Introduction
Human gastric cancer is the fourth most common
malignancy and the second leading cause of cancer-related deaths
worldwide (1). Adenocarcinoma is the
most common pathological type of gastric cancer, while lymphoma,
carcinoid, and sarcoma constitute <5% of the pathology (2). The pathogenesis of gastric cancer is
complicated owing to the interaction of multiple factors, including
Helicobacter pylori infection, environment, and heredity.
The environmental factors play critical roles in the pathogenesis
of gastric cancer; the major risk factors include smoking and diet
(3). However, determining the
molecular markers of gastric cancer is yet a great challenge.
Recently, the identification and characterization of
circular RNA (circRNA) have revolutionized the field of RNA.
CircRNAs have gained increasing attention in deciphering the
complicated mechanisms underlying the malignant processes such as
tumorigenesis, multidrug resistance, invasion, and metastasis.
Although circRNAs have been reported as early as 20 years ago
(4), they were mostly misinterpreted
as splicing artefacts or gene rearrangements. Following
high-throughput RNA sequencing and bioinformatics, thousands of
different circRNAs have been rediscovered in the recent several
years (5–8). Preliminary data revealed that circRNAs
were abundantly expressed and evolutionarily conserved across the
eukaryotes and functioned as miRNA sponges (5,6,9). Cdr1as (also known as ciRS-7), as the
maximally studied circRNA, was reported as the miR-7 sponge or
inhibitor (5,6).
ciRS-7 was highly expressed in a wide variety of
cancer cell lines, and ciRS-7/miR-7 network suggested a therapeutic
potential for carcinoma. This network might regulate the majority
of the cancer pathways such as p21-activated kinase 1 (Pak1)
(10), epidermal growth factor
receptor (EGFR) (11), activated
cdc42-associated kinase 1 (Ack1) (12), and phosphoinositide 3-kinase catalytic
subunit delta (PIK3CD) (13).
Increasing number of evidence indicated miR-7 as a potential tumor
suppressor in several human cancers. Xiong et al reported
that miR-7 selectively induced growth suppression and apoptosis of
non-small cell lung cancer (NSCLC) by targeting B-cell lymphoma-2
(BCL-2) in vitro (14).
Similarly, miR-7 was confirmed as a novel miRNA exhibiting tumor
suppression function in colon cancer (15). Although the majority of reports
supported the tumor-enhancing effect of circRNAs, the converse was
also reported. ciR-ITCH demonstrated an inhibitory effect on
esophageal squamous cell carcinoma, acting as a sponge of miR-7,
miR-17, and miR-214 via the regulation of the Wnt/β-catenin pathway
(16). Altogether, these results
suggested that the relationship between circRNA and cancer was
complicated and precise mechanisms needed further
investigation.
In this study, we presented the circRNAs' expression
profile in normal gastric tissue and gastric adenocarcinoma through
microarray technology in order to explore the function of circRNAs
in gastric cancer for early diagnosis and treatment of cancer.
Materials and methods
Patient samples
The present study was approved by the Research
Ethics Committee of the Affiliated Drum Tower Hospital of Nanjing
University, Medical School, and all patients provided informed
consent before the samples were collected. Gastric cancer was
confirmed by histopathological diagnosis. Finally, 15 patients
(eight men, seven women; mean age 64.1 years, range 48–81) were
enrolled and 15 pairs of gastric carcinoma tissues and normal
para-carcinoma samples were collected. All samples were rapidly
frozen in liquid nitrogen and stored at −80°C for subsequent
investigation. For circRNA microarray analysis, a total of 3
gastric carcinoma and 3 normal para-carcinoma gastric tissues
(control) were randomly selected for the study.
RNA extraction
Total RNA was extracted from the frozen tissue block
that was homogenized (IKA Werke GmbH & Co. KG, Staufen,
Staufen, Germany) and resuspended in TRIzol reagent (Invitrogen,
Carlsbad, CA, USA), according to the manufacturer's instructions.
Then, the total RNA was quantified using a NanoDrop ND-1000
spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
Additionally, the RNA integrity was assessed by denaturing agarose
gel electrophoresis.
RNA labeling and array
hybridization
The sample labeling and microarray hybridization
were performed by KangChen Biotech (Shanghai, China). Briefly, the
circRNA was treated with RNase R (Epicenter Biotechnologies, USA)
to remove the linear RNAs. Each sample was amplified and
transcribed into fluorescent cRNA using a random priming method
(Arraystar Super RNA Labeling kit; Arraystar Inc., Rockville, MD,
USA). Subsequently, these labeled cRNAs were purified by RNeasy
Mini kit (cat. no. 74106, Qiagen GmbH, Hilden, Germany Germany),
and the concentration and specific activity were measured by
NanoDrop (NanoDrop Technologies). Then, 1 µg of each labeled cRNA
was fragmented by adding 5 µl of 10X blocking agent and 1 µl of 25X
fragmentation buffer, followed by heating at 60°C for 30 min. Then,
25 µl of 2X hybridization buffer was added to dilute the labeled
cRNA, and 50 µl of the hybridization solution was dispensed into
the gasket slide and assembled on the circRNA expression microarray
slide. These slides were incubated for 17 h at 65°C in a
hybridization oven (Agilent). The hybridized arrays were washed,
fixed, and scanned using the Axon GenePix 4000B microarray scanner
(Molecular Devices, LLC, Sunnyvale, CA, USA).
Microarray analysis
The Arraystar Human Circular RNA Microarray V2.0
(Arraystar, Inc.) was designed for the purpose of profiling both
circRNAs and protein-coding RNAs in the human genome. The
differentially expressed circRNAs were identified through
fold-change filtering and standard Student's t-test. The circRNAs
are exhibiting a fold-change ≥2.0 and a P-value <0.05 were
selected as significantly differentially expressed circRNAs.
Quantitative reverse transcription
PCR
cDNA samples were prepared from total RNA of gastric
tissues by reverse transcription. In total, 20 circRNAs were
analyzed by SYBR Green I dye-based detection with specific primer
sequences. The primer sequences were shown in Table I. The 2−ΔΔCq method was
performed in 18 samples and applied for the quantification of the
relative expression of circRNAs that was normalized against the
expression of the housekeeping gene, GAPDH.
 | Table I.Primer sequences for reverse
transcription-polymerase chain reaction. |
Table I.
Primer sequences for reverse
transcription-polymerase chain reaction.
| Gene | Direction | Primer sequence |
|---|
| hsa_circ_0001017 | Forward |
AGTGCGAAGTAATCTATGCCAGC |
|
| Reverse |
AGCCATTCTTTGCTGGGCTC |
| hsa_circ_0001772 | Forward |
GCCAGAGGAGGAGCAGCTTTA |
|
| Reverse |
GCTCTTCATCTGACAAATCCGAC |
| hsa_circ_0002346 | Forward |
GTGCAAACCAGTTTTCGGCG |
|
| Reverse |
TCCAGTTCTCATCTTGTTGGCA |
| hsa_circ_0000072 | Forward |
TTGGCAGCAAATGGAGTTCGT |
|
| Reverse |
GTGCCTGCCACCATTTCCTTA |
| hsa_circ_0003221 | Forward |
GATGCGGGGCAATGCACTA |
|
| Reverse |
ACCAGTACCCAGGTGAGTCTT |
| hsa_circ_0001865 | Forward |
GCTCCACAGACTTCCCAGAGT |
|
| Reverse |
GGCAAGTTCCAACGTCTCCT |
| hsa_circ_0003441 | Forward |
ACCACAGTTCTTGGTGGTGAAG |
|
| Reverse |
TGACTTTGTCTGGAGAGCTTGTG |
| hsa_circ_0008285 | Forward |
GCTGTTAACGGGAAAGGTTGAA |
|
| Reverse |
GCGTCTGTTGAAGTCGTGGA |
| hsa_circ_0023923 | Forward |
AGCACATCAAAGCTGCCCAA |
|
| Reverse |
TGCACTGAATTAAGTCTCCCCA |
| hsa_circ_0000347 | Forward |
GAAAAAGAACCAATGCAAAGAAGGT |
|
| Reverse |
GCACTGAATTAAGTCTCTGCAACT |
| hsa_circ_0046881 | Forward |
AAGTCAGGCAGCTTTGCTGG |
|
| Reverse |
CACAGTTGGTTAGCCACAGC |
| hsa_circ_0023940 | Forward |
ATGCTCCTGTTCAAAGATGCCA |
|
| Reverse |
TTTGAAGACCACCACCCAACT |
|
hsa_circ_0023891 | Forward |
CCTGCTACTACACCAACAGGC |
|
| Reverse |
ACTGAATTAAGTCTGTGCTCCTGA |
|
hsa_circ_0002433 | Forward |
TGAGCGTTTTATTCAGTATTTGGCT |
|
| Reverse |
GCACTGAATTAAGTCTTGCAATCCA |
|
hsa_circ_0050278 | Forward |
AAGCCAGACCTGATCACTTGTC |
|
| Reverse |
TGTCAATGGTCCCTGTGGGT |
|
hsa_circ_0000154 | Forward |
ACCAACGTTGAGCAAGATGC |
|
| Reverse |
TTCTCCAGTGTCATTCCAACAGA |
|
hsa_circ_0075048 | Forward |
GGCCACATCGACAACTCCAT |
|
| Reverse |
GCTCGTTCACACTTGTTGATGC |
|
hsa_circ_0001824 | Forward |
TGCATCAGCTCCAGGGCAAT |
|
| Reverse |
TTGAAAGAAATGTGGCATGTGAGA |
|
hsa_circ_0000835 | Forward |
CAGCATGGTCATGGAGGATGG |
|
| Reverse |
ATGCTTGATGCCTATTGCCACT |
|
hsa_circ_0000825 | Forward |
GAAAAGCGCGCTAAAGCTGA |
|
| Reverse |
TCCATCTCAGCACGGAGTTCA |
|
hsa_circ_0009109 | Forward |
ATCTGGCTCAGATGACACCAA |
|
| Reverse |
TATGTTTGCTCGGTGCCCTG |
|
hsa_circ_0087855 | Forward |
ACTTCCACACCTGCATCCAT |
|
| Reverse |
TGCTTTCACCTGTCAGTTGCT |
|
hsa_circ_0001747 | Forward |
GACAAGCTGGTGTTGAAGGGT |
|
| Reverse |
AGCAGGCCTTTCGAGCTTTAG |
|
hsa_circ_0009061 | Forward |
CCAAGCATCAGGTGTGGAGG |
|
| Reverse |
TCTCTGTACTCTACTGTGCGGT |
|
hsa_circ_0000997 | Forward |
TGCACCACTGGATGTTGTTTACT |
|
| Reverse |
GTGGTCTCCACCTGTTTTGGAT |
|
hsa_circ_0001073 | Forward |
ACTTGTTCCAACTCAAGTGCTATAC |
|
| Reverse |
GTAGCAAAACAATGCCGCCG |
|
hsa_circ_0000085 | Forward |
TTTGGCAGACTTTTACCTGGTG |
|
| Reverse |
TGGATTGCTGCTTAAGCTTCCT |
|
hsa_circ_0088021 | Forward |
GCTGAACAGGTGCCTGAACT |
|
| Reverse |
CAATTCCAGGTCTGCTGCCG |
|
hsa_circ_0020353 | Forward |
GCAGACTCCTGCAAGTTCCC |
|
| Reverse |
GTGCTTATCCACAAGGGCCA |
Statistical analysis
The two groups were compared by the standard
Student's t-test for the evaluation of the microarray analysis. The
results were considered statistically significant at a P-value
<0.05. The false discovery rate (FDR) was calculated to correct
the P-value. Fold-change ≥2.0 and P-value <0.05 were used to
identify the differentially expressed circRNAs.
Results
circRNA expression profiles in gastric
cancer tissues relative to adjacent normal gastric tissues
Randomly, 3 gastric cancer and 3 adjacent normal
gastric tissues were selected for a standard circRNA microarray
independently. The circRNA expression patterns between gastric
cancer and the adjacent normal gastric tissues were found to be
significantly different. After scanning and normalization, a total
of 950 circRNAs were found to be differentially expressed in the
microarray (fold-change in expression ≥2.0, P<0.05), consisting
of 347 upregulated circRNAs and 603 downregulated circRNAs (data
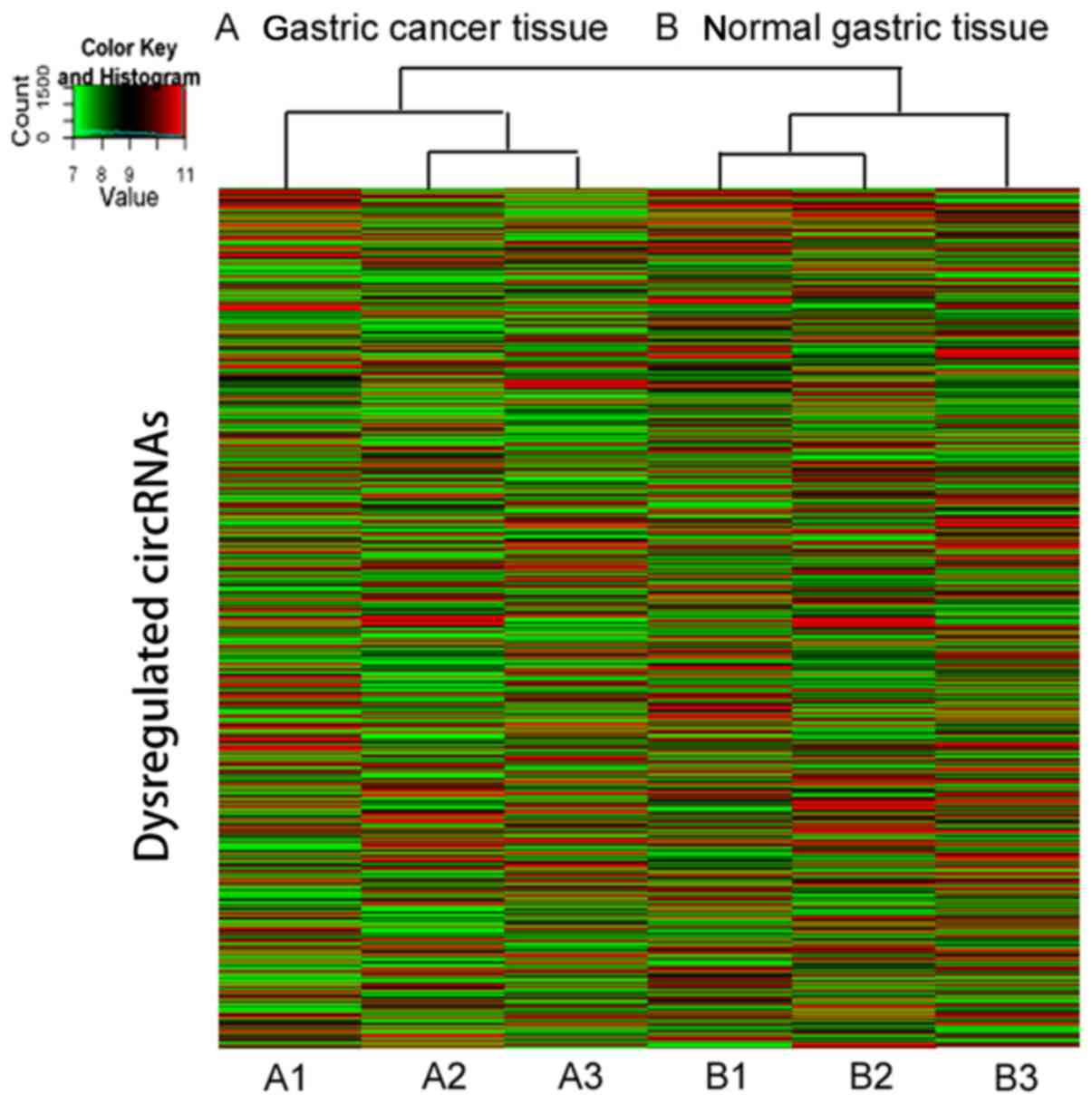
not shown). The hierarchical clustering of circRNA expression
described the variation in the expression between the groups of
gastric cancer and normal gastric tissues (Fig. 1). Furthermore, the variation in
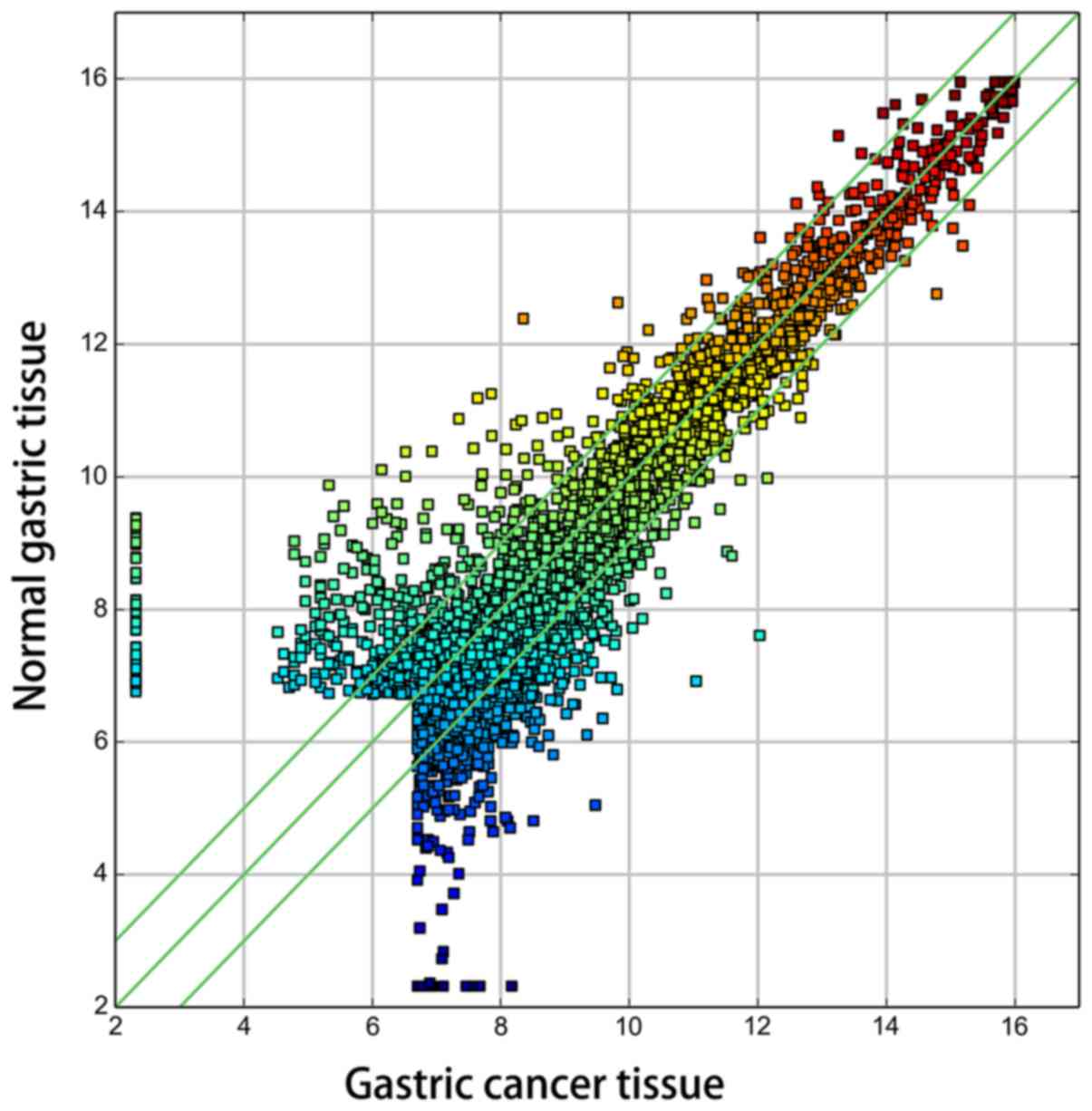
circRNA expression among the samples was assessed by Scatter-plot
visualization (Fig. 2).
Annotation of differentially expressed
circRNAs in gastric cancer tissues
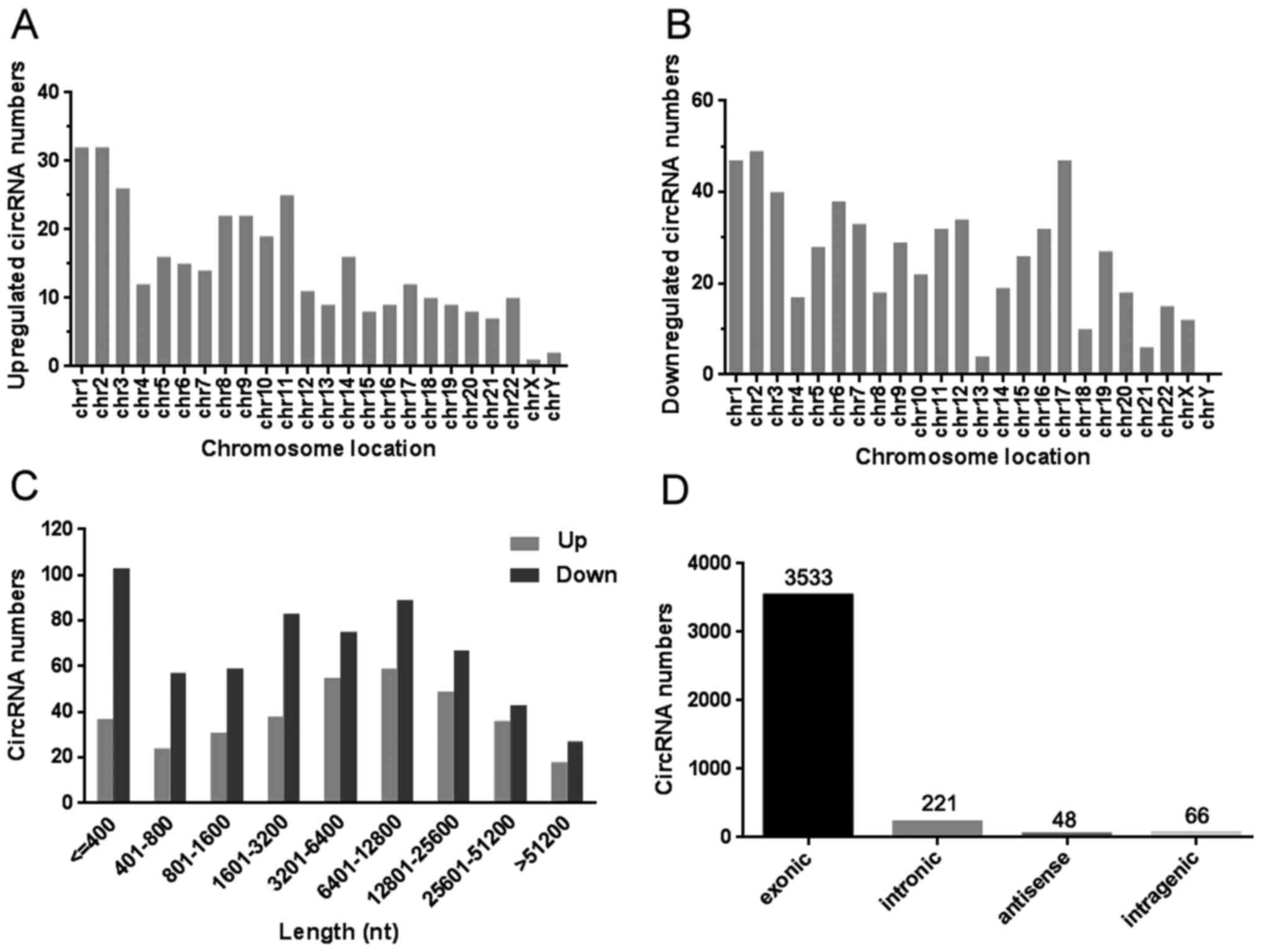
The general data, including the chromosome location,
length distribution and functional classification of these
differentially expressed circRNAs were summarized. Fig. 3A and B demonstrated that the up- and
downregulated circRNAs were located in human chromosomes. The
length data displayed two peaks that were distributed among these
dysregulated circRNAs in ≤400 bp and 1,601–25,600 bp (Fig. 3C). Fig.
3D revealed the relationship between the mentioned circRNAs and
their molecular targets including exonic, intronic, antisense and
intragenic. Nevertheless, the exonic targets occupied the vast
majority of all types of functional classification.
Reverse transcription-PCR validation
of some differentially expressed circRNAs
We set a threshold as log2 fold-change
>5 in upregulated circRNAs, >3 in downregulated circRNAs as
the previous study (17), and P-value
<0.05, and found 29 upregulated (Table II) and 28 downregulated
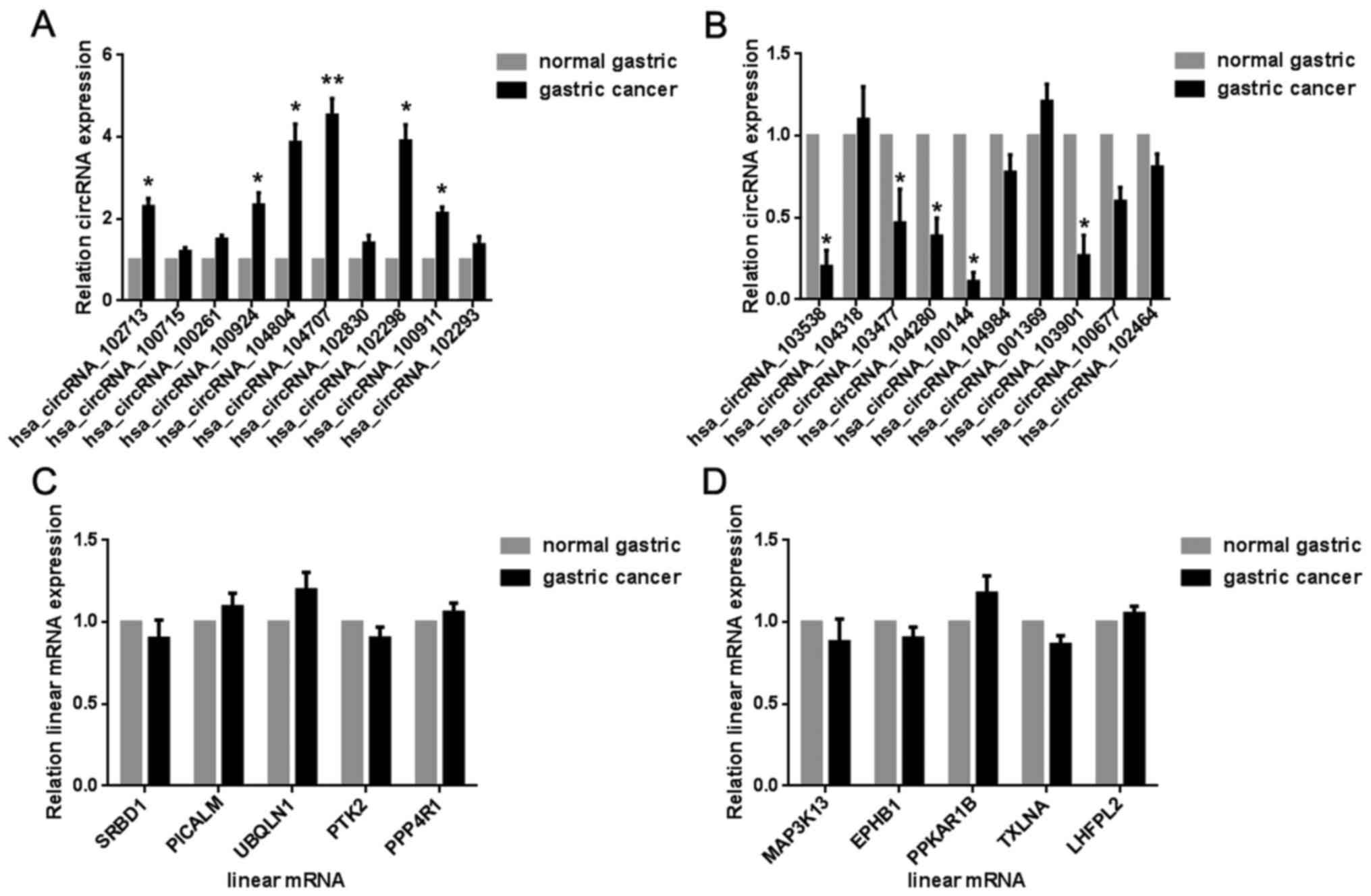
differentially expressed circRNAs (Table III). Next, we randomly selected 20
differentially expressed circRNAs, including 10 upregulated
(102713, 100715, 100261, 100924, 104804, 104707, 102830, 102298,
100911, and 102293) and 10 downregulated circRNAs (103538, 104318,
103477, 104280, 100144, 104984, 001369, 103901, 100677, and 102464)
for substantiation in the gastric tissue samples. The results of
the microarray were in agreement with those of the real-time PCR; 6
selected upregulated circRNAs (Fig.
4A) and 5 selected downregulated circRNAs (Fig. 4B) were verified. However, the
expressions of upregulated (Fig. 4C)
and downregulated circRNAs (Fig. 4D)
were not related to the expression of the host genes.
 | Table II.Upregulated circRNAs between gastric
cancer and normal gastric tissues. |
Table II.
Upregulated circRNAs between gastric
cancer and normal gastric tissues.
| circRNA | P-value | FC | Log2
FC | Regulation | circRNA type | Chrom | Strand | Best
transcript | Gene symbol |
|---|
|
hsa_circRNA_104804 | <0.001 | 117.219 | 6.873 | Up | Exonic | chr9 | − | uc004amv.3 | UBQLN1 |
|
hsa_circRNA_102678 | <0.001 | 125.119 | 6.967 | Up | Exonic | chr2 | + | uc002rpd.3 | CRIM1 |
|
hsa_circRNA_100261 | 0.011 | 41.319 | 5.368 | Up | Exonic | chr1 | − | uc001dex.4 | ANKRD13C |
|
hsa_circRNA_100927 | 0.002 | 70.919 | 6.148 | Up | Exonic | chr11 | − | uc001pbl.3 | PICALM |
|
hsa_circRNA_100924 | 0.001 | 88.219 | 6.463 | Up | Exonic | chr11 | − | uc001pbl.3 | PICALM |
|
hsa_circRNA_100922 | <0.001 | 101.119 | 6.659 | Up | Exonic | chr11 | − | uc001pbl.3 | PICALM |
|
hsa_circRNA_102713 | 0.010 | 44.119 | 5.463 | Up | Exonic | chr2 | − | uc002rus.3 | SRBD1 |
|
hsa_circRNA_102499 | 0.008 | 56.519 | 5.820 | Up | Exonic | chr19 | + | uc002npf.3 | ZNF85 |
|
hsa_circRNA_102315 | 0.007 | 52.219 | 5.706 | Up | Exonic | chr18 | + | uc002ktp.3 | MIB1 |
|
hsa_circRNA_100911 | 0.004 | 71.319 | 6.156 | Up | Exonic | chr11 | − | uc001pbl.3 | PICALM |
|
hsa_circRNA_104850 | 0.015 | 45.319 | 5.502 | Up | Exonic | chr9 | + | uc011lwa.2 | RAD23B |
|
hsa_circRNA_100925 | 0.002 | 75.269 | 6.233 | Up | Exonic | chr11 | − | uc001pbl.3 | PICALM |
|
hsa_circRNA_102293 | 0.010 | 49.919 | 5.641 | Up | Exonic | chr18 | + | uc002knq.2 | CCDC165 |
|
hsa_circRNA_102737 | <0.001 | 133.619 | 7.061 | Up | Exonic | chr2 | − | uc002sbj.3 | XPO1 |
|
hsa_circRNA_104868 | 0.021 | 34.519 | 5.109 | Up | Exonic | chr9 | − | uc010muc.1 | KIAA0368 |
|
hsa_circRNA_100381 | 0.013 | 48.619 | 5.603 | Up | Exonic | chr1 | + | uc001gev.3 | DCAF6 |
|
hsa_circRNA_104707 | <0.001 | 120.019 | 6.907 | Up | Exonic | chr8 | − | uc003yvs.3 | PTK2 |
|
hsa_circRNA_104532 | <0.001 | 132.219 | 7.046 | Up | Exonic | chr7 | + | uc003wme.3 | RBM33 |
|
hsa_circRNA_104492 | 0.012 | 45.319 | 5.502 | Up | Exonic | chr7 | + | uc003vqs.3 | MKLN1 |
|
hsa_circRNA_104689 | 0.009 | 53.719 | 5.747 | Up | Exonic | chr8 | − | uc003ysz.2 | ASAP1 |
|
hsa_circRNA_100380 | 0.010 | 56.219 | 5.812 | Up | Exonic | chr1 | + | uc001gev.3 | DCAF6 |
|
hsa_circRNA_104016 | 0.011 | 54.219 | 5.760 | Up | Exonic | chr5 | + | uc003mby.4 | ERGIC1 |
|
hsa_circRNA_100715 | 0.021 | 34.419 | 5.105 | Up | Exonic | chr10 | − | uc001lif.4 | CTBP2 |
|
hsa_circRNA_101270 | <0.001 | 108.219 | 6.757 | Up | Exonic | chr13 | + | uc001vib.4 | TDRD3 |
|
hsa_circRNA_102830 | 0.014 | 41.519 | 5.375 | Up | Exonic | chr2 | + | uc002twg.3 | ACVR2A |
|
hsa_circRNA_100241 | <0.001 | 123.419 | 6.947 | Up | Exonic | chr1 | − | uc001cyx.1 | OMA1 |
|
hsa_circRNA_104052 | 0.001 | 103.319 | 6.690 | Up | Exonic | chr6 | + | uc003mwi.3 | CDYL |
|
hsa_circRNA_102298 | 0.003 | 87.319 | 6.448 | Up | Exonic | chr18 | − | uc002kod.1 | PPP4R1 |
|
hsa_circRNA_100097 | 0.011 | 44.619 | 5.479 | Up | Exonic | chr1 | + | uc001bgi.2 | KDM1A |
 | Table III.Downregulated circRNAs between
gastric cancer and normal gastric tissues. |
Table III.
Downregulated circRNAs between
gastric cancer and normal gastric tissues.
| circRNA | P-value | FC | Log2
FC | Regulation | circRNA type | chrom | Strand |
Best_transcript | Gene symbol |
|---|
|
hsa_circRNA_103538 | <0.001 | −40.819 | −5.351 | Down | Exonic | chr3 | + | uc003fpi.3 | MAP3K13 |
|
hsa_circRNA_102016 | 0.038 | −8.032 | −3.005 | Down | Exonic | chr17 | − | uc002heo.1 | SSH2 |
|
hsa_circRNA_102464 | 0.006 | −17.362 | −4.117 | Down | Exonic | chr19 | + | uc002myp.3 | PKN1 |
|
hsa_circRNA_100109 | 0.036 | −8.135 | −3.024 | Down | Exonic | chr1 | + | uc001bmt.1 | ARID1A |
|
hsa_circRNA_103477 | 0.002 | −20.819 | −4.379 | Down | Exonic | chr3 | + | uc003eqt.3 | EPHB1 |
|
hsa_circRNA_100801 | 0.007 | −21.424 | −4.421 | Down | Exonic | chr11 | + | uc001mxq.4 | HSD17B12 |
|
hsa_circRNA_104984 | 0.028 | −12.154 | −3.603 | Down | Exonic | chrX | − | uc004czk.2 | MAP3K15 |
|
hsa_circRNA_104968 | 0.030 | −9.387 | −3.230 | Down | Exonic | chr9 | + | uc004coa.3 | EHMT1 |
|
hsa_circRNA_101585 | 0.034 | −11.713 | −3.550 | Down | Exonic | chr15 | − | uc010biv.1 | CELF6 |
|
hsa_circRNA_102540 | 0.035 | −10.064 | −3.331 | Down | Exonic | chr19 | + | uc002ohk.3 | SIPA1L3 |
|
hsa_circRNA_103568 | 0.038 | −9.790 | −3.291 | Down | Exonic | chr3 | − | uc003fxp.2 | DLG1 |
|
hsa_circRNA_100144 | <0.001 | −57.519 | −5.845 | Down | Exonic | chr1 | + | uc001bui.3 | TXLNA |
|
hsa_circRNA_105041 | 0.005 | −27.519 | −4.782 | Down | Exonic | chrX | − | uc004flx.1 | G6PD |
|
hsa_circRNA_103901 | <0.001 | −36.919 | −5.206 | Down | Exonic | chr5 | − | uc003kfo.3 | LHFPL2 |
|
hsa_circRNA_100061 | 0.004 | −19.228 | −4.265 | Down | Exonic | chr1 | − | uc001aub.3 | DHRS3 |
|
hsa_circRNA_104046 | 0.003 | −20.371 | −4.348 | Down | Exonic | chr6 | + | uc003mtz.3 | WRNIP1 |
|
hsa_circRNA_104351 | 0.040 | −9.361 | −3.226 | Down | Exonic | chr7 | − | uc011kbg.2 | GLI3 |
|
hsa_circRNA_104601 | 0.035 | −12.922 | −3.691 | Down | Exonic | chr8 | − | uc003xpe.3 | SLC20A2 |
|
hsa_circRNA_102489 | <0.001 | −35.219 | −5.138 | Down | Exonic | chr19 | + | uc002nkf.3 | UPF1 |
|
hsa_circRNA_102471 | 0.041 | −8.065 | −3.011 | Down | Exonic | chr19 | + | uc002nfj.1 | MYO9B |
|
hsa_circRNA_104280 | 0.002 | −21.819 | −4.447 | Down | Exonic | chr7 | − | uc003six.1 | PRKAR1B |
|
hsa_circRNA_101657 | 0.038 | −9.248 | −3.209 | Down | Exonic | chr15 | + | uc010urq.2 | IGF1R |
|
hsa_circRNA_104318 | 0.032 | −11.642 | −3.541 | Down | Exonic | chr7 | − | uc003sti.3 | ANKMY2 |
|
hsa_circRNA_102212 | 0.038 | −9.358 | −3.226 | Down | Exonic | chr17 | − | uc002jwc.1 | USP36 |
|
hsa_circRNA_100752 | 0.007 | −24.319 | −4.604 | Down | Exonic | chr11 | − | uc001maq.2 | OR51B5 |
|
hsa_circRNA_104190 | 0.002 | −23.098 | −4.529 | Down | Exonic | chr6 | − | uc003qez.2 | HBS1L |
|
hsa_circRNA_001369 | 0.024 | −10.828 | −3.436 | Down | Antisense | chr12 | − | NM_000020 | ACVRL1 |
|
hsa_circRNA_100677 | 0.005 | −21.387 | −4.418 | Down | Exonic | chr10 | − | uc009xxl.3 | PCGF6 |
Discussion
Human gastric cancer is one of the most commonly
known malignancies all over the world. A large number of studies
have shown that the occurrence of gastric cancer involves several
molecular mechanisms. However, the precise biological process of
gastric cancer is not yet clearly elucidated. Several circRNAs have
been recently discovered constituting a new specific class of
endogenous non-coding RNAs. Hsa_circ_001569 promoted colorectal
cancer in cell proliferation and invasion as a sponge of miR-145
(18). On the contrary,
hsa_circ_002059 was found to be significantly down-regulated in
gastric cancer as a typical circRNA, and its expression level was
correlated with tumor metastasis and TNM stage (19). Thus, circRNAs might play a major role
in the occurrence and development of gastric cancer; however, our
understanding about the correlation between circRNAs expression and
gastric cancer remains controversial due to the limited number of
studies. Hence, the expression profile of circRNAs in gastric
cancer necessitates further exploration with respect to the
potential mechanisms.
The specific repeated pattern of chromosomal
aberrations is not associated with tumorigenesis and progression in
gastric cancer. The current study conformed to the conclusion
considering that all the chromosomes can experience unequable
changes (20). However, there are
inconsistencies regarding the chromosomal location of dysregulated
circRNAs. Our experimental results revealed that chromosomal
abnormalities were mainly distributed on chromosomes 1, 2, 3, 6, 9,
11, and 17, while previous studies designated chromosomes 8, 12,
15, 17, and 20 in gastric cancer (21,22). This
phenomenon might be attributed to the following: firstly, the
experimental method to detect chromosomal dysregulation in the past
was FISH technology, while currently gene microarray is employed.
Secondly, the sample size in the current study was small due to the
high cost of gene microarray technology. Finally, the inherent
differences in gastric cancer, such as the degree of pathological
differentiation, stages, and grades, might also result in the
differential distribution of chromosomal abnormalities.
Here, we reverse transcription-PCR verified the
microarray analysis results. Recent evidence demonstrated that
circRNAs play a crucial role in fine-tuning the level of
miRNA-mediated regulation of gene expression via miRNA
sequestration (23,24). In addition, several of the predicted
binding sites of circRNAs on miRNAs are functional and appear to be
under less selective pressure as compared to the corresponding
miRNA binding sites in mRNAs (25).
However, in the current study, the expression of circRNAs did not
correlate with the expression of the host genes, suggesting an
independent regulation of transcription vs. circRNA formation. We
will verify the findings in further studies with larger sample
size. Combining with previous studies (26,27), we
currently propose that the circRNA-miRNA-mRNA axis may be the
putative mechanism promoting the growth of the tumor, although the
specific effect might not be deduced. Thus, further studies are
essential for an insight into the exact mechanism.
In conclusion, we reported the profile of
differentially expressed circRNAs between normal gastric and
gastric cancer tissues. The network of differentially expressed
numerous circRNAs was constructed and they found to be involved in
the development and metabolism of gastric cancer with our and
previous studies (27–29). Therefore, a further exploration of the
biological processes and molecular mechanisms of the dysregulated
circRNAs is imperative in order to clarify the pathogenesis of
gastric cancer or provide a new therapeutic target via the
regulation of the key circRNAs.
Acknowledgements
The authors would like to thank Dr Tianwei Gu for
his linguistic assistance.
Funding
The present study was supported by the Outstanding
Youth Project of Nanjing City (grant no. JQX14005) and the Medjaden
Academy & Research Foundation for Young Scientists (Grant no.
MJR20170029).
Availability of data and materials
All data generated or analyzed during the study are
included in this published article.
Authors' contributions
XZ and XL designed the research. YS, JZ, ZF, BZ, MC,
XL and XZ performed the experiments. YS, JZ, BZ and MC analyzed the
data. YS wrote the paper and ZF, XL and XZ critically revised the
manuscript for important intellectual content.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of the Affiliated Drum Tower Hospital of Nanjing
University, Medical School (Nanjing, China) and all patients
provided informed consent prior to their inclusion within the
study.
Consent for publication
All patients provided written informed consent for
the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kheir Bou T, Futoma-Kazmierczak E,
Jacobsen A, Krogh A, Bardram L, Hother C, Grønbæk K, Federspiel B,
Lund AH and Friis-Hansen L: miR-449 inhibits cell proliferation and
is down-regulated in gastric cancer. Mol Cancer. 10:292011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tan P and Yeoh KG: Genetics and molecular
pathogenesis of gastric adenocarcinoma. Gastroenterology.
149:1153–1162.e3. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lochhead P and El-Omar EM: Gastric cancer.
Br Med Bull. 85:87–100. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nigro JM, Cho KR, Fearon ER, Kern SE,
Ruppert JM, Oliner JD, Kinzler KW and Vogelstein B: Scrambled
exons. Cell. 64:607–613. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Guo JU, Agarwal V, Guo H and Bartel DP:
Expanded identification and characterization of mammalian circular
RNAs. Genome Biol. 15:4092014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang PL, Bao Y, Yee MC, Barrett SP, Hogan
GJ, Olsen MN, Dinneny JR, Brown PO and Salzman J: Circular RNA is
expressed across the eukaryotic tree of life. PLoS One.
9:e908592014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Reddy SD, Ohshiro K, Rayala SK and Kumar
R: MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase
1 and regulates its functions. Cancer Res. 68:8195–8200. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Webster RJ, Giles KM, Price KJ, Zhang PM,
Mattick JS and Leedman PJ: Regulation of epidermal growth factor
receptor signaling in human cancer cells by microRNA-7. J Biol
Chem. 284:5731–5741. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Saydam O, Senol O, Würdinger T, Mizrak A,
Ozdener GB, Stemmer-Rachamimov AO, Yi M, Stephens RM, Krichevsky
AM, Saydam N, et al: miRNA-7 attenuation in schwannoma tumors
stimulates growth by upregulating three oncogenic signaling
pathways. Cancer Res. 71:852–861. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fang Y, Xue JL, Shen Q, Chen J and Tian L:
MicroRNA-7 inhibits tumor growth and metastasis by targeting the
PI3K/AKT pathway in hepatocellular carcinoma. Hepatology.
55:1852–1862. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xiong S, Zheng Y, Jiang P, Liu R, Liu X
and Chu Y: MicroRNA-7 inhibits the growth of human non-small cell
lung cancer A549 cells through targeting BCL-2. Int J Biol Sci.
7:805–814. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang N, Li X, Wu CW, Dong Y, Cai M, Mok
MT, Wang H, Chen J, Ng SS, Chen M, et al: microRNA-7 is a novel
inhibitor of YY1 contributing to colorectal tumorigenesis.
Oncogene. 32:5078–5088. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li F, Zhang L, Li W, Deng J, Zheng J, An
M, Lu J and Zhou Y: Circular RNA ITCH has inhibitory effect on ESCC
by suppressing the Wnt/β-catenin pathway. Oncotarget. 6:6001–6013.
2015.PubMed/NCBI
|
|
17
|
Wu HJ, Zhang CY, Zhang S, Chang M and Wang
HY: Microarray expression profile of circular RNAs in heart tissue
of mice with myocardial infarction-induced heart failure. Cell
Physiol Biochem. 39:205–216. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y,
Yang S, Zeng Z, Liao W, Ding YQ and Liang L: Emerging roles of
circRNA_001569 targeting miR-145 in the proliferation and invasion
of colorectal cancer. Oncotarget. 7:26680–26691. 2016.PubMed/NCBI
|
|
19
|
Li P, Chen S, Chen H, Mo X, Li T, Shao Y,
Xiao B and Guo J: Using circular RNA as a novel type of biomarker
in the screening of gastric cancer. Clin Chim Acta. 444:132–136.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Luo H, Zhao X, Wan X, Huang S and Wu D:
Gene microarray analysis of the lncRNA expression profile in human
urothelial carcinoma of the bladder. Int J Clin Exp Med.
7:1244–1254. 2014.PubMed/NCBI
|
|
21
|
Cheng L and Zhang Q, Yang S, Yang Y, Zhang
W, Gao H, Deng X and Zhang Q: A 4-gene panel as a marker at
chromosome 8q in Asian gastric cancer patients. Genomics.
102:323–330. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Noguchi T, Wirtz HC, Michaelis S, Gabbert
HE and Mueller W: Chromosomal imbalances in gastric cancer.
Correlation with histologic subtypes and tumor progression. Am J
Clin Pathol. 115:828–834. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Andrés-León E, Núñez-Torres R and Rojas
AM: miARma-Seq: A comprehensive tool for miRNA, mRNA and circRNA
analysis. Sci Rep. 6:257492016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Caiment F, Gaj S, Claessen S and Kleinjans
J: High-throughput data integration of RNA-miRNA-circRNA reveals
novel insights into mechanisms of benzo[a]pyrene-induced
carcinogenicity. Nucleic Acids Res. 43:2525–2534. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mu XJ, Lu ZJ, Kong Y, Lam HY and Gerstein
MB: Analysis of genomic variation in non-coding elements using
population-scale sequencing data from the 1000 Genomes Project.
Nucleic Acids Res. 39:7058–7076. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li W, Zhong C, Jiao J, Li P, Cui B, Ji C
and Ma D: Characterization of hsa_circ_0004277 as a new biomarker
for acute myeloid leukemia via circular RNA profile and
bioinformatics analysis. Int J Mol Sci. 18:pii: E597. 2017.
|
|
27
|
Dang Y, Ouyang X, Zhang F, Wang K, Lin Y,
Sun B, Wang Y, Wang L and Huang Q: Circular RNAs expression
profiles in human gastric cancer. Sci Rep. 7:90602017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shao Y, Li J, Lu R, Li T, Yang Y, Xiao B
and Guo J: Global circular RNA expression profile of human gastric
cancer and its clinical significance. Cancer Med. 6:1173–1180.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sui W, Shi Z, Xue W, Ou M, Zhu Y, Chen J,
Lin H, Liu F and Dai Y: Circular RNA and gene expression profiles
in gastric cancer based on microarray chip technology. Oncol Rep.
37:1804–1814. 2017. View Article : Google Scholar : PubMed/NCBI
|