Introduction
Lung cancer is the most common type of cancer
worldwide (1), and 85% of lung cancer
cases are diagnosed as non-small cell lung cancer (NSCLC) (2). In China alone, ~7,333,000 patients were
diagnosed with NSCLC and, of these, ~6,102,000 patients succumbed
to mortality in 2015 (3). With
increased levels of environmental pollution, the incidence of NSCLC
and the mortality rate of patients with this disease have increased
significantly (1), resulting in
increased health and economic burdens on patients and society.
Although NSCLC treatments have improved with the development of
medical technology, certain clinically applied drugs targeting
late-stage NSCLC (2,4) are ineffective due to the presence of
multiple gene mutations and comprehensive pathogenesis (5,6).
Therefore, it is important to obtain a greater understanding of the
underlying mechanism of NSCLC development in order to improve the
efficacy of clinical treatment.
Long non-coding RNAs (lncRNAs), comprising >200
nucleotides, are a class of transcripts with no protein-coding
capacity (7). Previous studies have
demonstrated that lncRNAs perform diverse functions in cancer
cells, including proliferation, migration, invasion and apoptosis
(8,9).
The roles of lncRNAs have also been assessed in patients with
NSCLC. Shi et al (10)
identified that growth arrest specific 5 lncRNA serves critical
roles in the proliferation and apoptosis of NSCLC cells via
p53-dependent and -independent pathways and may serve as a
diagnostic marker for NSCLC. In addition, Yang et al
(11) demonstrated that PVT1 oncogene
lncRNA is significantly upregulated in patients with NSCLC and its
expression level is associated with lymph node metastasis and a
poorer prognosis. Kruer et al (12) also indicated that maternally expressed
3 lncRNA is involved in the regulation of lung cancer cell
proliferation via the retinoblastoma pathway. In addition, a
previous study indicated that urothelial cancer associated 1 lncRNA
may serve as an oncogene in NSCLC by targeting microRNA
(miR)-193a-3p (13), which has been
demonstrated to suppress the metastasis of NSCLC by downregulating
the Erb-B2 receptor tyrosine kinase 4/phosphoionositide-3-kinase
regulatory subunit 3/mechanistic target of the rapamycin/ribosomal
protein S6 kinase B2 signaling pathway (14). The results of these studies indicated
that lncRNAs serve important roles in the regulation of NSCLC
pathogenesis.
Metastasis-associated lung adenocarcinoma transcript
1 (MALAT-1), also known as MALTA1 or nuclear-enriched abundant
transcript 2, is the first identified lncRNA to be associated with
lung cancer. Since its discovery over a decade ago, a large number
of studies have demonstrated the associations between MALAT1 and
cancer progression (15–17). Recently, a study reported that MALAT1
may promote bone metastasis in patients with NSCLC (18). However, associations between MALAT1
and NSCLC clinical outcome or occurrence have rarely been addressed
(19). Therefore, the present study
aimed to elucidate the associations between MALAT1 and the clinical
characteristics of patients with NSCLC and to assess the potential
effect of MALAT1 on the progression of NSCLC.
Materials and methods
Patients and samples
A total of 120 patients with NSCLC, who had
undergone surgical resection at the Taizhou Hospital of Wenzhou
Medical University (Linhai, China) and were diagnosed by biopsy,
were enrolled in the present study between June 1, 2010 and
December 30, 2016. Patients were excluded if they exhibited any of
the following: Hepatitis infection, autoimmune disease, human
immunodeficiency virus infection or psychosis. During enrollment,
patient clinical data, including sex, age, vessel invasion,
clinical stage, tumor type, tumor differentiation, tumor diameter
and recurrence, were collected and are presented in Table I. Tumor-Node-Metastasis (TNM) stage
evaluation was performed according to 2014 8th edition of the TNM
classification (20).
 | Table I.Associations between MALAT1 expression
and the clinical characteristics of patients with non-small cell
lung cancer. |
Table I.
Associations between MALAT1 expression
and the clinical characteristics of patients with non-small cell
lung cancer.
|
| MALAT1 |
|
|
|---|
|
|
|
|
|
|---|
| Term | Low-expression | High-expression | χ2 | P-value |
|---|
| Sex |
|
|
|
|
| Male | 69 | 40 | 5.467 | 0.019 |
|
Female | 3 | 8 |
|
|
| Age, year |
|
|
|
|
| ≤50 | 25 | 24 | 0.025 | 0.838 |
|
>50 | 37 | 34 |
|
|
| Maximum diameter,
cm |
|
|
|
|
| ≤5 | 38 | 33 | 0.629 | 0.504 |
|
>5 | 24 | 25 |
|
|
| Tumor number |
|
|
|
|
|
Single | 21 | 19 | 0.012 | 0.905 |
|
Multiple | 43 | 37 |
|
|
| TNM stage |
|
|
|
|
|
I–II | 32 | 20 | 5.172 | 0.016 |
|
III–IV | 20 | 48 |
|
|
| Vessel
invasion |
|
|
|
|
| No | 35 | 45 | 6.483 | 0.032 |
|
Yes | 8 | 32 |
|
|
| Pathological
differentiation |
|
|
|
|
|
High-medium | 23 | 65 | 12.383 | 0.013 |
|
Low | 0 | 32 |
|
|
| Recurrence |
|
|
|
|
| No | 30 | 15 | 9.542 | 0.006 |
|
Yes | 24 | 51 |
|
|
During surgical resection, paired tumor tissues and
adjacent non-cancerous tissues were collected from patients with
NSCLC. Tissue sections were preserved at −80°C or in liquid
nitrogen for subsequent analysis. The present study was approved by
the Ethics Committee of Taizhou Hospital of Wenzhou Medical
University (Linhai, China) and written informed consent was
obtained from all patients prior to enrollment.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissue samples using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), according to the manufacturer's protocol. RNA
quality was assessed using the Nanodrop N-1000 (Agilent
Technologies, Inc., Santa Clara, CA, USA) and 2 µg total RNA was
reverse transcribed into cDNA using the Applied
Biosystems® TaqMan® mRNA Reverse
Transcription kit (Applied Biosystems; Thermo Fisher Scientific,
Inc.) according to manufacturer's protocol. qPCR was subsequently
performed using SYBR Green (Applied Biosystems; Thermo Fisher
Scientific, Inc.) on an ABI 7900 Real-Time PCR instrument (Applied
Biosystems; Thermo Fisher Scientific, Inc.) with the following
thermocycling conditions: 95°C for 30 sec, 40 cycles at 95°C for 5
sec and 60°C for 31 sec. GAPDH was used as the internal control.
The gene primers utilized were as follows: MALAT1 forward,
5′-AGGCGTTGTGCGTAGAGGA-3′ and reverse, 5′-GGATTTTACCAACCACTCGC-3′;
and GAPDH forward, 5′-AGAAGGCTGGGGCTCATTTG-3′ and reverse,
5′-AGGGGCCATCCACAGTCTTC-3′. Each experiment was performed in
triplicate and was quantified using the 2−ΔΔCq method
(21).
Cell culture and transfection
The NSCLC A549 cell line was purchased from ATCC
(Manassas, VA, USA) and cells cultured in Dulbecco's modified
Eagle's medium (DMEM; Invitrogen; Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (FBS) at 37°C in a
humidified atmosphere with 5% CO2.
Small interfering RNA (siRNA) targeting MALAT1 (10
nmol/l) and its corresponding control were transfected onto a
6-well plate with a cell confluence of 70% in each well using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. The sequences for siRNA
were designed as follows: si-MALAT1, forward,
5′-CACAGGGAAAGCGAGUGGUUGGUAATT-3′, and reverse,
5′-UUACCAACCACUCGCUUUCCCUGUGTT-3′; and si-control, forward,
5′-UUCUCCGAACGUGUCACGUTT-3′, and reverse
5′-ACGUGACACGUUCGGAGAATT-3′. The siRNA sequences were synthetized
by Shanghai GenePharma Co., Ltd (Shanghai, China). After 48 h,
stable cell strains were screened using puromycin (Thermo Fisher
Scientific, Inc.) and the expression of MALAT1 was evaluated using
RT-qPCR was performed as aforementioned using the human tissue.
Cell proliferation assay
Following the determination of the effect of
silenced MALAT1, transfected A549 cells were seeded onto a 96-well
plate at a density of 5×103 cells/well. Subsequently,
CellTiter 96 (Promega Corporation, Madison, WI, USA) was added to
each well, according to the manufacturer's protocol. Following
culture at 37°C, the absorbance of cells and the index of cellular
proliferation were measured at 490 nm using a Bio-Rad plate-reader
(Bio-Rad Laboratories, Inc., Hercules, CA, USA) every 24 h. A total
of 100 µl DMEM was utilized as a blank control. Each sample was
conducted with five repeats and experiments were independently
repeated twice.
Wound healing assay
For the wound healing assay, cells were seeded onto
6-cm plates and were cultured in DMEM supplemented with 10% FBS.
Once the cells had reached 80% confluency, they were scraped with a
200 µl tip to wound the monolayer and the starting width of the
wound (200 µl) was recorded. Cells were then cultured in DMEM
medium (Invitrogen; Thermo Fisher Scientific, Inc.) for 48 h.
Images of the migrated distance of cells were captured and measured
using a light microscope at magnification, ×100, 0 and 48 h after
scraping.
Invasion assay
For the invasion assay, 1×104 cells in
200 µl FBS-free DMEM were seeded into an upper Matrigel-coated
chamber (BD Biosciences, Franklin Lakes, NJ, USA). The lower
chamber was filled with 500 µl RPMI-1640 medium containing 10% FBS
to induce cell invasion. Following a 24-h incubation, cells on the
upper filter were removed using cotton swabs. Cells on the lower
filter, namely the invading cells, were fixed with 95% ethanol at
4°C for 1 h, stained with 0.5% crystal violet at room temperature
for 12 min, and monitored using an inverted microscope at a
magnification of ×200. For each sample, cells in ≥5 random
microscopic visual fields were counted and the average number was
used as the final result.
Statistical analyses
In the present study, SPSS 20.0 (IBM Corp., Armonk,
NY, USA) was used for statistical analyses. Continuous variables
are presented as the mean ± standard deviation. Comparisons between
two groups were analyzed using Student's t-test, and pair-wise
comparisons among multiple groups were determined using a one-way
analysis of variance followed by the Least Significant Difference
test. Associations between MALAT1 and clinical characteristics were
determined using the χ2 test. The receiver operative
characteristics (ROC) curve analysis in SPSS was utilized to create
a summary ROC curve in order to evaluate the predicative ability of
MALAT1 in NSCLC recurrence. P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression of MALAT1 in clinical NSCLC
tissues
Following the collection of clinical samples, the
expression levels of MALAT1 in tumor and adjacent non-cancerous
tissues were determined using RT-qPCR. The results demonstrated
that the expression of MALAT1 was significantly increased in NSCLC
tumor tissues when compared with non-tumor adjacent tissues
(P<0.05, data not shown). Additionally, the expression of MALAT1
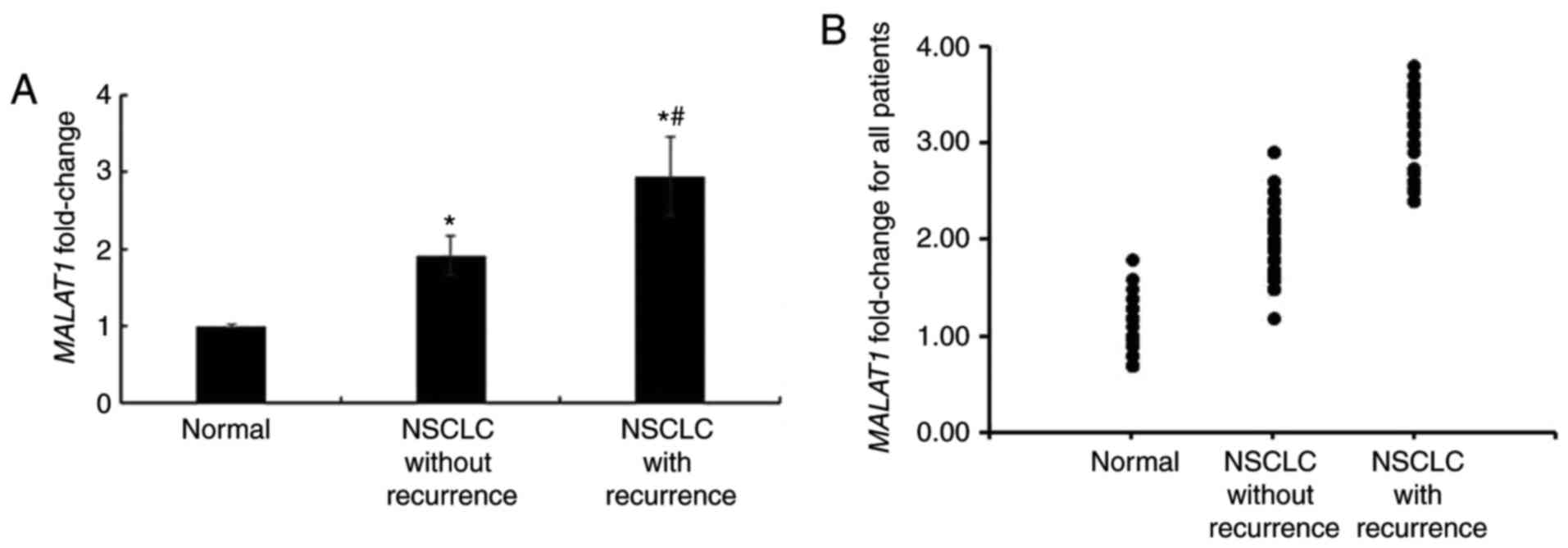
in patients with recurrent NSCLC was markedly higher than that in
NSCLC patients without recurrence (Fig.
1).
Association between MALAT1 and
clinical characteristics
Following the assessment of MALAT1 expression in
tissue samples, associations between MALAT1 and clinical
characteristics were evaluated (Table
I). The results demonstrated that a high expression of MALAT1
was significantly associated with female sex (P=0.019), TNM stage
(P=0.016), vessel invasion (P=0.032), pathological differentiation
(P=0.013) and recurrence (P=0.006) in patients with NSCLC. However,
no significant associations were identified between the expression
of MALAT1 and age, tumor maximum diameter or tumor number
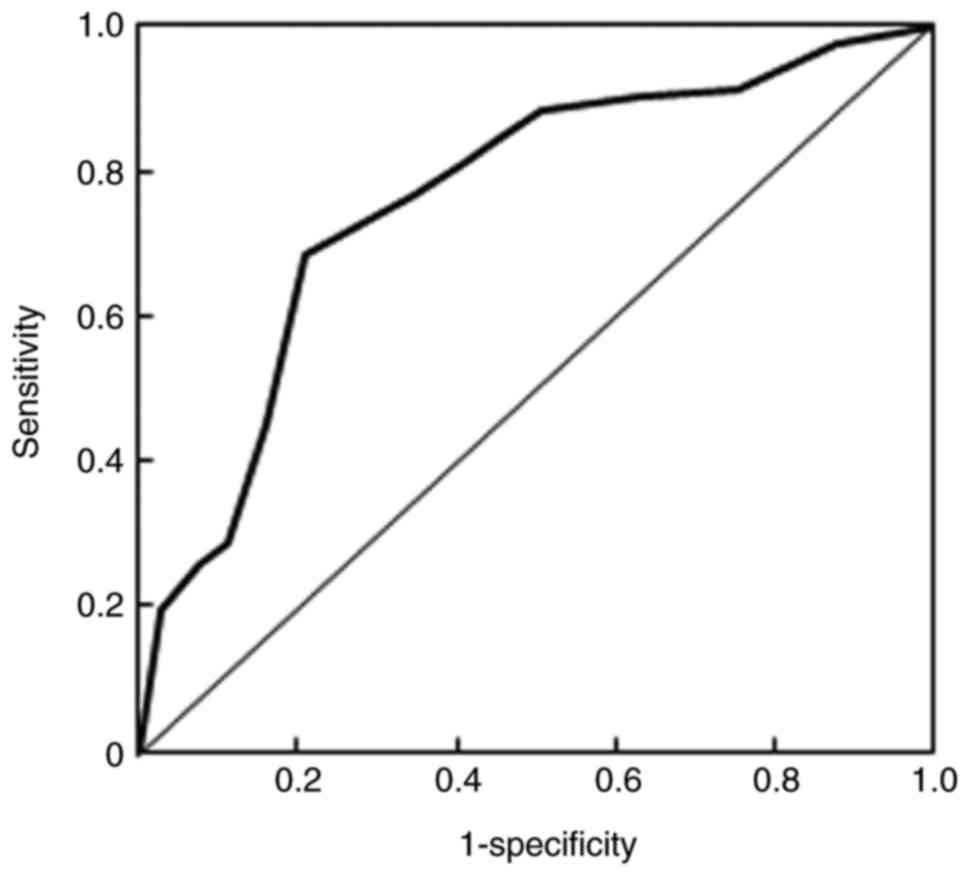
(P>0.05). In addition, the diagnostic value of MALAT1 in the
recurrence of NSCLC was also estimated (Fig. 2). The area under curve value was 0.76
and the Q index was 0.71, indicating that MALAT1 has potential
diagnostic value for the predication of NSCLC recurrence.
Effects of silenced MALAT1 on cell
proliferation
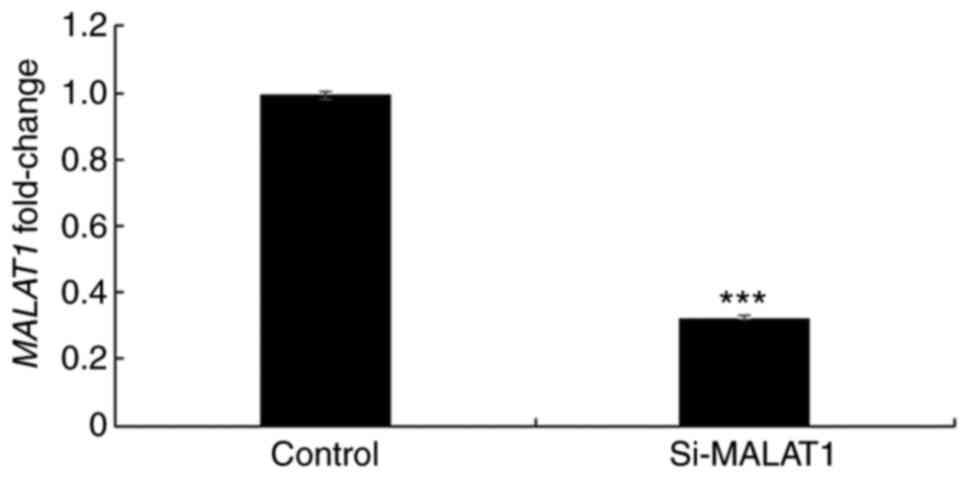
In order to further elucidate the underlying
mechanism of MALAT1, its expression was silenced using siRNA in
A549 cells and the efficacy was confirmed using RT-qPCR (Fig. 3). Once silencing had been successful,
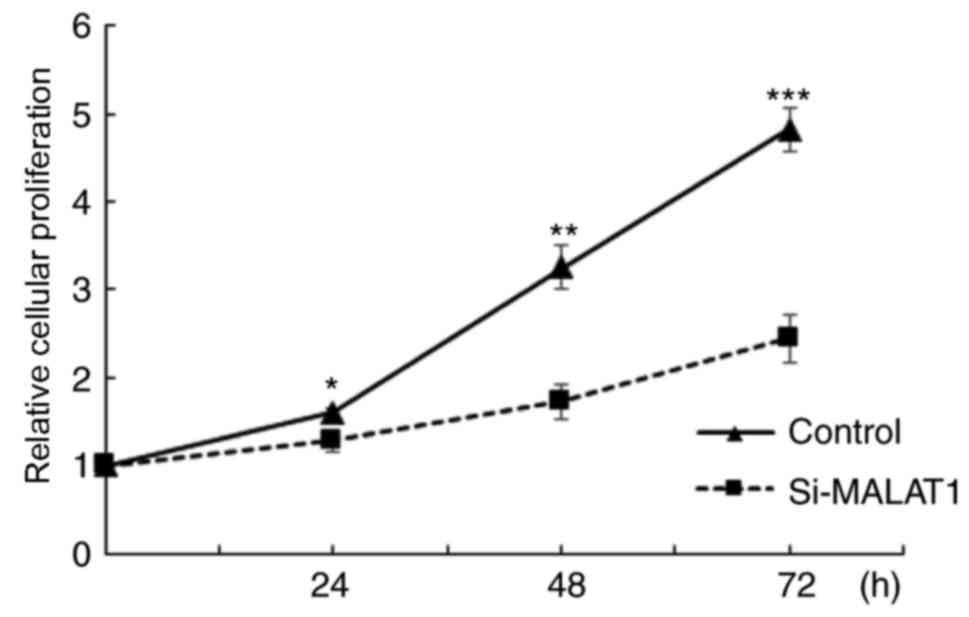
the influence of MALAT1 on cell proliferation was assessed. The
results demonstrated that A549 cellular proliferation was
significantly decreased in si-MALAT1 A549 cells compared with the
controls at all time points (P<0.05; Fig. 4). These results indicated that MALAT1
promoted the proliferation of A549 cells.
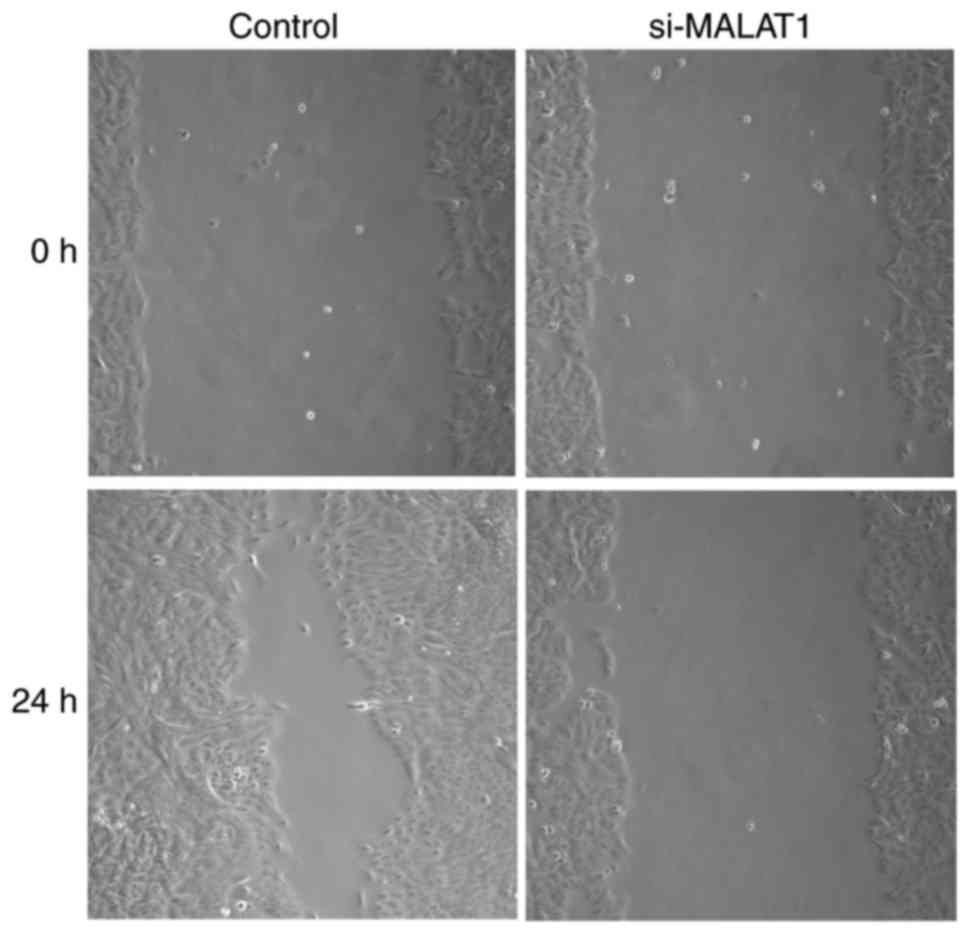
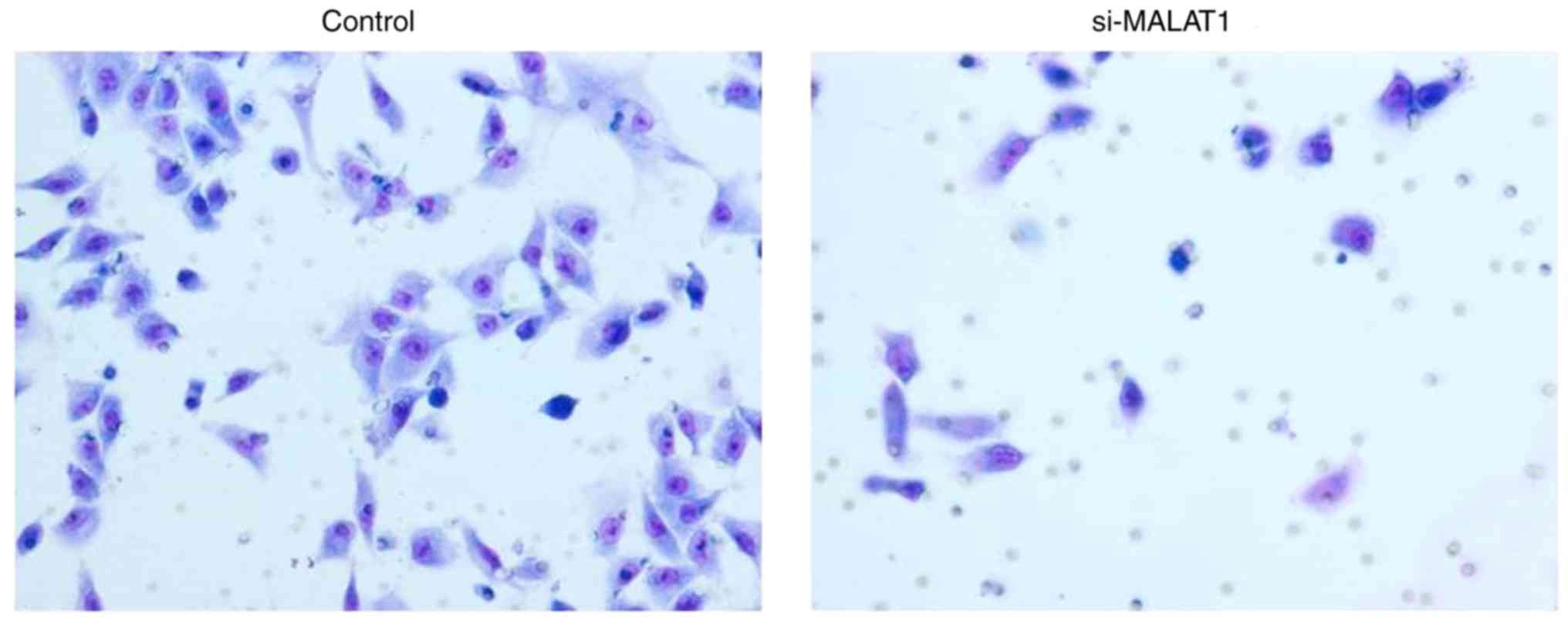
Effects of si-MALAT1 on cell migration
and invasion
The influence of si-MALAT1 on A549 cell migration
and invasion were assessed using wound healing and Matrigel assays,
respectively. The results of the wound healing assay demonstrated
that migratory abilities were markedly decreased in si-MALAT1 A549
cells compared with control cells (Fig.
5). In addition, Transwell migration demonstrated that the
invasive ability of si-MALAT1 A549 cells was also markedly
inhibited compared with control cells (Fig. 6). These results indicated that MALAT1
may improve the migration and invasion ability of A549 cells.
Discussion
The present study aimed to assess the associations
between MALAT1 expression and the clinical characteristics of
patients with NSCLC, and to reveal the potential underlying
mechanism of MALAT1 in NSCLC. The results of the present study
demonstrated that the expression of MALAT1 was significantly
increased in patients with NSCLC, particularly in those with
recurrent malignancy. Furthermore, the silencing of MALAT1 markedly
decreased the proliferability, migration and invasiveness of A549
cells. Therefore, MALAT1 served a crucial role in the occurrence of
NSCLC.
It has been demonstrated that lncRNA serves
important roles in various processes involved in cancer
pathogenesis, including proliferation, differentiation, migration,
invasion and apoptosis (22). As an
important lncRNA discovered a decade ago, the abnormal expression
of MALAT1 has been exhibited in certain malignancies (23). Lai et al (24) demonstrated that MALAT1 expression was
significantly increased in hepatocellular carcinoma clinical
tissues and cell lines, serving as a novel biomarker for tumor
recurrence following liver transplantation. Ying et al
(25) also revealed that MALAT1 is
abnormally upregulated in bladder cancer and that it promotes
cellular migration by inducing epithelial-mesenchymal transition.
Furthermore, Schmidt et al (26) observed that MALAT1 expression is
increased in NSCLC and interacts with B cell lymphoma 2 to regulate
lung cancer cell apoptosis. In addition, Liu et al (18) demonstrated that MALAT1 promotes bone
metastasis in patients with NSCLC; however, the study only compared
NSCLC tissues and NSCLC cell lines with and without bone
metastasis. The results of the present study indicated that there
was a significant increase in the expression of MALAT1 in NSCLC and
that the expression of MALAT1 in patients with NSCLC that exhibited
recurrence was markedly higher than that in those who did not.
These results therefore indicate that MALAT1 serves a crucial role
in the development and recurrence of NSCLC and that it may act as
an onco-lncRNA to regulate the migration, differentiation and
apoptosis of cancer cells as malignancies progress.
Considering the effect of MALAT1 in cancer,
associations between MALAT1 and the clinical characteristics and
survival rates of patients with NSCLC were assessed. Shen et
al (27) demonstrated that MALAT1
promotes brain metastasis by inducing epithelial-mesenchymal
transition in lung cancer. The results of the present study
demonstrated that the invasive and migratory abilities of
MALAT1-silenced A549 cells were significantly decreased. This may
be the reason for the significant association between a high
expression of MALAT1 and low pathological differentiation, as well
as vessel invasion, as identified in the present study.
Furthermore, a study undertaken by Tano et al (28) revealed that MALAT1 facilitates lung
adenocarcinoma cell motility by regulating the expression of
motility-related genes, including CTHRC1, CCT4, ROD1 and
HMMR. Therefore, these results indicated that MALAT1 serves
a crucial role in NSCLC metastasis.
However, an in vivo study further
demonstrated that MALAT1-deficient cells exhibit an impaired
migration and form fewer tumor nodules in a mouse xenograft
(29). In the present study, it was
demonstrated that si-MALAT1 also significantly suppresses A549 cell
proliferation and migration. In addition, Schmidt et al
(30) revealed that a high-expression
of MALAT1 indicates a poor prognosis in patients with NSCLC. These
observations may indicate that MALAT1 promotes the proliferation of
NSCLC cells. MALAT1 has also been revealed to serve a role in the
migration and invasion of breast cancer and osteosarcoma cells, and
was demonstrated to be significantly inhibited by 17β-Estradiol
(31,32). In line with this, significant
associations were identified between a high expression of MALAT1
and TNM stage, as well as recurrence. The results of the present
study also indicated that MALAT1 may be able to predict NSCLC
recurrence. These results indicate that MALAT1 serves an important
role in the progression of NSCLC and that it may serve as a
potential biomarker for the development and recurrence of
NSCLC.
The present study exhibited certain limitations. In
order to confirm the association between MALAT1 expression and
NSCLC recurrence, further studies are required. The present study
also only utilized a histological method to confirm NSCLC
diagnosis, and histological analysis of NSCLC tissue was not
conducted to assess the effects of MALAT1. Further study would
therefore be required in order to gain a greater understanding of
this.
In conclusion, to the best of our knowledge, the
present study was the first to demonstrate that MALAT1 was
associated with NSCLC recurrence. MALAT1 was revealed to be an
important onco-lncRNA involved in the pathogenesis of NSCLC.
Inhibiting the expression of MALAT1 significantly decreased the
proliferation, migration and invasion of tumor cells. According to
these results, MALAT1 may serve as a potential treatment target and
a possible diagnostic biomarker of NSCLC. However, the detailed
underlying mechanism of MALAT1 in NSCLC remains to be
elucidated.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
LL wrote the manuscript and conducted a majority of
the experiments. HL, YZ and SH collected and analyzed the data. HG
designed the study and approved the manuscript for submission.
Ethics approval and consent to
participate
Written informed consent was obtained from all
participants and the present study was approved by the Research
Committee of Taizhou Hospital of Wenzhou Medical University
(Linhai, China).
Consent for publication
Written informed consent was obtained from all
patients.
Competing interests
The authors declare that there are no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Friese-Hamim M, Bladt F, Locatelli G,
Stammberger U and Blaukat A: The selective c-Met inhibitor
tepotinib can overcome epidermal growth factor receptor inhibitor
resistance mediated by aberrant c-Met activation in NSCLC models.
Am J Cancer Res. 7:962–972. 2017.PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shaw AT, Gandhi L, Gadgeel S, Riely GJ,
Cetnar J, West H, Camidge DR, Socinski MA, Chiappori A, Mekhail T,
et al: Alectinib in ALK-positive, crizotinib-resistant,
non-small-cell lung cancer: A single-group, multicentre, phase 2
trial. Lancet Oncol. 17:234–242. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
White E: Exploiting the bad eating habits
of Ras-driven cancers. Genes Dev. 27:2065–2071. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wagner SA, Beli P and Serve H: Abstract
3888: Systematic characterization of aberrant signaling induced by
oncogenic fusions in non-small cell lung cancer. Cancer Res. 76 14
Suppl:S3888. 2016. View Article : Google Scholar
|
|
7
|
Liz J and Esteller M: lncRNAs and
microRNAs with a role in cancer development. Biochim Biophys Acta.
1859:169–176. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shi SJ, Wang LJ, Yu B, Li YH, Jin Y and
Bai XZ: LncRNA-ATB promotes trastuzumab resistance and
invasion-metastasis cascade in breast cancer. Oncotarget.
6:11652–11663. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shi X, Sun M, Liu H, Yao Y, Kong R, Chen F
and Song Y: A critical role for the long non-coding RNA GAS5 in
proliferation and apoptosis in non-small-cell lung cancer. Mol
Carcinog. 54 Suppl 1:E1–E12. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang YR, Zang SZ, Zhong CL, Li YX, Zhao SS
and Feng XJ: Increased expression of the lncRNA PVT1 promotes
tumorigenesis in non-small cell lung cancer. Int J Clin Exp Pathol.
7:6929–6935. 2014.PubMed/NCBI
|
|
12
|
Kruer TL, Dougherty SM, Reynolds L, Long
E, de Silva T, Lockwood WW and Clem BF: Expression of the lncRNA
maternally expressed gene 3 (MEG3) contributes to the control of
lung cancer cell proliferation by the Rb pathway. PloS One.
11:e01663632016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nie W, Ge HJ, Yang XQ, Sun X, Huang H, Tao
X, Chen WS and Li B: LncRNA-UCA1 exerts oncogenic functions in
non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett.
371:99–106. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yu T, Li J, Yan M, Liu L, Lin H, Zhao F,
Sun L, Zhang Y, Cui Y, Zhang F, et al: MicroRNA-193a-3p and −5p
suppress the metastasis of human non-small-cell lung cancer by
downregulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway.
Oncogene. 34:413–423. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li P, Zhang X, Wang H, Wang L, Liu T, Du
L, Yang Y and Wang C: MALAT1 is associated with poor response to
oxaliplatin-based chemotherapy in colorectal cancer patients and
promotes chemoresistance through EZH2. Mol Cancer Ther. 16:739–751.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang TH, Liang LZ, Liu XL, Wu JN, Su K,
Chen JY, Zheng QY, Huang HZ and Liao GQ: Long non-coding RNA MALAT1
interacts with miR-124 and modulates tongue cancer growth by
targeting JAG1. Oncol Rep. 37:2087–2094. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Malakar P, Shilo A, Mogilevsky A, Stein I,
Pikarsky E, Nevo Y, Benyamini H, Elgavish S, Zong X, Prasanth KV
and Karni R: Long non-coding RNA MALAT1 promotes hepatocellular
carcinoma development by SRSF1 upregulation and mTOR activation.
Cancer Res. 77:1155–1167. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu M, Sun W, Liu Y and Dong X: The role
of lncRNA MALAT1 in bone metastasis in patients with non-small cell
lung cancer. Oncol Rep. 36:1679–1685. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang R, Xia Y, Wang Z, Zheng J, Chen Y,
Li X, Wang Y and Ming H: Serum long non-coding RNA MALAT-1
protected by exosomes is upregulated and promotes cell
proliferation and migration in non-small cell lung cancer. Biochem
Biophys Res Commun. 490:406–414. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Goldstraw P, Chansky K, Crowley J,
Rami-Porta R, Asamura H, Eberhardt WE, Nicholson AG, Groome P,
Mitchell A, Bolejack V, et al: The IASLC lung cancer staging
project: Proposals for revision of the TNM stage groupings in the
forthcoming (Eighth) edition of the TNM classification for lung
cancer. J Thorac Oncol. 11:39–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-(Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang G, Lu X and Yuan L: LncRNA: A link
between RNA and cancer. Biochim Biophys Acta. 1839:1097–1109. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xue Y, Teng YQ, Zhou JD and Rui YJ:
Prognostic value of long noncoding RNA MALAT1 in various
carcinomas: Evidence from nine studies. Tumor Biol. 37:1211–1215.
2016. View Article : Google Scholar
|
|
24
|
Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY,
Zhang F, Wu LM, Chen LM and Zheng SS: Long non-coding RNA MALAT-1
overexpression predicts tumor recurrence of hepatocellular
carcinoma after liver transplantation. Med Oncol. 29:1810–1816.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ying L, Chen Q, Wang Y, Zhou Z, Huang Y
and Qiu F: Upregulated MALAT-1 contributes to bladder cancer cell
migration by inducing epithelial-to-mesenchymal transition. Mol
Biosyst. 8:2289–2294. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Schmidt LH, Görlich D, Spieker T, Rohde C,
Schuler M, Mohr M, Humberg J, Sauer T, Thoenissen NH, Huge A, et
al: Prognostic impact of Bcl-2 depends on tumor histology and
expression of MALAT-1 lncRNA in non-small-cell lung cancer. J
Thorac Oncol. 9:1294–1304. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shen L, Chen L, Wang Y, Jiang X, Xia H and
Zhuang Z: Long noncoding RNA MALAT1 promotes brain metastasis by
inducing epithelial-mesenchymal transition in lung cancer. J
Neurooncol. 121:101–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tano K, Mizuno R, Okada T, Rakwal R,
Shibato J, Masuo Y, Ijiri K and Akimitsu N: MALAT-1 enhances cell
motility of lung adenocarcinoma cells by influencing the expression
of motility-related genes. FEBS Lett. 584:4575–4580. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gutschner T, Hämmerle M, Eissmann M, Hsu
J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al:
The noncoding RNA MALAT1 is a critical regulator of the metastasis
phenotype of lung cancer cells. Cancer Res. 73:1180–1189. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Schmidt LH, Spieker T, Koschmieder S,
Schäffers S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D,
Marra A, et al: The long noncoding MALAT-1 RNA indicates a poor
prognosis in non-small cell lung cancer and induces migration and
tumor growth. J Thorac Oncol. 6:1984–1992. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhao Z, Chen C, Liu Y and Wu C:
17β-Estradiol treatment inhibits breast cell proliferation,
migration and invasion by decreasing MALAT-1 RNA level. Biochem
Biophys Res Commun. 445:388–393. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fang D, Yang H, Lin J, Teng Y, Jiang Y,
Chen J and Li Y: 17β-estradiol regulates cell proliferation, colony
formation, migration, invasion and promotes apoptosis by
upregulating miR-9 and thus degrades MALAT-1 in osteosarcoma cell
MG-63 in an estrogen receptor-independent manner. Biochem Biophys
Res Commun. 457:500–506. 2015. View Article : Google Scholar : PubMed/NCBI
|