Introduction
Gliomas are a type of tumor formed by neoplastic
transformation of neural stem cells, progenitor cells and
differentiated glial cells, including astrocytes, oligodendrocytes
and ependymal cells (1). Neoplastic
cells may spread diffusely to normal brain tissues and damage
normal neurological functions, which is the reason why malignant
gliomas are detrimental to human health (2). In China, gliomas constitute 44.69% of
primary intracranial tumors and 1–3% of generalized malignancies
(3). According to the World Health
Organization, malignant glioma causes the second highest amount of
mortalities in sufferers <34 years old, and the third highest
amount of mortalities in sufferers aged 35–54 years old (4). It is estimated that the survival time of
the majority of glioma sufferers is ~1 year. Although there are
currently various treatments available, including excision,
chemotherapy and radiotherapy, the characteristic of strong
invasiveness has severely influenced the effectiveness of glioma
treatment.
MicroRNAs (miRNAs/miRs) are an important molecular
mediator of cell genetic changes, and are directly or indirectly
associated with the occurrence and development of a number of tumor
types, including glioma, when the miRNAs are abnormally expressed
in the tumors (5).
Human Tudor-staphylococcal nuclease (SN), also known
as P100, is a multi-functional protein with overexpression in
various malignant tumor types, including breast cancer, prostate
cancer, colorectal cancer and melanoma (6–10).
Previous studies have indicated that Tudor-SN has a close
association with lipid metabolism, and that the expression of
lipoprotein in liver cells can be affected through adjustment of
lipid metabolism-associated genes (11,12).
Inactivation of alkylglycerone phosphate synthase (AGPS) can lower
the ether ester level in the tumor cell and cancer pathogenicity,
while overexpression of AGPS can increase the ether ester level,
viability and migration potential of various tumor cells, including
231MFP, C8161 melanoma, PC3 prostate cancer and primary breast
cancer cells, and advance the growth and migration of the tumor
cells (13,14). The aforementioned studies demonstrated
that Tudor-SN and AGPS serve an important role in tumor
development.
In the present study, the role of Tudor-SN and AGPS
in the proliferation and migration of glioma U87MG cells and the
association of Tudor-SN with AGPS in this process was
investigated.
Materials and methods
Cell lines and cell culture
Human glioma U87 cells were purchased from the Cell
Bank of the Chinese Academy of Sciences (Shanghai, China) and
cultured in Dulbecco's modified Eagle's medium (Corning Life
Sciences, Manassas, VA, USA) with 10% fetal bovine serum (Corning
Life Sciences) at 37°C, with an atmosphere containing 5%
CO2. The AGPS and Tudor-SN silencing U87 cell line
[Tudor-SN short hairpin (shRNA) group] was established by the Basic
Medical College, Tianjin Medical University (Tianjin, China).
A total of 3×105 cells/well were seeded
onto a 6-well plate and cultured at 37°C for 24 h. A total of 2.5
µg AGPS shRNA plasmid, 2.5 µg Tudor-SN shRNA plasmid (Santa Cruz
Biotechnology, Inc., Dallas, TX, USA), 2.5 µg NF-κB p65 expression
plasmid (Santa Cruz Biotechnology, Inc.) and 2.5 µg miR-127 siRNA
plasmid (OBIO Biotechnology, Inc., Shanghai, China) were
transfected using GeneJuice® (Merck KGaA, Darmstadt,
Germany), according to the manufacturer's protocol, for another 6
h. Fresh Dulbecco's modified Eagle's medium (Corning Life Sciences,
Manassas, VA, USA) was then added and the cells were harvested
after 72 h for all experiments.
Cell proliferation assay
A total of 3,000 cells/well (negative control and
AGPS shRNA, n=5) were seeded into a 96-well plate and cultured at
37°C for 72 h. The BrdU cell proliferation kit (ab126556, Abcam,
Cambridge, UK) was used to determine the optical density value to
reflect the cell proliferation, according to the manufacturer's
instructions. Briefly, 20 µl of BrdU label (negative BrdU control
for determining assay background) was added at 37°C for 12 h, and
cells were fixed by 200 µl/well fixing solution (3.7% formaldehyde
in PBS) at room temperature for 30 min, then washed for 3 times
using PBS, and 100 µl/well anti-BrdU monoclonal detector antibody
(1:2,000, supplied in the BrdU cell proliferation kit) was added
and incubated for 1 h at room temperature. Then, they were washed 3
times using PBS, and 100 µl/well peroxidase-conjugated goat
anti-mouse IgG antibody (1:2,000, also supplied in the BrdU cell
proliferation kit) was added and incubated for 30 min at room
temperature. Subsequently the cells were washed 3 times using PBS,
and 100 µl/well TMB peroxidase substrate was added and incubated
for 30 min at room temperature in the dark. Finally, 100 µl of stop
solution (also supplied in the BrdU cell proliferation kit) was
added, and the OD value was measured every 24 h using the
Multiskan™ Spectrum at 450 nm (Thermo Fisher Scientific,
Inc., Waltham, MA, USA).
Cell migration assay
A total of 3×106 cells/well were seeded
into the insert of the Transwell kit (Cell Biolabs, Inc., San
Diego, CA, USA) with 200 µl 10% bovine serum albumin (BSA, Santa
Cruz Biotechnology, Inc., Santa Cruz, CA, USA) in the upper chamber
and 600 µl Dulbecco's modified Eagle's medium with 10% fetal bovine
serum, cultured at 37°C for 72 h, according to the manufacturer's
instructions. Non-migratory cells were then washed off using PBS,
and migratory cells were stained by 0.1% crystal violet at 37°C for
10 min and counted using a light microscope (Olympus Corporation,
Tokyo, Japan) to determine the cell migration (magnification,
×200).
Western blotting assay
Cells were co-transfected with AGPS shRNA plasmid,
and Tudor-SN shRNA plasmid, NF-κB p65 expression plasmid (retrieval
experiment) or miR-127 siRNA plasmid (retrieval experiment) in
order to explore the affect of Tudor-SN, NF-κB p65 and miR-127 on
the AGPS, p-mTOR and mTOR by western blotting assay.
A total of 3×105 cells/well were seeded
onto a 6-well plate and cultured for 24 h. Cells were lysed and
total proteins were extracted by centrifugation at 12,000 × g for
10 min at 4°C with protein extraction buffer (Bioo Scientific,
Austin, TX, USA). Protein was measured by Bradford assay (Beyotime
Institute of Biotechnology, Haimen, China) and 50 ng protein were
separated via 12% SDS-PAGE and transferred onto a polyvinylidene
fluoride membrane. The membrane was then blocked using 1% BSA for 1
h at 37°C. The membrane was incubated with antibodies against AGPS
(sc-374201; 1:2,000 dilution), phosphorylated mechanistic target of
rapamycin (sc-293133, p-mTOR; 1:1,000 dilution) and mTOR (sc-8319;
1:1,500 dilution) (all Santa Cruz Biotechnology, Inc., Dallas, TX,
USA) overnight at 4°C, and then incubated for 1 h at 37°C with
mouse peroxidase-labeled anti-rabbit immunoglobulin G (cat no.
sc-2357; 1:2,000 dilution; Santa Cruz Biotechnology, Inc.).
Following this, the membrane was washed with PBS plus 0.05% Tween20
three times. The membrane was visualized using Immobilon Western
chemiluminescent horseradish peroxidase substrate (EMD Millipore,
Billerica, MA, USA). β-actin (A5441; 1:5,000 dilution;
Sigma-Aldrich; Merck KGaA) was used as the control.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) assay
Cells were co-transfected with AGPS shRNA plasmid,
and Tudor-SN shRNA plasmid, NF-κB p65 expression plasmid (retrieval
experiment) or miR-127 siRNA plasmid (retrieval experiment) in
order to explore the effect of Tudor-SN, NF-κB p65 and miR-127 on
the AGPS by RT-qPCR.
A total of 3×105 cells/well were seeded
onto a 6-well plate and cultured for 24 h. Cells were lysed and
total RNA was extracted using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA was then
reverse transcribed using PrimeScript™ RT reagent Kit (Takara
Biotechnology, Dalian, China) and mRNA, circular RNA (circRNA) and
long non-coding RNA (lncRNA) expression of the target genes were
detected using a qPCR assay (ABI7500; Applied Biosystems; Thermo
Fisher Scientific, Inc.) and the 2−∆∆Cq method using a
qRT-PCR SYBR® Kit (Takara Biotechnology) (15). The RT-qPCR primers used were as
follows: AGPS, forward, 5′-ACCAGATTCCCTGGAGTTCA-3′ and reverse,
5′-GAACCACCAGGTCCTCGATA-3′; circ-ubiquitin-associated protein 2
(UBAP2) forward, 5′-AGCCTCAGAAGCCAACTCCTTTG-3′ and reverse,
5′-TCAGGTTGAGATTTGAAGTCAAGAT-3′; circ-zinc finger protein 292
(ZNF292) forward, 5′-GCTCAAGAGACTGGGGTGTG-3′ and reverse,
5′-AGTGTGTGTTCTGGGGCAAG-3′; circ-homeodomain-interacting protein
kinase 3 (HIPK3) forward, 5′-TATGTTGGTGGATCCTGTTCGGCA-3′ and
reverse, 5′-TGGTGGGTAGACCAAGACTTGTGA-3′; H19 imprinted maternally
expressed transcript (non-protein coding) (H19) forward,
5′-ATCGGTGCCTCAGCGTTCGG-3′ and reverse, 5′-CTGTCCTCGCCGTCACACCG-3′;
colon cancer-associated transcript 1 (non-protein coding) (CCAT1)
forward, 5′-CATTGGGAAAGGTGCCGAGA-3′ and reverse,
5′-ACGCTTAGCCATACAGAGCC-3′; hepatocellular carcinoma upregulated
long non-coding RNA (HULC) forward, 5′-CAGGAAGAGTCGTCACGAGAACCAG-3′
and reverse, 5′-CTTCTTGCTTGATGCTTTGGTCTGT-3′; and β-actin forward,
5′-AGGCACCAGGGCGTGAT-3′ and reverse, 5′-GCCCACATAGGAATCCTTCTGAC-3′.
β-actin was used as the control. The PCR conditions were as
follows: Denaturation at 95°C for 10 min, followed by 40 cycles of
95°C for 15 sec, 60°C for 60 sec and a final elongation at 95°C for
15 sec, followed by 60°C for 60 sec and 95°C for 15 sec.
Statistical analysis
SPSS version 11.0 (SPSS Inc., Chicago, IL, USA) was
used for statistical analysis. Data are presented as the mean ±
standard deviation. The statistical analysis was performed using
analysis of variance with Tukey's post-hoc test. P≤0.05 was
considered to indicate a statistically significant difference.
Results
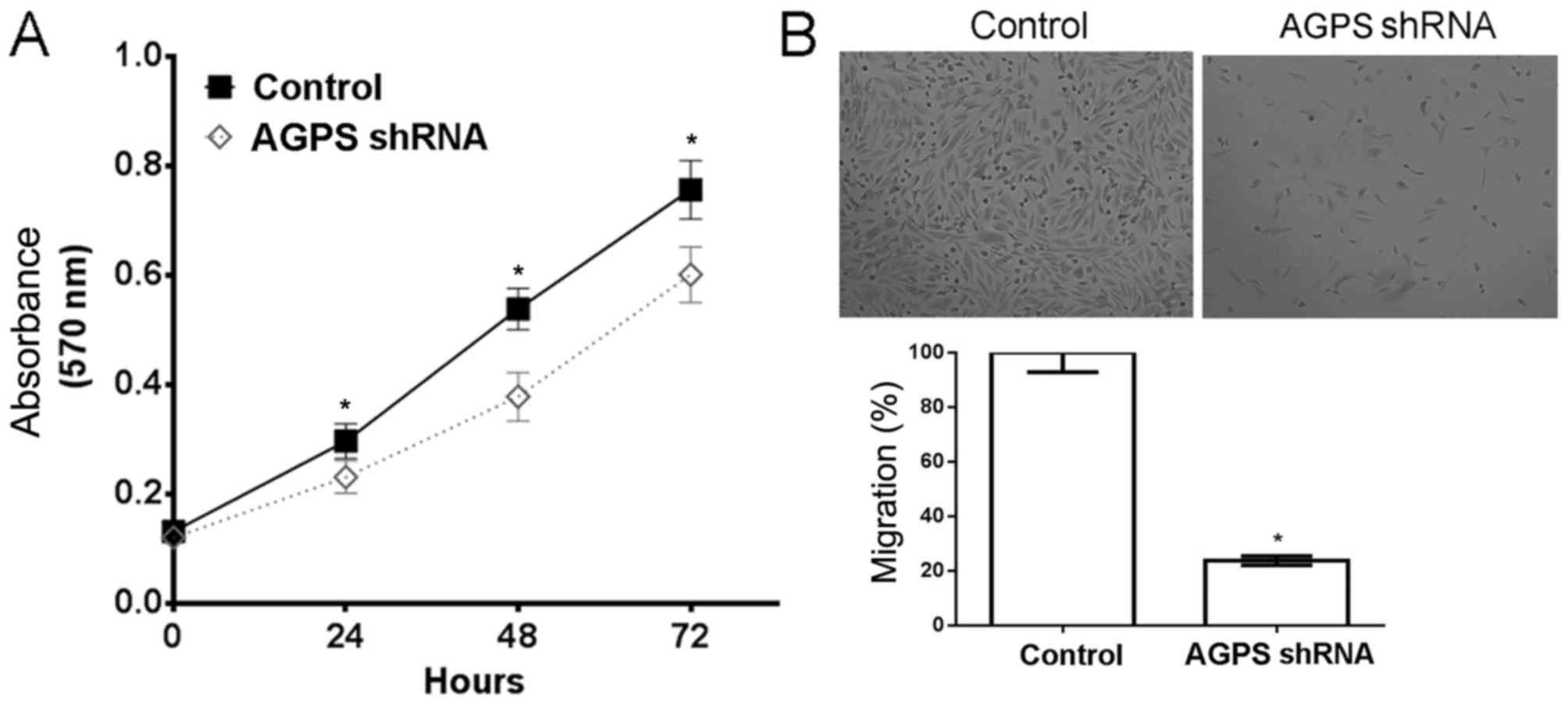
AGPS silencing reduces proliferation
and migration in human glioma U87MG cells
The cell proliferation assay demonstrated that the
potential for proliferation was significantly reduced by AGPS
silencing in human glioma U87MG cells (P<0.05) (Fig. 1A). Furthermore, the Transwell assay
indicated that the potential for migration was reduced by 23.7% as
a result of AGPS silencing in human glioma U87MG cells, compared
with that in the control group (Fig.
1B).
AGPS silencing regulates the
expression of circRNAs and lncRNAs in human glioma U87MG cells
The RT-qPCR assay demonstrated that AGPS silencing
significantly downregulated the expression of the circRNAs
circUBAP2, circZNF292 and circHIPK3, and the lncRNAs H19, CCAT1 and
HULC (Fig. 2).
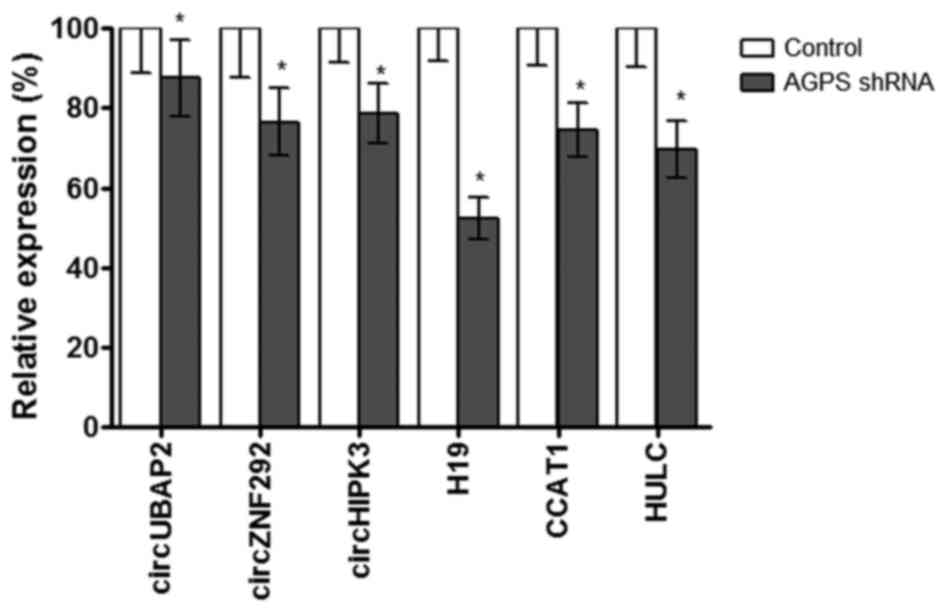 | Figure 2.AGPS silencing regulates the
expression of circRNAs and lncRNAs in human glioma U87MG cells. The
reverse transcription-quantitative polymerase chain reaction assay
demonstrated that AGPS silencing regulated the expression of
circRNAs (circUBAP2, circZNF292 and circHIPK3) and lncRNAs (H19,
CCAT1 and HULC) in human glioma U87MG cells. *P≤0.05, compared with
control group. AGPS, alkylglycerone phosphate synthase; shRNA,
short hairpin RNA; circ, circular; UBAP, ubiquitin-associated
protein 2; ZNF292, zinc finger protein 292; HIPK3,
homeodomain-interacting protein kinase 3; H19, H19 imprinted
maternally expressed transcript (non-protein coding); CCAT1, colon
cancer-associated transcript 1 (non-protein coding); HULC,
hepatocellular carcinoma upregulated long non-coding RNA. |
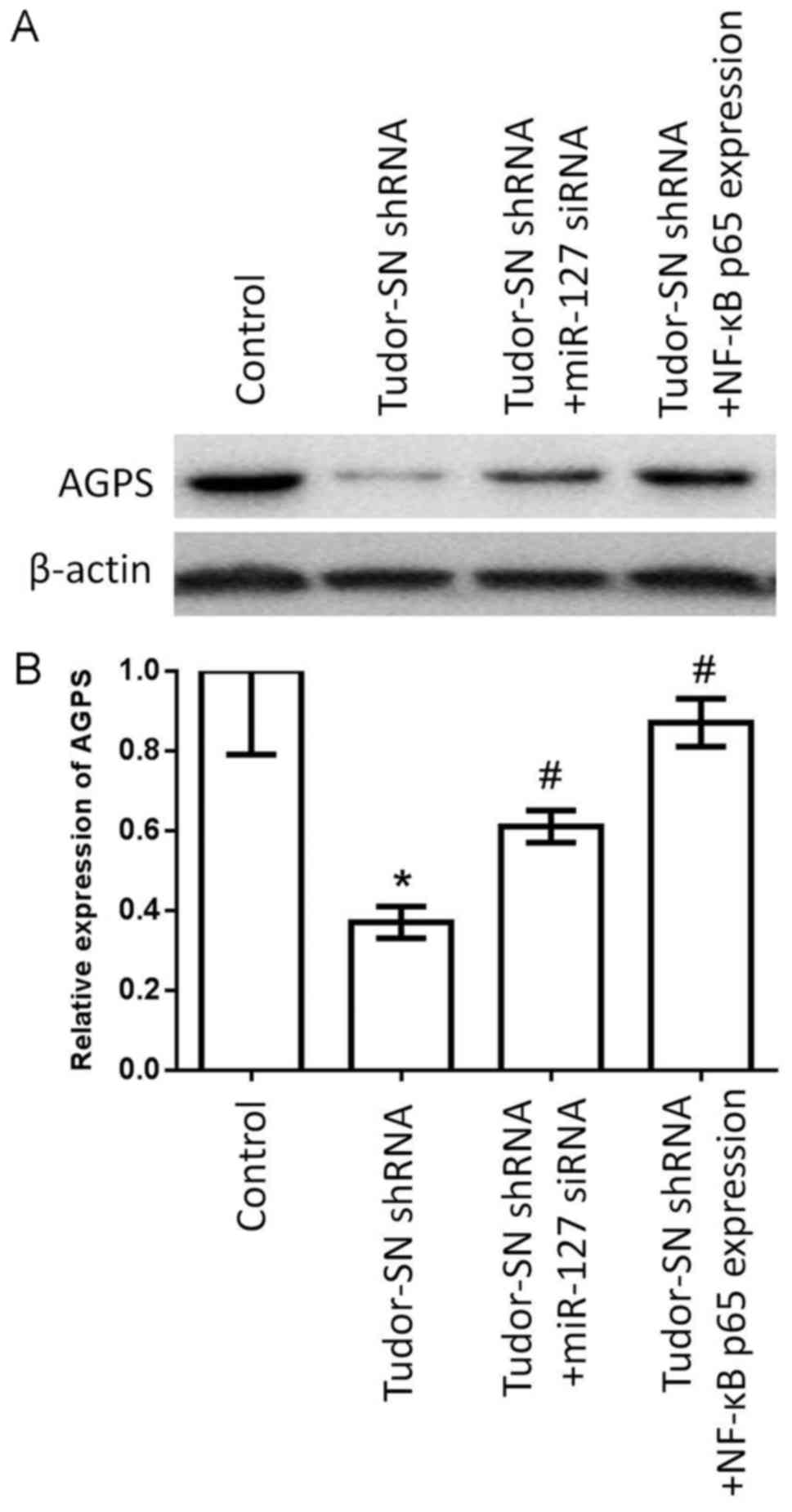
Silencing Tudor-SN decreases the
expression of AGPS by NF-κB and miR-127 in human glioma U87MG
cells
The western blotting and RT-qPCR assay indicated
that silencing Tudor-SN decreased the expression of AGPS.
Furthermore, it was determined that the expression of AGPS was
partially restored by NF-κB and miR-127 retrieval experiments
(Fig. 3). Therefore, it was
considered that the effect of Tudor-SN-regulated AGPS may be
partially dependent on NF-κB and miR-127.
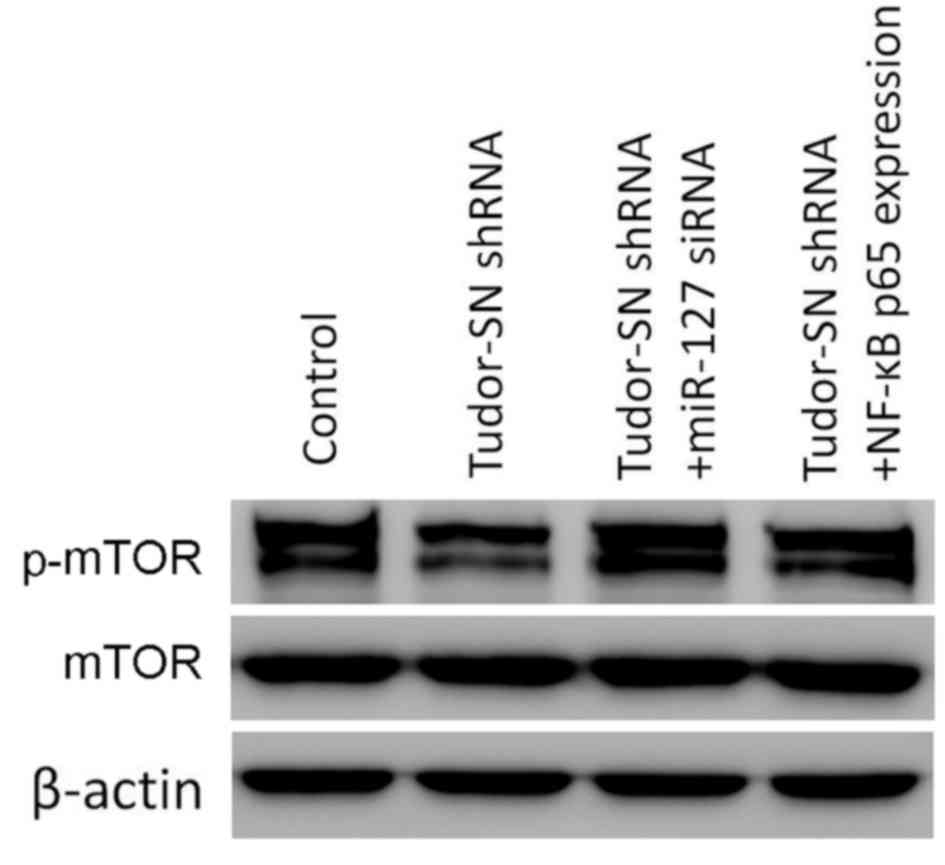
Silencing Tudor-SN decreases the
activity of the mTOR signaling pathway via NF-κB and miR-127 in
human glioma U87MG cells
The western blotting assay demonstrated that
Tudor-SN silencing decreased the activity of the mTOR signaling
pathway. Furthermore, it was determined that the activity of the
mTOR signaling pathway was partially restored by NF-κB and miR-127
retrieval experiments (Fig. 4).
Therefore, it was considered that the effect of the
Tudor-SN-regulated mTOR signaling pathway may be partially
dependent on NF-κB and miR-127.
Discussion
AGPS and Tudor-SN serve a key role in the adjustment
of tumor angiogenesis, and are expressed highly in multiple tumor
tissue types and are associated with the prognosis of patients
(16). In the present study, it was
determined that AGPS silencing or suppression inhibited the
proliferation and migration of glioma U87MG cells, validating the
biological function of AGPS in glioma (17). circRNAs and lncRNAs are considered to
serve a key role in tumor progression, and their altered expression
can improve or suppress the potential for proliferation and
migration in glioma. Therefore, circRNAs and lncRNAs are considered
to be important targets for glioma therapeutics (18,19). The
present study determined that the silencing of AGPS downregulated
the expression of the circRNAs circUBAP2, cicZNF292 and circHIPK3,
and the lncRNAs H19, CCAT1 and HULC. All the aforementioned
circRNAs and lncRNAs have been reported to be oncogenes (20–24).
It was also determined that Tudor-SN silencing
suppressed the expression of AGPS; however, the mechanism
underlying Tudor-SN-regulated AGPS in human glioma was not clear.
Tudor-SN is a type of multi-functional protein that is widely
expressed in tumor cells. Tudor-SN has the ability to activate
various transcription factors, including NF-κB, and is involved in
adjusting the expression of miRNAs (11).
miRNAs serve an important role in the progression of
cancer, and a number of miRNAs are tumor suppressors, such as
miR-127 (25). Inhibiting Tudor-SN
promotes the expression of miR-127, and miR-127 has the ability to
inhibit the migration of tumor cells (26); therefore, Tudor-SN is considered to
have the ability to adjust the expression of AGPS in glioma cells
through miR-127 adjustment and further control of the biological
function of glioma cells. The present study determined that AGPS
expression decreased following the inhibition of Tudor-SN.
Following a retrieval experiment to inhibit miR-127,
the expression of AGPS was partially recovered, similar to that of
NF-κB. The aforementioned results confirmed the alteration of AGPS
expression through control of NF-κB and miR-127 by Tudor-SN in
glioma cells. In the present study, it was also determined that
Tudor-SN regulates the activity of the mTOR signaling pathway via
miR-127 and NF-κB, indicating that Tudor-SN may regulate the
expression of AGPS via the mTOR signaling pathway.
The data demonstrated that silencing AGPS reduced
the potential for the proliferation and migration of glioma U87MG
cells, and use of NF-κB and miR-127 may be the manner in which
Tudor-SN regulates AGPS expression via the m-TOR signaling pathway,
laying a theoretical foundation and experimental basis for further
investigation of the pathogenesis and therapeutics of malignant
gliomas.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 31501159), the
Tianjin Public Health Key Research Project (grant no. 15KG108), the
Tianjin Science and Technology Key Project on Chronic Diseases
Prevention and Treatment (grant no. 16ZXMJSY00020), the Tianjin
Research Program of Application Foundation and Advanced Technology
(grant no. 16JCYBJC27200) and the Special Program of Talents
Development for Excellent Youth Scholars in Tianjin, China
(TJTZJH-QNBJRC-2-9).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
YZhu was responsible for study conception and
design. YZha, YL, JJ and YQ were responsible for acquisition of
data. HH and Y-GC performed analysis of the data.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhang S, Xie R, Zhao T, Yang X, Han L, Ye
F, Lei T and Wan F: Neural stem cells preferentially migrate to
glioma stem cells and reduce their stemness phenotypes. Int J
Oncol. 45:1989–1996. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ellison D, Love S, Chimelli L, Harding BN,
Lowe JS, Vinter HV, Brandner S and Yong WH: NeuropathologyA
reference text of CNS pathology. 3rd ed. Edinburgh: Elsevier/Mosby;
2013
|
|
3
|
Zhang Z, Li C, Tan Q, Xie C, Yang Y, Zhan
W, Han F, Sharma HS and Sharma A: Curcumin suppresses tumor growth
and angiogenesis in human glioma cells through modulation of
vascular endothelial growth factor/angiopoietin-2/thrombospondin-1
signaling. CNS Neurol Disord Drug Targets. 16:346–350. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Birk HS, Han SJ and Butowski NA: Treatment
options for recurrent high-grade gliomas. CNS Oncol. 6:61–70. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li Y, Xu J, Chen H, Bai J, Li S, Zhao Z,
Shao T, Jiang T, Ren H, Kang C and Li X: Comprehensive analysis of
the functional microRNA-mRNA regulatory network identifies miRNA
signatures associated with glioma malignant progression. Nucleic
Acids Res. 41:e2032013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dewert N, Amschler K, Lorenz V and Schön
MP: The IKKα-dependent non-canonical pathway of NF-κB activation is
constitutively active and modulates progression-related functions
in a subset of human melanomas. Arch Dermatol Res. 308:733–742.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fashe T, Saarikettu J, Isomäki P, Yang J
and Silvennoinen O: Expression analysis of Tudor-SN protein in
mouse tissues. Tissue Cell. 45:21–31. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kochan DZ, Ilnytskyy Y, Golubov A, Deibel
SH, McDonald RJ and Kovalchuk O: Circadian disruption-induced
microRNAome deregulation in rat mammary gland tissues. Oncoscience.
2:428–442. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tsuchiya N, Ochiai M, Nakashima K, Ubagai
T, Sugimura T and Nakagama H: SND1, a component of RNA-induced
silencing complex, is up-regulated in human colon cancers and
implicated in early stage colon carcinogenesis. Cancer Res.
67:9568–9576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yeo SK, French R, Spada F and Clarkson R:
Opposing roles of Nfkb2 gene products p100 and p52 in the
regulation of breast cancer stem cells. Breast Cancer Res Treat.
162:465–477. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao X, Duan Z, Liu X, Wang B, Wang X, He
J, Yao Z and Yang J: MicroRNA-127 is downregulated by Tudor-SN
protein and contributes to metastasis and proliferation in breast
cancer cell line MDA-MB-231. Anat Rec (Hoboken). 296:1842–1849.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Armengol S, Arretxe E, Enzunza L, Llorente
I, Mendibil U, Navarro-Imaz H, Ochoa B, Chico Y and Martínez MJ:
SREBP-2-driven transcriptional activation of human SND1 oncogene.
Oncotarget. 8:108181–108194. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fung S, Xu C, Hamel E, Wager-Miller JB,
Woodruff G, Miller A, Sanford C, Mackie K and Stella N: Novel
indole-based compounds that differentiate alkylindole-sensitive
receptors from cannabinoid receptors and microtubules:
Characterization of their activity on glioma cell migration.
Pharmacol Res. 115:233–241. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhu Y, Liu XJ, Yang P, Zhao M, Lv LX,
Zhang GD, Wang Q and Zhang L: Alkylglyceronephosphate synthase
(AGPS) alters lipid signaling pathways and supports chemotherapy
resistance of glioma and hepatic carcinoma cell lines. Asian Pac J
Cancer Prev. 15:3219–3226. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jin Y, Wu W, Zhang W, Zhao Y, Wu Y, Ge G,
Ba Y, Guo Q, Gao T, Chi X, et al: Involvement of EGF receptor
signaling and NLRP12 inflammasome in fine particulate
matter-induced lung inflammation in mice. Environ Toxicol.
32:1121–1134. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhu Y, Liu A, Zhang X, Qi L, Zhang L, Xue
J, Liu Y and Yang P: The effect of benzyl isothiocyanate and its
computer-aided design derivants targeting alkylglycerone phosphate
synthase on the inhibition of human glioma U87MG cell line. Tumor
Biol. 36:3499–3509. 2015. View Article : Google Scholar
|
|
18
|
Bian EB, Li J, Xie YS, Zong G, Li J and
Zhao B: LncRNAs: New players in gliomas, with special emphasis on
the interaction of lncRNAs with EZH2. J Cell Physiol. 230:496–503.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang P, Qiu Z, Jiang Y, Dong L, Yang W, Gu
C, Li G and Zhu Y: Silencing of cZNF292 circular RNA suppresses
human glioma tube formation via the Wnt/β-catenin signaling
pathway. Oncotarget. 7:63449–63455. 2016.PubMed/NCBI
|
|
20
|
Zhang H, Wang G, Ding C, Liu P, Wang R,
Ding W, Tong D, Wu D, Li C, Wei Q, et al: Increased circular RNA
UBAP2 acts as a sponge of miR-143 to promote osteosarcoma
progression. Oncotarget. 8:61687–61697. 2017.PubMed/NCBI
|
|
21
|
Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang
C, Liu D, Wang M, Wang L, Zeng F and Jiang G: CircHIPK3 sponges
miR-558 to suppress heparanase expression in bladder cancer cells.
EMBO Rep. 18:1646–1659. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
He TD, Xu D, Sui T, Zhu JK, Wei ZX and
Wang YM: Association between H19 polymorphisms and osteosarcoma
risk. Eur Rev Med Pharmacol Sci. 21:3775–3780. 2017.PubMed/NCBI
|
|
23
|
Zhu H, Zhao H, Zhang L, Xu J, Zhu C, Zhao
H and Lv G: Dandelion root extract suppressed gastric cancer cells
proliferation and migration through targeting lncRNA-CCAT1. Biomed
Pharmacother. 93:1010–1017. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fan YH, Wu MJ, Jiang Y, Ye M, Lu SG, Wu L
and Zhu XG: Long non-coding RNA HULC as a potential prognostic
biomarker in human cancers: A meta-analysis. Oncotarget.
8:21410–21417. 2017.PubMed/NCBI
|
|
25
|
Jiang H, Jin C, Liu J, Hua D, Zhou F, Lou
X, Zhao N, Lan Q, Huang Q, Yoon JG, et al: Next generation
sequencing analysis of miRNAs: MiR-127-3p inhibits glioblastoma
proliferation and activates TGF-β signaling by targeting SKI.
OMICS. 18:196–206. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gutierrez-Beltran E, Denisenko TV,
Zhivotovsky B and Bozhkov PV: Tudor staphylococcal nuclease:
Biochemistry and functions. Cell Death Differ. 23:1739–1748. 2016.
View Article : Google Scholar : PubMed/NCBI
|