) MME, (B) CXCL12, (C)
CDH1, (D) PROM1 and (E) ALB. a,
Transcriptional level (from GEPIA database, one-way ANOVA was used
for differential analysis) and b, immunohistochemistry images
obtained from translational level analyses of the Human Protein
Atlas database, for which no scale bar is available. *P<0.05 and
***P<0.001. ALB, albumin; ccRCC, clear cell renal cell
carcinoma; CDH1, cadherin 1; CXCL12, chemokine (C-X-C
motif) ligand 12; MME, metallo-endopeptidase; ROM1,
prominin 1. |
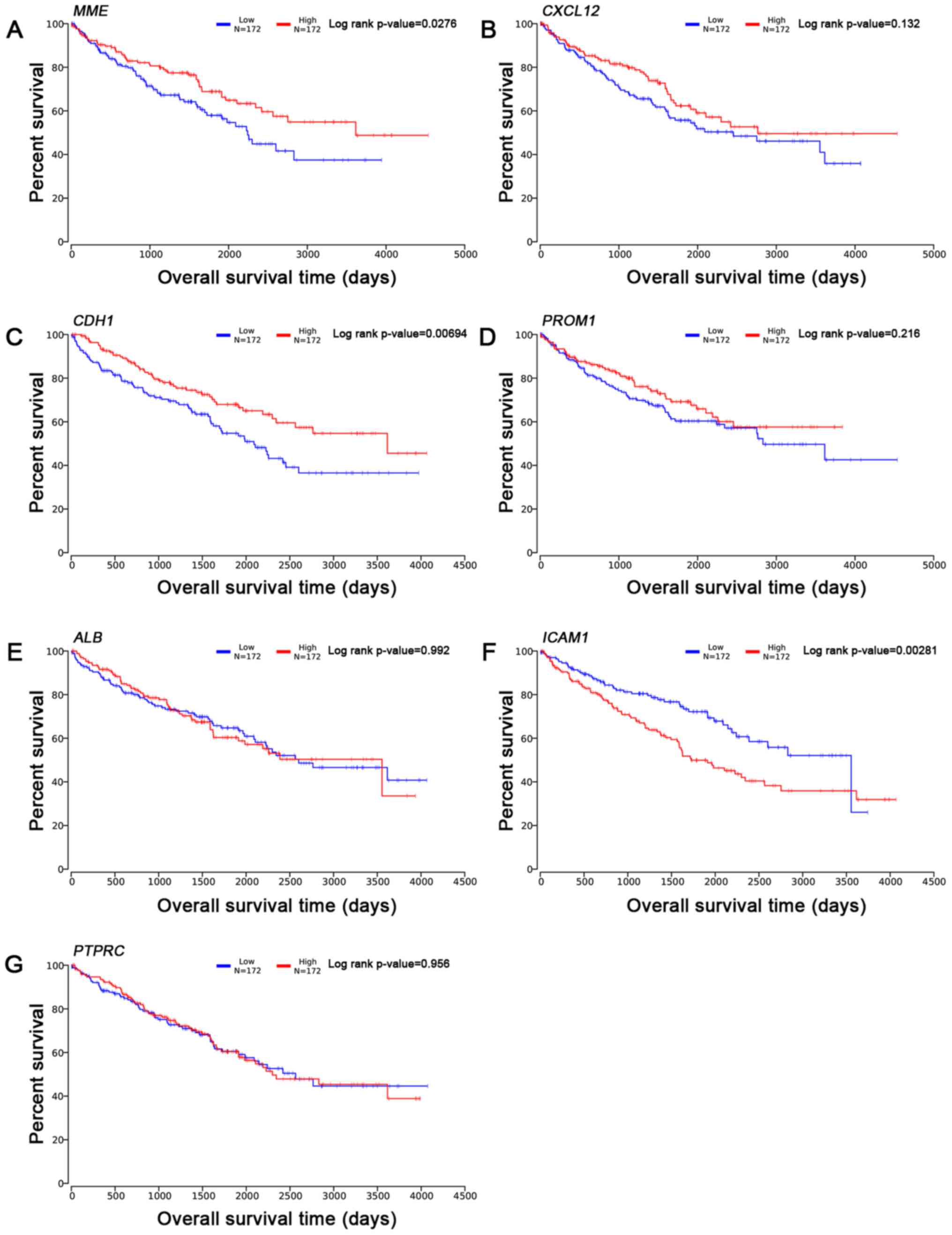 | Figure 6.Overall survival time analysis (from
OncoLnc database). Overall survival time analysis of 7 hub genes as
follows: (A) MME, (B) CXCL12, (C) CDH1, (D)
PROM1, (E) ALB, (F) ICAM1 and (G)
PTPRC. ALB, albumin; CDH1, cadherin 1;
CXCL12, chemokine (C-X-C motif) ligand 12; MME,
metallo-endopeptidase; ROM1, prominin 1; PTPRC,
protein tyrosine phosphatase receptor type C; ICAM1,
intercellular adhesion molecule 1. |
Discussion
In the present study, a total of 805 DEGs including
403 up- and 402 downregulated genes were selected. The present
study demonstrated that the most significant functions and pathways
identified were associated with biological metabolism. Previous
studies have indicated that molecular metabolisms served a
comprehensive role in the process of tumor generation and
development (20,21). It is known that HIF and the hypoxic
response serve a key role in the pathways that are involved in
tumorigenesis (22). In addition to
hypoxia, increased reactive oxygen species (ROS) production is
another aberrant metabolic condition encountered by the majority of
tumor cells (23), the balance
between this oxidative stress and antioxidant systems has important
effects at different stages of tumorigenesis, particularly in the
initial stage (24). Metabolic
disturbances, including ROS, are observed in normal physiological
processes and in pathologies across a range of tissues, including
in cancerous tissues; thus, it is considered that almost all core
metabolic pathways are associated with cancer development (25). In addition, previous studies have
reported that carbon metabolism and antioxidant response are
associated with the development of ccRCC (26–28).
The present study demonstrated that the MAPK
signaling pathway was associated with ccRCC. It is known that MAPK
signaling pathways serve essential roles in cell proliferation and
differentiation: Previous studies have demonstrated its activation
in tumorigenesis and the metastasis of multiple human malignancies,
including RCC (29,30). Huang et al (31) also proved this phenomenon in ccRCC,
and its inhibition using anthrax lethal toxin was able to suppress
ccRCC growth in vivo.
A PPI network with DEGs was constructed and listed
the top degree hub genes including the following: MME, ALB,
CXCL12, CDH1, PROM1, ICAM1, and PTPRC. MME, a
downregulated gene in the present study, encodes a glycoprotein
identified in a variety of normal and malignant tissues. This
glycoprotein is particularly abundant in kidney, where it is
present on the brush border of proximal tubules and on glomerular
epithelium (32). Amălinei et
al (33) reported that the
generation of angiostatin induced by MME was responsible for
the prevention of tumor growth. A previous study also demonstrated
that among children with B-lineage acute lymphoblastic leukemia,
MME(+) infants tend to have a better prognosis than those with
MME(−) infants (34). The
aforementioned study, combined with the survival curve and
immunohistochemistry staining results in the current study,
indicate that MME may be a tumor suppressor gene in ccRCC.
The second hub gene was ALB and it has been reported that
low serum albumin level indicate a shorter progression-free
survival time in patients with metastatic RCC (35). Although the present analysis indicated
that ALB may serve a suppressive role in the ccRCC
development, this was not confirmed by the survival analysis.
CXCL12, the third hub gene, encodes a stromal cell-derived α
chemokine that serve a role in tumor growth and metastasis
(36). CXCL12 was identified
as a downregulated gene in the present analysis. Furthermore, Ping
et al (37) reported that
decreased CXCL12 in the tumor microenvironment may
facilitate lymphoid malignant cells metastasis by limiting the
interactions between malignant cells and surrounding cells in
lymphocytic leukemia. CDH1, as a member of the cadherin
superfamily, may serve a critical role in apoptotic process and
cell-cell adhesion (38). A previous
study demonstrated that CDH1 expression was lower in gastric
cancer and this decrease contributed to intestinal-type gastric
carcinogenesis (39). Another study
reported that attenuated expression of CDH1 had a key role
in tumor invasion and metastasis (40). PROM1 was another downregulated
gene in the present study, which encodes a pentaspan transmembrane
glycoprotein. D'Alterio et al (41) reported PROM1 expression was low
in ccRCC. PROM1 has been observed to contribute to cell
differentiation in numerous tissue types, including glomerular
visceral (42) and tubule epithelial
(43). However, the biological
function of PROM1 remains unclear. In RCC, PROM1
progenitor cells were able to differentiate into endothelial cells
enhancing vascularization and tumor growth, which consequently
promoted the development of RCC (44,45).
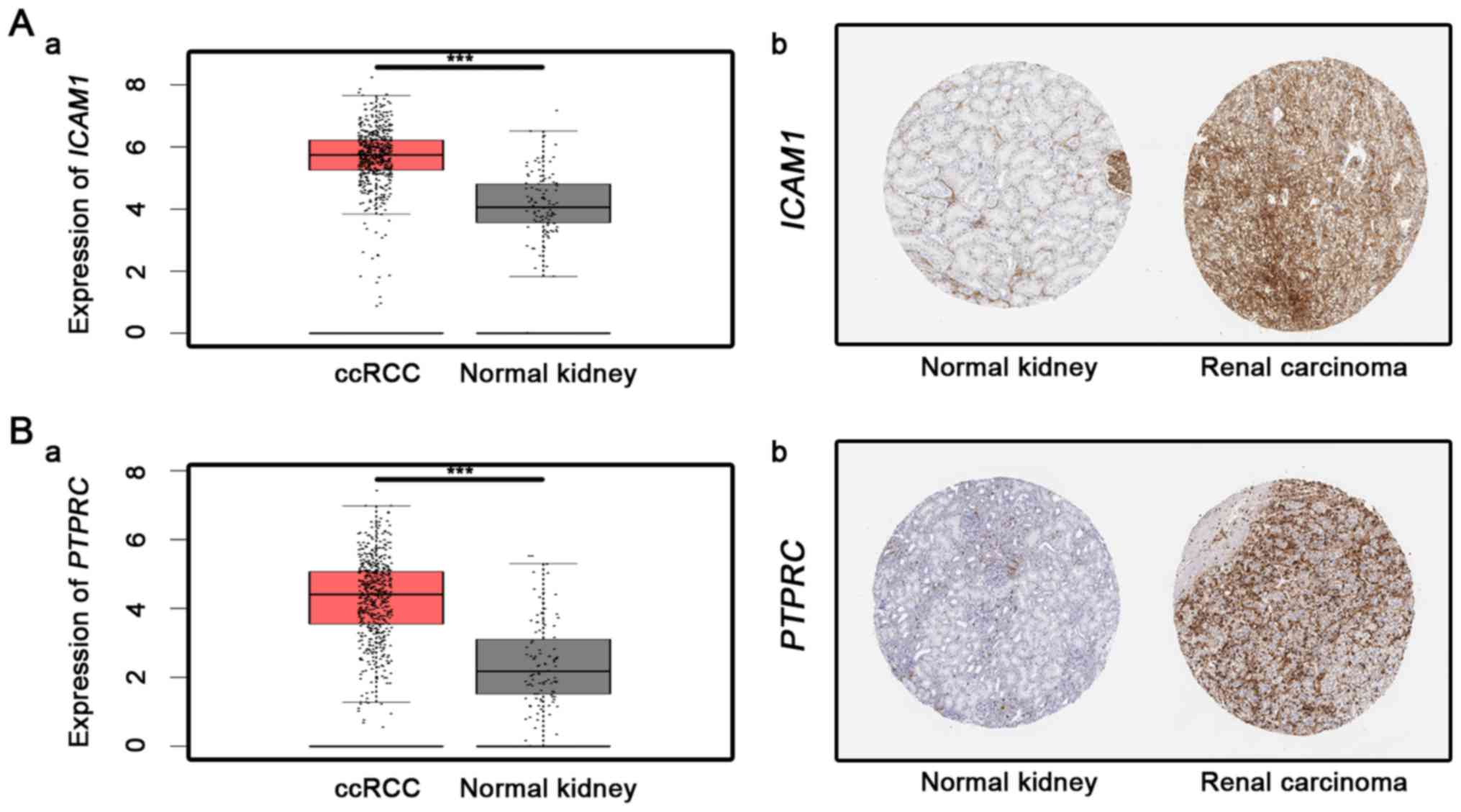
ICAM1 was the most significantly upregulated
hub genes in the present analysis and is a cell adhesion molecule
of the immunoglobulin superfamily. ICAM-1 expression has
been reported to be upregulated in several cancer types, including
thyroid carcinoma, hepatocellular carcinoma, oral cancer, RCC and
bladder cancer (46–50). In ccRCC, ICAM1 expression was
upregulated following treatment with a series of cytokines,
including tumor necrosis factor α, interferon-γ and phorbol
myristate acetate (51). Furthermore,
as an independent predictor for the prognosis of ccRCC, its high
expression in tumor cells indicates a shorter survival time
(52). The results of studies are in
accordance with the present analysis. PTPRC was another
upregulated hub gene, encoding a member of the PTP family (53). PTPs are known to be signaling
molecules that regulate a variety of cellular processes, including
cell growth, differentiation, mitosis and oncogenic transformation
(54). Clark et al (55) reported that galectin-3 served an
anti-apoptotic role in diffuse large B-cell lymphoma via binding to
PTPRC product.
Although our study shows that the seven Hub genes
were associated with the occurrence or development of ccRCC, the
survival analysis by OncoLnc database showed that altered MME,
CDH1 and ICAM1 were associated with the prognosis of ccRCC, but
CXCL12, PROM1, ALB and PTPRC were not. On one hand,
the prognosis might not be affected by the differentially expressed
hub genes (CXCL12, PROM1, ALB and PTPRC). On the
other hand, they might involve in the carcinogenesis of ccRCC
through the interconnection with other genes, and might not be
sufficient as an independent risk factor in the progression of
ccRCC.
In conclusion, the present study used various
bioinformatics analysis tools to identify 7 novel hub genes
(MME, ALB, CXCL12, CDH1, PROM1, ICAM1 and PTPRC),
which may serve key roles in the tumorigenesis of human ccRCC.
These genes may serve as novel biomarkers of ccRCC. However, the
lack of in vivo and in vitro experiments is a
limitation of the present study, further experiments are required
to confirm the present findings, and confirm the role of these
candidate genes in ccRCC.
Acknowledgements
The authors would like to thank Ms. Shanshan Zhang
and Ms. Danni Shan (Zhongnan Hospital of Wuhan University, Wuhan,
China) for their technical assistance.
Funding
The present study was supported in part by the
Zhongnan Hospital of Wuhan University Science, Technology and
Innovation Seed Fund (grant no. cxpy20160010) and Natural Sciences
Foundation of Hubei Province (grant no. 2014CFA006). The funders
had no role in the design of the study, the collection, analysis,
and interpretation of data, or in writing the manuscript.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
JW, LY, YX and XW conceived and designed the study.
JW, LY, XL, GW and YX collected the data and performed the
analysis. JW, LY, YZ, KQ. and YX analyzed the results. JW, YX and
XW contributed analysis tools. JW, LY, YX and XW contributed to the
writing of the manuscript. All authors reviewed the manuscript.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sanfilippo KM, McTigue KM, Fidler CJ,
Neaton JD, Chang Y, Fried LF, Liu S and Kuller LH: Hypertension and
obesity and the risk of kidney cancer in 2 large cohorts of US men
and women. Hypertension. 63:934–941. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Frew IJ and Moch H: A clearer view of the
molecular complexity of clear cell renal cell carcinoma. Annu Rev
Pathol. 10:263–289. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Papale M, Vocino G, Lucarelli G,
Rutigliano M, Gigante M, Rocchetti MT, Pesce F, Sanguedolce F, Bufo
P, Battaglia M, et al: Urinary RKIP/p-RKIP is a potential
diagnostic and prognostic marker of clear cell renal cell
carcinoma. Oncotarget. 8:40412–40424. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Neely BA, Wilkins CE, Marlow LA,
Malyarenko D, Kim Y, Ignatchenko A, Sasinowska H, Sasinowski M,
Nyalwidhe JO, Kislinger T, et al: Proteotranscriptomic analysis
reveals stage specific changes in the molecular landscape of
Clear-cell renal cell carcinoma. PLoS One. 11:e01540742016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gowrishankar B, Ibragimova I, Zhou Y,
Slifker MJ, Devarajan K, Al-Saleem T, Uzzo RG and Cairns P:
MicroRNA expression signatures of stage, grade, and progression in
clear cell RCC. Cancer Biol Ther. 15:329–341. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gossage L, Eisen T and Maher ER: VHL, the
story of a tumour suppressor gene. Nat Rev Cancer. 15:55–64. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Escudier B, Szczylik C, Porta C and Gore
M: Treatment selection in metastatic renal cell carcinoma: Expert
consensus. Nat Rev Clin Oncol. 9:327–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mandriota SJ, Turner KJ, Davies DR, Murray
PG, Morgan NV, Sowter HM, Wykoff CC, Maher ER, Harris AL, Ratcliffe
PJ, et al: HIF activation identifies early lesions in VHL kidneys:
Evidence for site-specific tumor suppressor function in the
nephron. Cancer Cell. 1:459–468. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kulasingam V and Diamandis EP: Strategies
for discovering novel cancer biomarkers through utilization of
emerging technologies. Nat Clin Pract Oncol. 5:588–599. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang M, Chen BL, Huang JB, Meng YN, Duan
XJ, Chen L, Li LR and Chen YP: Angiogenesis-related genes may be a
more important factor than matrix metalloproteinases in
bronchopulmonary dysplasia development. Oncotarget. 8:18670–18679.
2017.PubMed/NCBI
|
|
11
|
Xiao J, Liu A, Lu X, Chen X, Li W, He S,
He B and Chen Q: Prognostic significance of TCF21 mRNA expression
in patients with lung adenocarcinoma. Sci Rep. 7:20272017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Peterson LE: CLUSFAVOR 5.0: Hierarchical
cluster and principal-component analysis of microarray-based
transcriptional profiles. Genome Biol. 3:SOFTWARE00022002.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ito K and Murphy D: Application of ggplot2
to pharmacometric graphics. CPT Pharmacometrics Syst Pharmacol.
2:e792013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Szklarczyk D, Franceschini A, Kuhn M,
Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork
P, et al: The STRING database in 2011: Functional interaction
networks of proteins, globally integrated and scored. Nucleic Acids
Res. 39:(Database Issue). D561–D568. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mutwil M, Usadel B, Schütte M, Loraine A,
Ebenhöh O and Persson S: Assembly of an interactive correlation
network for the Arabidopsis genome using a novel heuristic
clustering algorithm. Plant Physiol. 152:29–43. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Interactive human protein atlas launches.
Cancer Discov. 5:3392015. View Article : Google Scholar
|
|
19
|
Anaya J: OncoLnc: Linking TCGA survival
data to mRNAs, miRNAs, and lncRNAs. Peer J Computer Sci. 2:e672016.
View Article : Google Scholar
|
|
20
|
Tian Y, Du W, Cao S, Wu Y, Dong N, Wang Y
and Xu Y: Systematic analyses of glutamine and glutamate
metabolisms across different cancer types. Chin J Cancer.
36:882017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Meng X, Zhong J, Liu S, Murray M and
Gonzalez-Angulo AM: A new hypothesis for the cancer mechanism.
Cancer Metastasis Rev. 31:247–268. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mimeault M and Batra SK: Hypoxia-inducing
factors as master regulators of stemness properties and altered
metabolism of cancer- and metastasis-initiating cells. J Cell Mol
Med. 17:30–54. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gorrini C, Harris IS and Mak TW:
Modulation of oxidative stress as an anticancer strategy. Nat Rev
Drug Discov. 12:931–947. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Harris IS, Treloar AE, Inoue S, Sasaki M,
Gorrini C, Lee KC, Yung KY, Brenner D, Knobbe-Thomsen CB, Cox MA,
et al: Glutathione and thioredoxin antioxidant pathways synergize
to drive cancer initiation and progression. Cancer Cell.
27:211–222. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cairns RA and Mak TW: Fire and water:
Tumor cell adaptation to metabolic conditions. Exp Cell Res.
356:204–208. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hakimi AA, Reznik E, Lee CH, Creighton CJ,
Brannon AR, Luna A, Aksoy BA, Liu EM, Shen R, Lee W, et al: An
integrated metabolic atlas of clear cell renal cell carcinoma.
Cancer Cell. 29:104–116. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li B, Qiu B, Lee DS, Walton ZE, Ochocki
JD, Mathew LK, Mancuso A, Gade TP, Keith B, Nissim I, et al:
Fructose-1,6-bisphosphatase opposes renal carcinoma progression.
Nature. 513:251–255. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sudarshan S, Karam JA, Brugarolas J,
Thompson RH, Uzzo R, Rini B, Margulis V, Patard JJ, Escudier B and
Linehan WM: Metabolism of kidney cancer: From the lab to clinical
practice. Eur Urol. 63:244–251. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chang L and Karin M: Mammalian MAP kinase
signalling cascades. Nature. 410:37–40. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Johnson GL and Lapadat R:
Mitogen-activated protein kinase pathways mediated by ERK, JNK, and
p38 protein kinases. Science. 298:1911–1912. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huang D, Ding Y, Luo WM, Bender S, Qian
CN, Kort E, Zhang ZF, VandenBeldt K, Duesbery NS, Resau JH, et al:
Inhibition of MAPK kinase signaling pathways suppressed renal cell
carcinoma growth and angiogenesis in vivo. Cancer Res. 68:81–88.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Noordmans GA, Caputo CR, Huang Y, Sheehan
SM, Bulthuis M, Heeringa P, Hillebrands JL, van Goor H and
Korstanje R: Genetic analysis of mesangial matrix expansion in
aging mice and identification of Far2 as a candidate gene. J Am Soc
Nephrol. 24:1995–2001. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Amălinei C, Căruntu ID, Giuscă SE and
Bălan RA: Matrix metalloproteinases involvement in pathologic
conditions. Rom J Morphol Embryol. 51:215–228. 2010.PubMed/NCBI
|
|
34
|
Maguer-Satta V, Besancon R and
Bachelard-Cascales E: Concise review: Neutral endopeptidase (CD10):
A multifaceted environment actor in stem cells, physiological
mechanisms, and cancer. Stem Cells. 29:389–396. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Stenman M, Laurell A and Lindskog M:
Prognostic significance of serum albumin in patients with
metastatic renal cell carcinoma. Med Oncol. 31:8412014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ray P, Lewin SA, Mihalko LA, Lesher-Perez
SC, Takayama S, Luker KE and Luker GD: Secreted CXCL12 (SDF-1)
forms dimers under physiological conditions. Biochem J.
442:433–442. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ping L, Ding N, Shi Y, Feng L, Li J, Liu
Y, Lin Y, Shi C, Wang X, Pan Z, et al: The Bruton's tyrosine kinase
inhibitor ibrutinib exerts immunomodulatory effects through
regulation of tumor-infiltrating macrophages. Oncotarget.
8:39218–39229. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen I, Mathews-Greiner L, Li D,
Abisoye-Ogunniyan A, Ray S, Bian Y, Shukla V, Zhang X, Guha R,
Thomas C, et al: Transcriptomic profiling and quantitative
high-throughput (qHTS) drug screening of CDH1 deficient hereditary
diffuse gastric cancer (HDGC) cells identify treatment leads for
familial gastric cancer. J Transl Med. 15:922017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Khouzam RA, Molinari C, Salvi S, Marabelli
M, Molinaro V, Orioli D, Saragoni L, Morgagni P, Calistri D and
Ranzani GN: Digital PCR identifies changes in CDH1 (E-cadherin)
transcription pattern in intestinal-type gastric cancer.
Oncotarget. 8:18811–18820. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
van Roy F and Berx G: The cell-cell
adhesion molecule E-cadherin. Cell Mol Life Sci. 65:3756–3788.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
D'Alterio C, Cindolo L, Portella L,
Polimeno M, Consales C, Riccio A, Cioffi M, Franco R, Chiodini P,
Carteni G, et al: Differential role of CD133 and CXCR4 in renal
cell carcinoma. Cell Cycle. 9:4492–4500. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ronconi E, Sagrinati C, Angelotti ML,
Lazzeri E, Mazzinghi B, Ballerini L, Parente E, Becherucci F, Gacci
M, Carini M, et al: Regeneration of glomerular podocytes by human
renal progenitors. J Am Soc Nephrol. 20:322–332. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sagrinati C, Netti GS, Mazzinghi B,
Lazzeri E, Liotta F, Frosali F, Ronconi E, Meini C, Gacci M,
Squecco R, et al: Isolation and characterization of multipotent
progenitor cells from the Bowman's capsule of adult human kidneys.
J Am Soc Nephrol. 17:2443–2456. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bruno S, Bussolati B, Grange C, Collino F,
Graziano ME, Ferrando U and Gamussi G: CD133+ renal progenitor
cells contribute to tumor angiogenesis. Am J Pathol. 169:2223–2235.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bussolati B, Bruno S, Grange C,
Buttiglieri S, Deregibus MC, Cantino D and Camussi G: Isolation of
renal progenitor cells from adult human kidney. Am J Pathol.
166:545–555. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Roland CL, Harken AH, Sarr MG and Barnett
CC Jr: ICAM-1 expression determines malignant potential of cancer.
Surgery. 141:705–707. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Usami Y, Ishida K, Sato S, Kishino M,
Kiryu M, Ogawa Y, Okura M, Fukuda Y and Toyosawa S: Intercellular
adhesion molecule-1 (ICAM-1) expression correlates with oral cancer
progression and induces macrophage/cancer cell adhesion. Int J
Cancer. 133:568–578. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Buitrago D, Keutgen XM, Crowley M,
Filicori F, Aldailami H, Hoda R, Liu YF, Hoda RS, Scognamiglio T,
Jin M, et al: Intercellular adhesion molecule-1 (ICAM-1) is
upregulated in aggressive papillary thyroid carcinoma. Ann Surg
Oncol. 19:973–980. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tanabe K, Campbell SC, Alexander JP,
Steinbach F, Edinger MG, Tubbs RR, Novick AC and Klein EA:
Molecular regulation of intercellular adhesion molecule 1 (ICAM-1)
expression in renal cell carcinoma. Urol Res. 25:231–238. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Tanabe K, Alexander JP, Steinbach F,
Campbell S, Novick AC and Klein EA: Retroviral transduction of
intercellular adhesion molecule-1 enhances endothelial attachment
of bladder cancer. Urol Res. 25:401–405. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Tomita Y, Nishiyama T, Watanabe H,
Fujiwara M and Sato S: Expression of intercellular adhesion
molecule-1 (ICAM-1) on renal-cell cancer: Possible significance in
host immune responses. Int J Cancer. 46:1001–1006. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Shi X, Jiang J, Ye X, Liu Y, Wu Q and Wang
L: Prognostic prediction and diagnostic role of intercellular
adhesion molecule-1 (ICAM1) expression in clear cell renal cell
carcinoma. J Mol Histol. 45:427–434. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Williamson AJ, Pierce A, Jaworska E, Zhou
C, Aspinall-O'Dea M, Lancashire L, Unwin RD, Abraham SA, Walker MJ,
Cadecco S, et al: A specific PTPRC/CD45 phosphorylation event
governed by stem cell chemokine CXCL12 regulates primitive
hematopoietic cell motility. Mol Cell Proteomics. 12:3319–3329.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Stanford SM, Ahmed V, Barrios AM and
Bottini N: Cellular biochemistry methods for investigating protein
tyrosine phosphatases. Antioxid Redox Signal. 20:2160–2178. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Clark MC, Pang M, Hsu DK, Liu FT, de Vos
S, Gascoyne RD, Said J and Baum LG: Galectin-3 binds to CD45 on
diffuse large B-cell lymphoma cells to regulate susceptibility to
cell death. Blood. 120:4635–4644. 2012. View Article : Google Scholar : PubMed/NCBI
|