Introduction
Malignant melanoma (MM) is an aggressive tumor
derived from melanocytes, which has a high degree of malignancy
with rapid progression and early metastasis. Patients with this
tumor have a poor prognosis and low survival rate (1,2). Despite
the majority of MMs originating from the skin (cutaneous
epithelia), they can arise in almost any part of the body.
Cutaneous melanoma, which comprises 91.2% of all melanoma cases, is
always associated with sun exposure (3). However, mucosal melanoma is another type
of MM that is observed in regions with low sun exposure. Mucosal
melanoma is rare, with the head and neck being the most common
sites, accounting for 1.4% of all melanomas (4). In total, ~6.5% of mucosal melanomas may
arise in the oral cavity and differ from their cutaneous
counterparts in terms of pathogenesis, biological behavior and
prognosis (5).
Oral mucosal melanoma (OMM) is estimated to account
for 0.2–8% of all MM cases, and appears to be more common in Asia,
including Japan and India, as compared with Western areas (6,7). In recent
years, the incidence of mucosal melanoma has increased
significantly, particularly in Asia. The prognosis of OMM is worse
compared with cutaneous melanoma, with a greater tendency to
metastasize and a 5-year survival rate of only 8–15% (5,7). In
contrast to cutaneous melanoma, which is associated with sun
exposure among other factors, the precise etiology of OMM has not
been defined due to various different influencing factors including
ingested tobacco, ingested alcohol and inhaled environmental
carcinogens, including smoke and formaldehyde (8). Although the use of tobacco, mechanical
trauma and denture use have been reported as possible risk factors
for OMM, the genetic etiology has not been extensively
investigated, and the pathogenesis remains unclear (7,9).
Considering its highly aggressive biological
behavior, the pathogenesis of melanoma has been examined in
numerous studies to identify more effective treatment approaches
(10–12). In 2002, Davies et al (13) reported that ~50% of all melanomas
harbored an activating mutation in BRAF. Among all
BRAF mutations, the V600E mutation accounted for >90% of
cases. This study led to a melanoma genomic revolution, and the
genetic etiology and potential therapeutic targets of this disease
have become a focus for research. A number of BRAF
inhibitors have been applied to clinical practice and have been
demonstrated to inhibit melanoma proliferation (11,12).
Melanoma arising in different parts of the body may
be associated with diverse molecular genetic profiles, suggesting
that those tumors may represent a distinct pathogenesis. In recent
years, an increasing number of studies have been focused on KIT,
BRAF, NRAS and other mutations. The KIT mutation is more
common in mucosal and acral melanomas than in cutaneous melanomas,
and the percentage of KIT mutations detected in mucosal
melanoma has been widely recognized to be 10–35% (14). The BRAF mutation is the most
common mutation in cutaneous melanoma with a high incidence.
However, several studies have demonstrated a low incidence of
BRAF mutation in melanomas arising from non-hair-bearing
skin, mucosa and internal organs that are totally sun protected,
while the BRAF mutation is also rare in uveal melanomas
(15–17). Furthermore, mutations in NRAS
have been detected in 15–25% of melanoma patients (18). The frequency of all these mutations
vary according to different methods of sourcing the analyzed tumor
and differences in processing methods, although there appears to be
no significant difference between cutaneous and mucosal
melanoma.
While the genetic etiology of cutaneous melanoma has
been widely reported, few studies have described the distribution
pattern of mutations in OMM and investigated the cause of its
variety. Thus, the aim of the present study was to detect the
presence of KIT, BRAF and NRAS mutations in 9 OMM
patients. In addition, exome sequencing was conducted to identify
the genes associated with OMM.
Materials and methods
Tumor samples and clinical
background
Information on the characteristics of the patients
included into the present study is listed in Table I. In total, 9 patients with a
histologically proven diagnosis of OMM were selected from the
Department of Oral and Maxillofacial Surgery, Peking University
Hospital of Stomatology (Beijing, China) between July 2009 and
November 2014. Diagnosis was based on the criteria provided by the
World Health Organization on the classification of head and neck
tumors (19). The study was reviewed
and approved by the Ethics Committee of the Peking University
Hospital of Stomatology (Beijing, China). Written informed consent
was obtained from all subjects prior to tissue collection.
 | Table I.Patient information. |
Table I.
Patient information.
| Patient | Age, years | Sex | Location | Lymph node
metastasis | Recurrent
melanoma |
|---|
| 1 | 61 | M | Upper gingiva | + | − |
| 2 | 69 | M | Upper gingiva | + | − |
| 3 | 57 | M | Upper gingiva | + | − |
| 4a | 56 | M | Upper and lower
gingiva | + | − |
| 5 | 44 | M | Lower gingiva | + | − |
| 6 | 47 | M | Lower gingiva | + | − |
| 7 | 61 | F | Buccal mucosa | + | + |
| 8 | 51 | M | Upper lip
vermilion | + | + |
| 9 | 42 | F | Lower gingiva | + | + |
Genomic DNA extraction and mutation
analysis
The tumor and para-carcinoma tissues were
transported to the Center Laboratory (Peking University Hospital of
Stomatology) within 30 min of surgery in 4°C Krebs-Ringer Hepes
(KRH) solution (containing 120 mM NaCl, 5.4 mM KCl, 1 mM
CaCl2, 0.8 mM MgCl2, 11.1 mM glucose and 20
mM HEPES, pH 7.4) and aerated with 95% O2. For DNA
isolation, tissues were minced and homogenized in 80 µl PBS using
Tissue Ruptor (IKA, Staufen, Germany). Then, the genomic DNA was
extracted using a QIAamp DNA Mini kit (Qiagen, Hilden, Germany),
according to the manufacturer's protocols. The concentration of the
extracted DNA was determined using the NanoDrop 8000
spectrophotometer (Thermo Fisher Scientific, Inc., Waltham, MA,
USA). Exons 11 and 15 of the BRAF gene (NG_007873.3), exons
1 and 2 of the NRAS gene (AH001530.2), and exons 11 and 13
of the KIT gene (NG_007456.1) were amplified by polymerase
chain reaction (PCR) with six pairs of primers that covered the
entire coding region of OMM. Primer sequences from previous studies
used for PCR amplification are presented in Table II (20,21). PCR
assays were performed in a 30 µl reaction with KOD-plus enzyme
(Toyobo Life Science, Osaka, Japan), as previously described
(22). Following an initial
denaturation step at 95°C for 5 min, 30 cycles of amplification
consisting of denaturation at 94°C for 1 min, annealing at 55°C for
30 sec and extension at 72°C for 45 sec were conducted, followed by
a final extension step at 72°C for 10 min. The PCR products were
separated by 1% agarose gel electrophoresis and purified using an
AxyPrepDNA gel Extraction kit (Axygen Scientific, Inc., Union City,
CA, USA). Products were then sequenced using an ABI 3730XL DNA
analyzer (Applied Biosystems; Thermo Fisher Scientific, Inc.).
Mutations were analyzed using Polymorphism Phenotyping (version 2;
http://genetics.bwh.harvard.edu/pph2/dbsearch.shtml)
and the probability of missense mutations being damaging based on
scores allocated to a combination of properties (including
single-nucleotide polymorphisms (SNPs), maps coding SNPs to gene
transcripts, extraction of protein sequence annotations and
structural attributes, and construction of conservation profiles)
was estimated, with a score closer to 1 indicating a greater
incidence of missense mutation (23).
 | Table II.Primer sequences of KIT, BRAF
and NRAS exons used in polymerase chain reaction. |
Table II.
Primer sequences of KIT, BRAF
and NRAS exons used in polymerase chain reaction.
| Gene | Exon | Size (bp) | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| KIT | 11 | 289 |
TGTTCTCTCTCCAGAGTGCTCTAA |
AAACAAAGGAAGCCACTGGA |
| KIT | 13 | 294 |
CATCAGTTTGCCAGTTGTGC |
AGCAAGAGAGAACAACAGTCTGG |
| BRAF | 11 | 204 |
CTCTCAGGCATAAGGTAATG |
CACTTTCCCTTGTAGACTGTT |
| BRAF | 15 | 209 |
CCTAAACTCTTCATAATGCTT |
ATAGCCTCAATTCTTACCAT |
| NRAS | 1 | 174 |
CGCCAATTAACCCTGATTACT |
CACTGGGCCTCACCTCTA |
| NRAS | 2 | 196 |
CCCCTTACCCTCCACAC |
AGGTTAATATCCGCAAATGAC |
Exome sequencing and variant
calling
The genomic DNAs of patients no. 6 and 10 (tumor and
para-carcinoma tissue), as presented in Table I, were randomly fragmented into
150–200 bp. Next, library construction, exome capture and
sequencing were performed according to the protocol of the Sure
SelectXT Target Enrichment System for Illumina Paired-End
Sequencing Library, Illumina HiSeq and MiSeq Multiplexed Sequencing
Platforms (Illumina, Inc., San Diego, CA, USA). Sequencing by
synthesis was performed on a HiScanSQ sequencer (Illumina, Inc.)
using the paired-end method. BAM files summarizing Burrows-Wheeler
alignments that were mapped to the hg19 reference sequence were
generated using the Genome Analysis Toolkit (GATK) (22,23). The
raw reads with all characteristics that were not confidently
aligned, exhibited a ratio of N >10%, a length <25 bp without
adapter and a quality score of <20 by Phred-scaled mapping were
excluded from further analysis. The single nucleotide variants
(SNVs), insertions and deletions were detected and annotated by the
GATK and Annovar software (release date Feburay 1st, 2016;
http://www.openbioinformatics.org/annovar/) (24).
Results
KIT, BRAF and NRAS mutations in
patients with OMM
Among the 9 patients included into the present
study, there were 7 males and 2 females with a median age of 54
years (age range, 42–69 years). Out of the 9 melanoma samples, 6
were obtained from the gingiva, 1 from the hard palate, 1 in the
upper lip vermilion and 1 in the buccal mucosa (Table I).
Among the 9 primary oral mucosa melanoma samples
that were analyzed in the present study, 3 patients (33.3%)
exhibited KIT, BRAF or NRAS mutations. KIT,
BRAF and NRAS mutations were each identified in 1/9
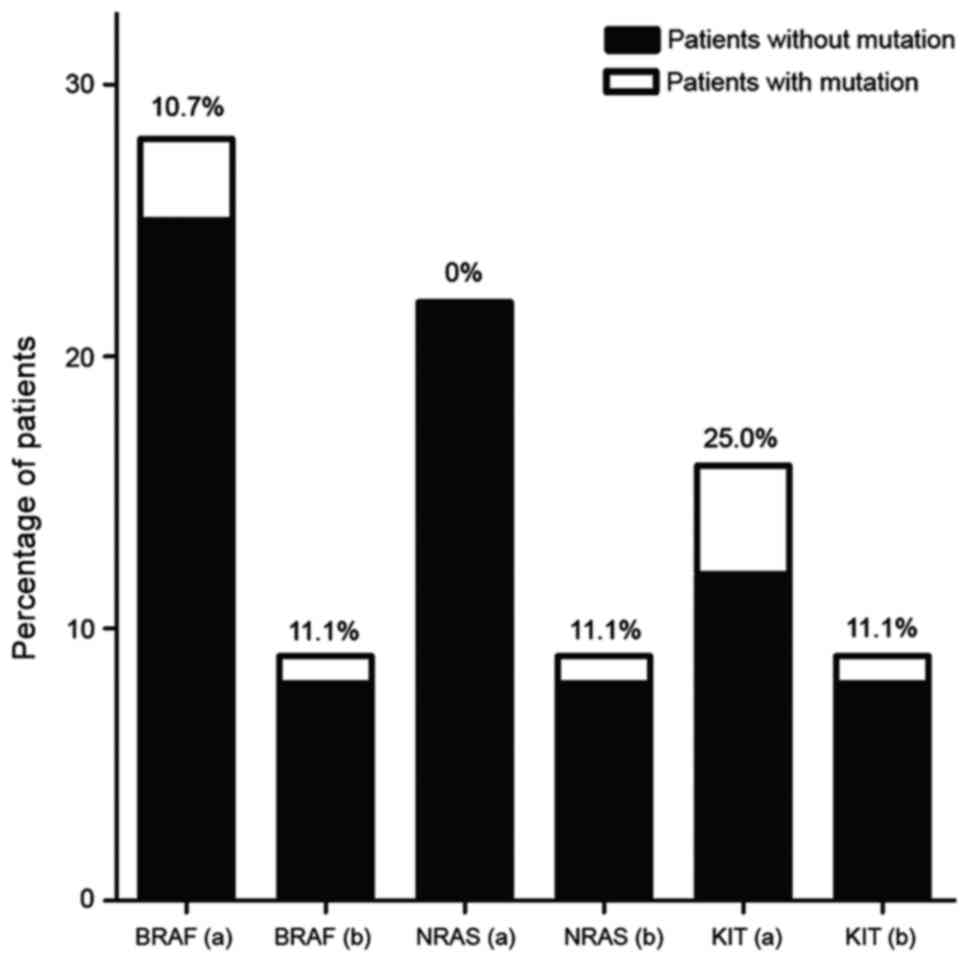
patients (11.1%; Fig. 1).
Furthermore, a novel mutation was identified, namely L589M in exon
11 of the KIT gene, which had not been previously detected
in melanomas or any other tumor types. The alteration in the
BRAF gene was detected in exon 15 (D594G), which has been
described in arcal melanoma (25).
The NRAS mutation involved codon 59, and although a mutation
at this position has been previously identified (25), a novel amino acid substitution (A59T)
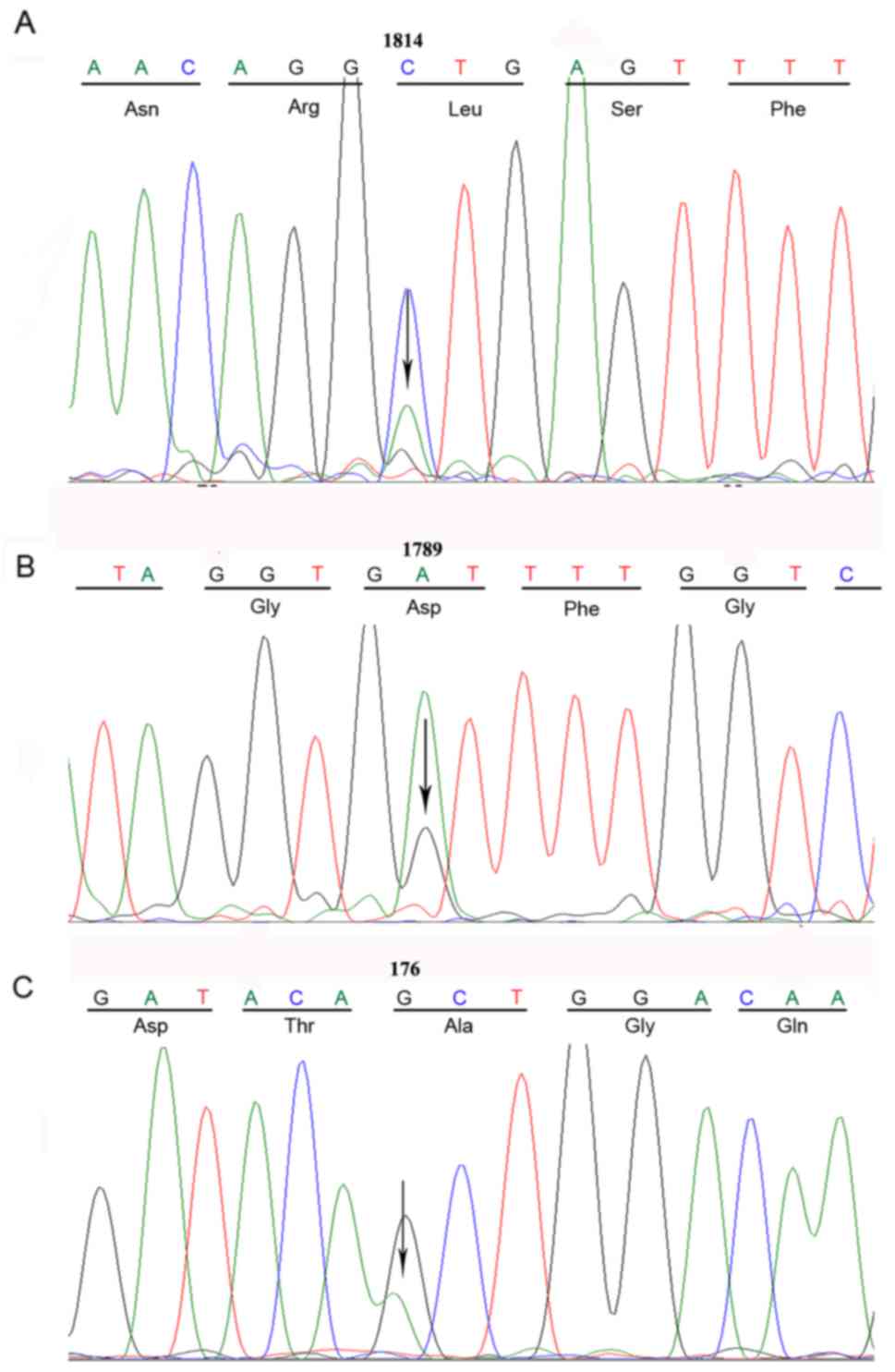
was detected in the current analysis (Table III and Fig. 2). No mutations in the adjacent tissues
were observed.
 | Table III.KIT, BRAF and NRAS gene
mutations in oral mucosal melanoma. |
Table III.
KIT, BRAF and NRAS gene
mutations in oral mucosal melanoma.
| Case no. | Age/sex | Gene | Exon no. | Nucleotide
definition | Amino acid
definition | Functional
effect | Polyphen-2
score |
|---|
| 9 | 61/F | KIT | 11 | c.1814C>A | p.L589M | Missense | 0.981 |
| 1 | 61/M | BRAF | 15 | c.1789A>G | p.D594G | Missense | 0.983 |
| 7 | 44/M | NRAS | 2 | c.176G>A | p.A59T | Missense | 0.745 |
Comparison between mutation analysis
and literature review results
The present study performed a systematic search of
studies published between July 1969 and March 2015 (https://www.ncbi.nlm.nih.gov/pubmed). Additional
information was identified by searching the reference lists of the
included publications. Inclusion criteria for the literature review
were as follows: i) Studies that included the key words ‘oral and
melanoma and (mutation OR mutated)’; ii) studies that examined gene
mutations in patients with oral malignant mucosal melanoma; iii)
studies that were only published in English. A preferred reporting
items for systematic reviews and meta-analyses flowchart for
literature attrition was also included (26). Only a single entry was made if the
same mutation or patient was reported across multiple studies.
Separate entries were made for each patient when the same mutation
was detected in unrelated patients. In the systematic review
conducted in the present study, a total of five studies (5,6,14,27,28) on
gene mutations in OMM were identified, which satisfied the
eligibility criteria. The distribution pattern of mutated genes is
summarized in Fig. 1. The KIT,
NRAS and BRAF mutations were detected in 3/28 (10.7%),
0/22 (0.0%), and 4/16 (25.0%) patients with OMM, respectively
(5,6,14,25,26). In
these articles, the 11 mutations observed were distributed on 5
exons (including exons 1, 2, 11, 13 and 15), of which there were 4
synonymous mutations and 6 missense mutations (Table IV). Of the 6 KIT mutations, 2
cases harbored the same mutation in exon 13, leading to a
replacement in position 642 of a lysine by a glutamic acid (K642E)
(6,29). In addition, 2 synonymous mutations
(S645S and P585P) were detected in these 2 cases; however, they did
not result in amino acid changes (28). A single mutation occurred at codon 557
in exon 11 and resulted in the replacement of a tryptophan by a
glutamic acid L-arginine (W557R), which has already been previously
described in gastrointestinal stromal tumors (29). Furthermore, the V569G mutation was
identified in 1 case (28), which has
not otherwise been previously reported. In regards to BRAF,
the classical V600E substitution that is typically identified in
cutaneous melanoma was detected in 2 patients previously (5,13). There
was also another mutation located at codon 600, namely the V600L
mutation, resulting in the replacement of the valine by leucine
(6). No missense mutation of
NRAS was reported in a previous study investigating OMM, but
two synonymous mutations, K166K and F66F, were
demonstrated in 2 patients (6).
 | Table IV.KIT, BRAF and NRAS
genes in oral mucosal melanoma mutations from the literature. |
Table IV.
KIT, BRAF and NRAS
genes in oral mucosal melanoma mutations from the literature.
| Authors, year | Gene | Exon no. | Amino acid
definition | Functional
effect | (Refs.) |
|---|
| Cohen et al,
2008 | KIT | 11 | W557R, V569G | Missense | 27 |
| Cohen et al,
2008 |
| 13 | K642E | Missense | 27 |
|
|
|
| S645S, P585P | Synonymous |
|
| Soma et al,
2014 | BRAF | 15 | V600L, V600E | Synonymous | 5 |
| Buery et al,
2011 |
|
|
|
| 6 |
| Buery et al,
2011 | NRAS | 2 | K166K, F66F | Synonymous | 6 |
FMNL2 mutations detected by exome
sequencing
Exome sequencing was used to identify the potential
pathogenic gene mutation in 6/9 patients in the present study,
which were revealed not to be mutations in the KIT, BRAF or
NRAS genes. However, only 2 of the 6 enrolled patients met
the sequencing requirements. The exome sequencing in these 2 cases
resulted in 2.1×108 reads, yielding >99% exome
coverage, with the exome average depth of each sample being between
58 and 70%. A total of 37 SNVs, as well as 5 insertion and deletion
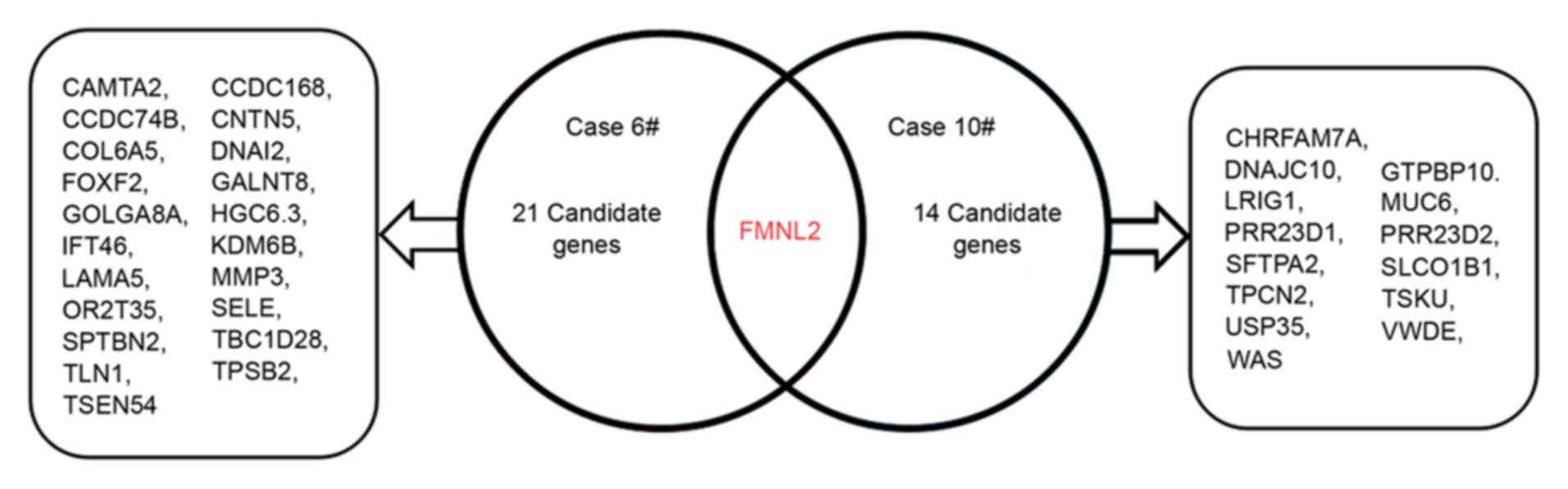
mutations, were detected in the 4 samples, while 22 mutations were
observed in patient no. 6 and 15 mutations in patient no. 10. These
mutations were only detected in the pathologic tissue, but not in
the para-carcinoma tissue. More detailed information is listed in
Fig. 3. These results revealed that
OMM has an increased number of somatic mutations. Among the 37
SNVs, there was a FMNL2 gene detected in the 2 patients
(Fig. 3), which demonstrated a
non-frame-shift insertion mutation, and these insertion mutations
involved the area of exon 15. While this mutation was predicted to
have no influence on the protein level, it may serve a role in
transcription.
Discussion
The RAS/RAF/mitogen-activated protein kinase
kinase/extracellular signal-regulated kinase (ERK), also known as
the mitogen-activated protein kinase (MAPK)/ERK pathway is the most
common cell-signaling pathway in the progression of melanoma by
regulating cell proliferation, differentiation and survival
(30). The BRAF gene, encoding
a serine/threonine protein kinase, is a well-known downstream
molecule participating in this signal cascade. The V600E
(Val600Glu) mutation that accounts for >90% of all BRAF
mutations in cutaneous melanoma has been demonstrated to be
activated by the RAS-guanosine triphosphate (GTP) protein, leading
to ERK activation and stimulating the growth of melanoma cells
(13,31). Another molecule that leads to the
activation of the MAPK pathway is NRAS, leading to
accumulation of RAS-GTP and affecting various downstream molecules,
including phosphoinositide 3-kinase and RAF kinases (32,26). Codon
61 is the major position for NRAS alterations in melanoma,
such as Q61H, Q61K and Q61L (32).
Furthermore, the KIT gene, one of the receptor tyrosine
kinases, is an important receptor for melanocyte proliferation and
migration (33). The MAPK pathway may
also be triggered by the activation of a KIT mutation, which
participates in melanoma development through the induction of
signaling proteins (34).
The KIT and BRAF mutations detected in
the present study are consistent with the results of the literature
review, while the mutation rate of NRAS was slightly
different. This discrepancy in the results may be due to the
inclusion of adjacent tissues in the samples used in previous
studies. In addition, the small sample sizes utilized in the
present study and systematic review may be another cause of this
difference. However, in general, the results of the current study
corresponded with the findings of previous studies included in the
systematic review. The BRAF and NRAS mutations are
the main genetic mutations in cutaneous melanoma cases, with a high
incidence rate of 30–70 and 15–25%, respectively (15–18). In
contrast, a low incidence of BRAF and NRAS mutations
has been described in mucosal melanoma patients. It appears that
the BRAF mutation is quite rare in mucosal melanoma, whereas
the NRAS mutation has a higher frequency in comparison with
BRAF in this melanoma, although its percentage remains
<20%. With regard to the mutation frequency in OMM, 1/9 patients
(11.1%) presented a BRAF mutation, while the same incidence
(1/9; 11.1%) of NRAS mutations was detected in the present
study. According to the results of the systematic review, 3/28 OMM
patients (10.7%) were reported to exhibit a BRAF mutation,
while no missense mutation of NRAS was reported among the 22
OMM patients (0.0%) included in previous studies (6,25). Thus,
it is clear that the BRAF mutation in OMM occurs with a low
incidence of <10%, which is consistent with the observations of
previous studies on mucosal melanoma (20,35,36). The
distinction of BRAF mutations existing between cutaneous
melanoma and OMM may therefore reveal a different molecular
pathogenesis between these two melanoma types, and the different
frequency of the NRAS mutation in mucosal melanoma and OMM
may indicate that OMM has a distinctive pathogenesis and is a
separate subtype. Furthermore, the novel NRAS mutation
(namely A59T) detected in the current study may provide a molecular
target for further investigation. Additionally, missense mutations
of NRAS (p.S39F) with a relatively low peak on the
sequencing map were identified in 6 patients, which may be
associated with the tumor heterogeneity phenomenon.
Prevalence differences in KIT mutations
between melanomas were observed at different sites (20,35), with
KIT mutations occurring less frequently in head and neck
melanomas. This observation was also supported by the study of
Beadling et al (36), who
reported a higher incidence of KIT mutations in melanomas of
the vulva, anorectum and vagina when compared with melanomas
occurring at the head and neck sites. In addition, Schoenewolf
et al (37) identified a high
frequency of KIT mutations (45%) in vulvovaginal melanomas
and no mutation in 12 sinonasal melanomas. On the basis of these
previous findings, the frequency of KIT mutations in head
and neck melanomas does not appear to be consistent, and this may
be associated with differences in the sample quantity and the
geographical region of patients. The two novel mutations detected
by sequencing in the present study, namely p.L589M of KIT
and p.A59Tof NRAS, have not been reported in primary mucosa
melanoma. These results indicate that oral mucosa melanoma has
different characteristics as compared with other mucosa melanomas.
Although the significance of these novel mutations has yet to be
revealed, the newly identified mutated sites may suggest a
potential path to examine the mechanism underlying OMM
pathogenesis.
In the present study cohort, 1 patient with a
KIT mutation was observed, as well as 1 patient with an
NRAS mutation and 1 patient with a BRAF mutation. The
mutation rate of all the 3 genes was 1/9 (11.1%), and this result
was consistent with previous OMM studies identified in the
systematic review (5,6,14,28,29),
indicating that mutations of the KIT, NRAS and BRAF
are associated with the occurrence of OMM. When compared with
cutaneous melanoma, the distribution of KIT and BRAF
mutations is markedly different in OMM (15,18). In
contrast to its role in cutaneous melanoma, the mutated KIT
serves a more significant role in comparison with the BRAF
gene in the occurrence of OMM (13,31).
However, on the basis of these results, the mutation rates of all
the 3 genes are not sufficiently enough to support a definite
relevance to OMM pathogenesis, as the highest rate was 26.7% for
KIT (28). Therefore, it
appears that none of the three mutations were the main genetic
factors leading to OMM.
In order to reveal other factors involved in the
pathogenesis of OMM, exome sequencing of candidate genes was
performed in the current study, and the FMNL2 gene was
considered as a possible key molecule. This gene, as an encoding
formin-associated protein gene, is essential in morphogenesis,
cytokines is and cell polarity. The protein is strongly expressed
in the central nervous system and numerous epithelia, whereas a
lack of expression may cause weak immunoreactivity in certain
mesenchymal cell types (38).
Furthermore, it has been reported that the gene is associated with
melanoma and colorectal tumors (38,39). In
cultured melanoma cells, FMNL-2 co-localizes with F-actin at the
tips of cellular protrusions, which supports the hypothesis that
the gene is associated with the formation of actin filament in
cellular protrusions during cellular migration. In consequence, the
FMNL2 mutation may be regarded as a potential cause of the
oral melanoma in these patients.
In conclusion, despite a small cohort and a lack of
extensive experimental repeat, the present study also revealed that
KIT, NRAS and BRAF mutations may be associated with
the occurrence of OMM, and that the FMNL2 mutations may be
regarded as a potential cause of OMM. The present study may be of
benefit for elucidating the underlying mechanism of OMM, however,
further studies are required to validate these conclusions.
Acknowledgements
The present study was supported by grants from the
National Natural Science Foundation of China (grant nos. 11275621,
81200762 and 81200799).
Glossary
Abbreviations
Abbreviations:
|
MM
|
malignant melanoma
|
|
OMM
|
oral mucosal melanoma
|
|
MAPK
|
mitogen-activated protein kinase
|
References
|
1
|
Balch CM, Gershenwald JE, Soong SJ,
Thompson JF, Atkins MB, Byrd DR, Buzaid AC, Cochran AJ, Coit DG,
Ding S, et al: Final version of 2009 AJCC melanoma staging and
classification. J ClinOncol. 27:6199–6206. 2009. View Article : Google Scholar
|
|
2
|
Nikolaou V and Stratigos AJ: Emerging
trends in the epidemiology of melanoma. Br J Dermatol. 170:11–19.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gandini S, Sera F, Cattaruzza MS, Pasquini
P, Picconi O, Boyle P and Melchi CF: Meta-analysis of risk factors
for cutaneous melanoma: II. Sun exposure. Eur J Cancer. 41:45–60.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mihajlovic M, Vlajkovic S, Jovanovic P and
Stefanovic V: Primary mucosal melanomas: A comprehensive review.
Int J Clin Exp Pathol. 5:739–753. 2012.PubMed/NCBI
|
|
5
|
Soma PF, Pettinato A, Agnone AM, Donia C,
Improta G and Fraggetta F: Oral malignant melanoma: A report of two
cases with BRAF molecular analysis. Oncol Lett. 8:1283–1286. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Buery RR, Siar CH, Katase N, Gunduz M,
Lefeuvre M, Fujii M, Inoue M, Setsu K and Nagatsuka H: NRAS and
BRAF mutation frequency in primary oral mucosal melanoma. Oncol
Rep. 26:783–787. 2011.PubMed/NCBI
|
|
7
|
Meleti M, Leemans CR, Mooi WJ, Vescovi P
and van der Waal I: Oral malignant melanoma: A review of the
literature. Oral Oncol. 43:116–121. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rapini RP, Golitz LE, Greer RO Jr,
Krekorian EA and Poulson T: Primary malignant melanoma of the oral
cavity. A review of 177 cases. Cancer. 55:1543–1551. 1985.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gu GM, Epstein JB and Morton TH Jr:
Intraoral melanoma: Long-term follow-up and implication for dental
clinicians. A case report and literature review. Oral Surg Oral Med
Oral Pathol Oral Radiol Endod. 96:404–413. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Eggermont AM and Robert C: Melanoma: Smart
therapeutic strategies in immuno-oncology. Nat Rev Clin Oncol.
11:181–182. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sabbatino F, Wang Y, Wang X, Ferrone S and
Ferrone CR: Emerging BRAF inhibitors for melanoma. Expert Opin
Emerg Drugs. 18:431–443. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Romano E, Schwartz GK, Chapman PB,
Wolchock JD and Carvajal RD: Treatment implications of the emerging
molecular classification system for melanoma. Lancet Oncol.
12:913–922. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Davies H, Bignell GR, Cox C, Stephens P,
Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W,
et al: Mutations of the BRAF gene in human cancer. Nature.
417:949–954. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Colombino M, Lissia A, Franco R, Botti G,
Ascierto PA, Manca A, Sini MC, Pisano M, Paliogiannis P, Tanda F,
et al: Unexpected distribution of cKIT and BRAF mutations among
southern italian patients with sinonasal melanoma. Dermatology.
226:279–284. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wong CW, Fan YS, Chan TL, Chan AS, Ho LC,
Ma TK, Yuen ST and Leung SY: Cancer Genome Project: BRAF and NRAS
mutations are uncommon in melanomas arising in diverse internal
organs. J Clin Pathol. 58:640–644. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cohen Y, Rosenbaum E, Begum S, Goldenberg
D, Esche C, Lavie O, Sidransky D and Westra WH: Exon 15 BRAF
mutations are uncommon in melanomas arising in nonsun-exposed
sites. Clin Cancer Res. 10:3444–3447. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Edwards RH, Ward MR, Wu H, Medina CA,
Brose MS, Volpe P, Nussen-Lee S, Haupt HM, Martin AM, Herlyn M, et
al: Absence of BRAF mutations in UV protected mucosal melanomas. J
Med Genet. 41:270–272. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Uhara H, Ashida A, Koga H, Ogawa E,
Uchiyama A, Uchiyama R, Hayashi K, Kiniwa Y and Okuyama R: NRAS
mutations in primary and metastatic melanomas of Japanese patients.
Int J Clin Oncol. 19:544–548. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Thompson L: World health organization
classification of tumours: Pathology and genetics of head and neck
tumours. Ear Nose Throat J. 85:742006.PubMed/NCBI
|
|
20
|
Curtin JA, Busam K, Pinkel D and Bastian
BC: Somatic activation of KIT in distinct subtypes of melanoma. J
Clin Oncol. 24:4340–4346. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ellerhorst JA, Greene VR, Ekmekcioglu S,
Warneke CL, Johnson MM, Cooke CP, Wang LE, Prieto VG, Gershenwald
JE, Wei Q and Grimm EA: Clinical correlates of NRAS and BRAF
mutations in primary human melanoma. Clin Cancer Res. 17:229–235.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qu J, Yu F, Hong Y, Guo Y, Sun L, Li X,
Zhang J, Zhang H, Shi R, Chen F and Li T: Underestimated PTCH1
mutation rate in sporadic keratocysticodontogenic tumors. Oral
Oncol. 51:40–45. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Adzhubei I, Jordan DM and Sunyaev SR:
Predicting functional effect of human missense mutations using
polyphen-2. Curr Protoc Hum Genet: Chapter 7. Unit7.20. 2013.
|
|
24
|
Wang K, Li M and Hakonarson H: ANNOVAR:
Functional annotation of genetic variants from next-generation
sequencing data. Nucleic Acids Res. 38:e1642010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Si L, Kong Y, Xu X, Flaherty KT, Sheng X,
Cui C, Chi Z, Li S, Mao L and Guo J: Prevalence of BRAF V600E
mutation in Chinese melanoma patients: Large scale analysis of BRAF
and NRAS mutations in a 432-case cohort. Eur J Cancer. 48:94–100.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Moher D, Liberati A, Tetzlaff J and Altman
DG: PRISMA Group: Preferred reporting items for systematic reviews
and meta-analyses: The PRISMA statement. PLoS Med. 6:e10000972009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cohen Y, Goldenberg-Cohen N, Akrish S,
Shani T, Amariglio N, Dratviman-Storobinsky O, Kaplan I, Barshack I
and Hirshberg A: BRAF and GNAQ mutations in melanocytic tumors of
the oral cavity. Oral Surg Oral Med Oral Pathol Oral Radiol.
114:778–784. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rivera RS, Nagatsuka H, Gunduz M, Cengiz
B, Gunduz E, Siar CH, Tsujigiwa H, Tamamura R, Han KN and Nagai N:
C-kit protein expression correlated with activating mutations in
KIT gene in oral mucosal melanoma. Virchows Arch. 452:27–32. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Antonescu CR, Viale A, Sarran L,
Tschernyavsky SJ, Gonen M, Segal NH, Maki RG, Socci ND, DeMatteo RP
and Besmer P: Gene expression in gastrointestinal stromal tumors is
distinguished by KIT genotype and anatomic site. Clin Cancer Res.
10:3282–3290. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Peyssonnaux C and Eychène A: The
Raf/MEK/ERK pathway: New concepts of activation. Biol Cell.
93:53–62. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Platz A, Egyhazi S, Ringborg U and Hansson
J: Human cutaneous melanoma; a review of NRAS and BRAF mutation
frequencies in relation to histogenetic subclass and body site. Mol
Oncol. 1:395–405. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Malumbres M and Barbacid M: RAS oncogenes:
The first 30 years. Nat Rev Cancer. 3:459–465. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Alexeev V and Yoon K: Distinctive role of
the cKit receptor tyrosine kinase signaling in mammalian
melanocytes. J Invest Dermatol. 126:1102–1110. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lennartsson J, Jelacic T, Linnekin D and
Shivakrupa R: Normal and oncogenic forms of the receptor tyrosine
kinase kit. Stem Cells. 23:16–43. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Omholt K, Grafström E, Kanter-Lewensohn L,
Hansson J and Ragnarsson-Olding BK: KIT pathway alterations in
mucosal melanomas of the vulva and other sites. Clin Cancer Res.
17:3933–3942. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Beadling C, Jacobson-Dunlop E, Hodi FS, Le
C, Warrick A, Patterson J, Town A, Harlow A, Cruz F III, Azar S, et
al: KIT gene mutations and copy number in melanoma subtypes. Clin
Cancer Res. 14:6821–6828. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Schoenewolf NL, Bull C, Belloni B,
Holzmann D, Tonolla S, Lang R, Mihic-Probst D, Andres C and Dummer
R: Sinonasal, genital and acrolentiginous melanomas show distinct
characteristics of KIT expression and mutations. Eur J Cancer.
48:1842–1852. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gardberg M, Talvinen K, Kaipio K, Iljin K,
Kampf C, Uhlen M and Carpén O: Characterization of
diaphanous-related formin FMNL2 in human tissues. BMC Cell Biol.
11:552010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhu XL, Zeng YF, Guan J, Li YF, Deng YJ,
Bian XW, Ding YQ and Liang L: FMNL2 is a positive regulator of cell
motility and metastasis in colorectal carcinoma. J Pathol.
224:377–388. 2011. View Article : Google Scholar : PubMed/NCBI
|