Introduction
As the second most common cancer type, lung cancer
has long been the leading cause of cancer-related mortality
worldwide. Despite advancements and improvements in early-stage
diagnosis, immune therapy, surgical and medical treatments, the
5-year survival rate of lung cancer patients remains poor (1). With regard to the treatment of
early-stage non-small lung cancer cell (NSCLC), great improvements
in local control have been made over the last decade in operable
and inoperable patients (2,3); however, ~20% of patients continue to
develop distant metastasis (4,5). This has
encouraged further research focusing on the molecular mechanisms of
NSCLC invasion leading to metastasis, and further understanding in
this regard could to be utilized to refine patient selection for
already existing therapies.
Long noncoding RNAs (lncRNAs) comprise a class of
noncoding RNAs that are >200 nucleotides in length, of which the
majority of biological functions are unknown (6). Previous research has identified lncRNAs
to be involved in crucial processes of cancer malignancy, including
metastasis and tumorigenesis (7). One
of the most well-studied lncRNAs, metastasis-associated lung
adenocarcinoma transcript 1 (MALAT-1), is a highly conserved
nuclear lncRNA. It functions as a molecular decoy, serving as a
structural link between ribonucleoproteins (7). Through the development of a MALAT-1
knockout model in human lung tumor cells, Ji et al (8) discovered that MALAT-1 significantly
regulates a set of metastasis-associated genes, rather than
regulating alternative splicing. Consequently, migration was
dramatically impaired in murine xenografts of MALAT-1-deficient
cells. This was further demonstrated by blocking MALAT-1 using
antisense oligonucleotides after tumor implantation.
Hypoxia represents a major microenvironmental
condition that directly impacts the regulation of hundreds of
protein-coding genes involved in dealing with the limitation of
oxygen supply (9). Not only
protein-coding genes, lncRNAs are not presented to be tightly
regulated by hypoxia. Lelli et al (9) identified a strong induction of MALAT-1
in the organs (including the kidneys, testes and lungs) of mice
exposed to inspiratory hypoxia. This raised the question of whether
hypoxia induces lncRNA in tumor cells and thus regulates malignant
processes.
PTB-associated splicing factor protein (PSF)
contains a DNA-binding domain that is responsible for
epigenetically regulating its target genes, such as the
proto-oncogene G antigen 6 (GAGE6), and two RNA-binding domains
(RBDs) that bind to lncRNAs, such as MALAT-1, to release PSF from
its downstream proto-oncogene and activate transcription (10–12).
MALAT-1 was demonstrated to epigenetically bind to the RBD of PSF
and release it from the promoter region of GAGE6, thus inhibiting
its proto-oncogene activity in melanoma cells (13). In lung cancer cells, MALAT-1 was
reported to be positively associated with tumor malignant behaviors
including proliferation, colony formation and migration (14). In A549 cells, the regulatory role of
hypoxia-induced MALAT-1 in the processes of epigenetic regulation
of PSF remains unknown.
In the present study, the differential expression of
MALAT-1 the exposure to hypoxic conditions of A549 cells was
detected. Subsequent analysis of the regulatory role of MALAT-1
upregulation in cell proliferation, migration and invasion via
epigenetic regulation of the transcriptional activity of PSF on its
downstream target gene, GAGE6, was performed. The present study
aimed to provide a novel therapeutic target for metastasis in lung
cancer.
Materials and methods
Cell culture and in vitro hypoxia
exposure
The lung adenocarcinoma cell line A549 was obtained
from ATCC (Manassas, VA, USA) and cultured routinely in
high-glucose Dulbecco's modified Eagle's medium (DMEM) supplemented
with 10% newborn bovine serum (NBS; Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) in a 5% CO2
incubator at 37°C. In order to avoid possible interference with
serum steroids, A549 cells were pre-incubated with 10%
dextran-coated charcoal (DCC). Cells were then placed in an
anaerobic system (Forma Scientific, Marietta, OH, USA) filled with
1% O2, 5% CO2, and 94% N2 for
hypoxic exposure. Cells were incubated in 5% CO2
incubator were considered as normoxia control. Following hypoxic
exposure, the medium was replaced with fresh DMEM supplemented with
10% NBS; and 24 h later, cells were suspended by trypsin and
collected for further analysis.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Cells were lysed using TRIzol reagent (Thermo Fisher
Scientific, Inc.) following the manufacturer's protocol, and 0.5 µg
total RNA was reverse transcribed using Ribo™
mRNA/lncRNA qRT-PCR Starter kit (Guangzhou RiboBio Co., Ltd.,
Guangzhou, China) according to the manufacturer's protocol. qPCR
was performed in triplicate using 2X SYBR Green PCR Master Mix
(Thermo Fisher Scientific, Inc.) with an ABI7500 system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The primer sequences
used were as follows: MALAT-1, 5′-GACTTCAGGTCTGTCTGTTCT-3′
(forward) and 5′-CAACAATCACTACTCCAAGC-3′ (reverse); nuclear
paraspeckle assembly transcript 1 (NEAT1),
5′-ATGCCACAACGCAGATTGAT-3′ (forward) and 5′-CGAGAAACGCACAAGAAGG-3′
(reverse); β-actin, 5′-CTTAGTTGCGTTACACCCTTTCTTG-3′ (forward) and
5′-CTGTCACCTTCACCGTTCCAGTTT-3′ (reverse); humanin (HN),
5′-AAGAACAGGTTTCTCTCTGTCCT-3′ (forward) and
5′-AGCTTTCCCTCCAATCTACTACT-3′ (reverse); and HOX transcript
antisense RNA (HOTAIR), 5′-GAGAGAGGGAGCCCAGAGTT-3′ (forward) and
5′-GCTTGGGTGTAATTGCTGGT-3′ (reverse). At the first step, cDNA was
denatured at 95°C for 5 min; for each cycle, denatured at 95°C for
10 sec, annealing and elongation at 60°C for 60 sec, repeat 40
cycles. In order to quantify gene expression, the 2−
(∆∆Cq) method was used (15).
β-actin expression levels were used as an internal control. RT-qPCR
reactions were performed in triplicate.
Semi-quantitative PCR
complementary DNA (cDNA) was synthesized using Ribo
mRNA/lncRNA qRT-PCR Starter kit (Guangzhou RiboBio Co., Ltd.,
Guangzhou, China) according to the manufacturer's protocol. The PCR
for β-actin mRNA was performed using the following reaction
conditions: 20 cycles of 95°C for 10 sec, 60°C for 60 sec. The
quantification of the MALAT-1 gene was performed using the
following PCR conditions: 26 cycles of 95°C for 10 sec, 60°C for 60
sec. The PCR products were fractionated using 2% agarose gel and
stained with ethidium bromide (EB; Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany).
Cell proliferation assay
Cell proliferation was detected by using a Cell
Counting Kit-8 (CCK-8; Sigma-Aldrich; Merck KGaA) according to the
manufacturer's protocol. A total of 1×104 cells/well
were suspended in 100 µl medium, plated into 96-well plate,
supplemented with 10% FBS and cultured for 1–5 days. After the
incubation period, 20 µl CCK-8 solution was added to each well and
incubated for a further 1 h at 37°C. Absorbance was then measured
at 450 nm using a Synergy 2 Multi-Mode Microplate Reader (BioTek
Instruments, Inc., Winooski, VT, USA).
Wound healing assay
To detect cell migration in vitro, cells were
allowed to form a confluent monolayer in a 6-well plate prior to
scraping a conventional 10-µl pipette tip across the monolayer to
form a ‘wound’. To avoid noise in the results due to proliferation,
medium containing 1% FBS was used for the subsequent incubation.
Cell migration was imaged using under a X71 (U-RFL-T) fluorescence
microscope (Olympus Corporation, Tokyo, Japan) at the same location
at 0 and 24 h and the number of invading cells was recorded.
Transwell chamber invasion assay
Cells were grown to ~80% confluence and incubated in
serum-free medium for 24 h. Cells were then trypsinized and
2×104 cells in serum-free medium were added to the upper
compartments of a Transwell chamber (Corning Inc., New York, NY,
USA) pre-coated with 1 mg/ml Matrigel (BD Biosciences, Franklin
Lakes, NJ, USA). The lower chamber was filled with medium
containing 10% FBS. Cells were incubated for 24 h at 37°C, after
which the non-invaded cells were removed from the upper surface.
The invaded cells were then fixed in 4% paraformaldehyde and
stained with 0.5% crystal violet in PBS. Stained cells were
visualized under an X71 (U-RFL-T) fluorescence microscope (Olympus
Corporation) at amplification of ×100. Cells in five randomly
selected fields were counted, and the average number was
recorded.
MALAT-1 knockdown
A small interfering RNA (siRNA) targeted to MALAT-1
at position 7,211–7,229 (5′-GAAGGAGCTTCCAGTTGAA-3′) or scrambled
siRNA (5′-GGATACGGAGTACTATAGC-3′), which was considered as a
negative control was synthesized (Sangon Biotech Co., Ltd.,
Shanghai, China). For RNA interference, 10 pmol siRNA duplex and 2
µl Lipofectamine RNAi MAX (Thermo Fisher Scientific, Inc.) in 200
µl Opti-MEM I were added to 3×104 cells and incubated
for 3 h at 37°C with 5% CO2. Next, the medium was
replaced by DMEM supplemented with 10% FBS and the transfected
cells were incubated for another 24 h.
Chromatin immunoprecipitation
(ChIP)
Cells were cultured were seeded in 10-cm dishes to
form an 80% confluent monolayer. For each sample, 1×107
cells were harvested. ChIP for PSF was conducted using a murine
monoclonal antibody against PSF (cat. no. ab11825, Abcam,
Cambridge, UK) or a non-specific rabbit IgG (control, cat. no.
ab171870, Abcam) conjugated to secondary Dynal magnetic beads
(Thermo Fisher Scientific, Inc.). Cells were cross-linked with 1%
formaldehyde for 15 min at room temperature and sonicated at 30%
for 10 sec and left on ice for 1 min. The sonication step was
repeated 10 times. Next, 10 µg of either antibody or control was
added into 100 µl lysate for a 4-h incubation at 4°C with 10 µl
protein-A beads (Thermo Fisher Scientific, Inc.). The beads were
washed three times with washing buffer (150 mM NaCl, 5 mM
MgCl2, 10 mM HEPES, pH 7.0, 1% NP-40) and eluted in 300
µl 3 M NaCl elution buffer for 3 h at 65°C. The ChIP products were
analyzed by RT-qPCR following the aforementioned protocol.
Dihydrofolate Reductase (DHFR) 5′ untranslated region (UTR)
was used as a negative control sequence. The primers for detecting
the PSF-reacting region on the GAGE6 promoter were as follows:
5′-GCCTTCTGCAAAGAAGTCTTGCGC-3′ (forward) and
5′-ATGCGAATTCGAGGCTGAGGCAGACAAT-3′ (reverse); DHFR 5′UTR,
5′-CTGATGTCCAGGAGGAGAAAGG-3′ (forward) and
5′-AGCCCGACAATGTCAAGGACTG-3′ (reverse).
RNA immunoprecipitation (RIP)
Cells (1×106) were lysed for 10 min on
ice in a lysis buffer [150 mM NaCl, 5 mM MgCl2, 10 mM
HEPES, pH 7.0, 1% NP-40, 100 U/ml RNase Inhibitor (Merck KGaA;
Sigma-Aldrich)]. The cell lysate was diluted (1:4) with binding
buffer containing 100 mM Tris-HCL, pH 7.4, 300 mM NaCl, 2 mM
MgCl2 and 100 U/ml RNase Inhibitor. Next, the PSF
antibody (cat. no. ab11825; 1:200; Abcam) or control IgG (cat. no.
ab171870; Abcam) was added to protein-A beads (Thermo Fisher
Scientific, Inc.) and incubated for 4 h at 4°C. The beads were
washed three times with binding buffer and resuspended in 100 µl
releasing buffer [100 mM Tris-HCl pH 7.4, 30 µg/ml proteinase K
(Merck KGaA; Sigma-Aldrich)] to release the ribonucleoprotein
complex. TRIzol reagent was used to extract RNA from the
immunoprecipitates.
Western blot analysis
Proteins from total cell lysates were extracted
using radioimmunoprecipitation assay buffer (Thermo Fisher
Scientific, Inc.) and quantified using BCA protein assay kit (Merck
KGaA; Sigma-Aldrich). A total of 20 µg total protein was
fractionated on 8% SDS-PAGE gel and transferred to polyvinylidene
fluoride membranes (Life Technologies; Thermo Fisher Scientific,
Inc.). Membranes were blocked in PBS containing 0.1% Tween-20 and
5% skim milk powder. Membranes were then incubated with specific
primary antibodies (1:2,000) overnight at 4°C. The following
antibodies were used: PSF antibody (cat. no. ab11825; Abcam),
β-actin antibody (1:2,000; cat. no. ab8226; Abcam). Then the
secondary antibody anti-mouse IgG conjugated with horseradish
peroxidase (HRP) from Abcam (1:5,000; cat. no. ab97040) was
incubated with membrane for 2 h at room temperature. Proteins were
visualized using an Enhanced Chemiluminescence kit (SuperSignal
West Pico substrate; Pierce; Thermo Fisher Scientific, Inc.).
Densitometry on bands was made using the NIH Image J 1.45S software
(National Institutes of Health, Bethesda, MD, USA).
Data analysis and statistics
Data are presented as the mean ± standard error of
the mean (SEM). All experiments were repeated a minimum of three
times. Differences were analyzed using a one-way ANOVA followed by
Tukey's post-hoc test with SPSS 13.0 software (SPSS, Inc., Chicago,
IL, USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
Hypoxia induces the expression of
MALAT-1 lncRNA
To examine the overall impact on the expression
profiles of lncRNAs, this study established the expression levels
of two lncRNAs, including MALAT-1, NEAT1, and one mRNA, HN. To
exclude global expression changes, the β-actin levels in normoxia-
and hypoxia-exposed A549 cells were determined by RT-qPCR after 24
h. The results showed that, without significantly altering the
NEAT1, HN and HOTAIR expression levels, there was a marked
upregulation of MALAT-1 in hypoxia-exposed cells when compared with
normoxia-exposed cells (Fig. 1A). To
investigate the hypoxia-induced upregulation of MALAT-1, MALAT-1
expression levels were detected at 0, 12, 24, 48 and 72 h following
the induction of hypoxic conditions. The results demonstrated that
upregulated MALAT-1 initially appeared at 0 and reached a peak at
12 h, after which the levels gradually decreased (Fig. 1B). This indicates that induction of
hypoxic conditions upregulates MALAT-1 expression for a relatively
long period of time in A549 cells.
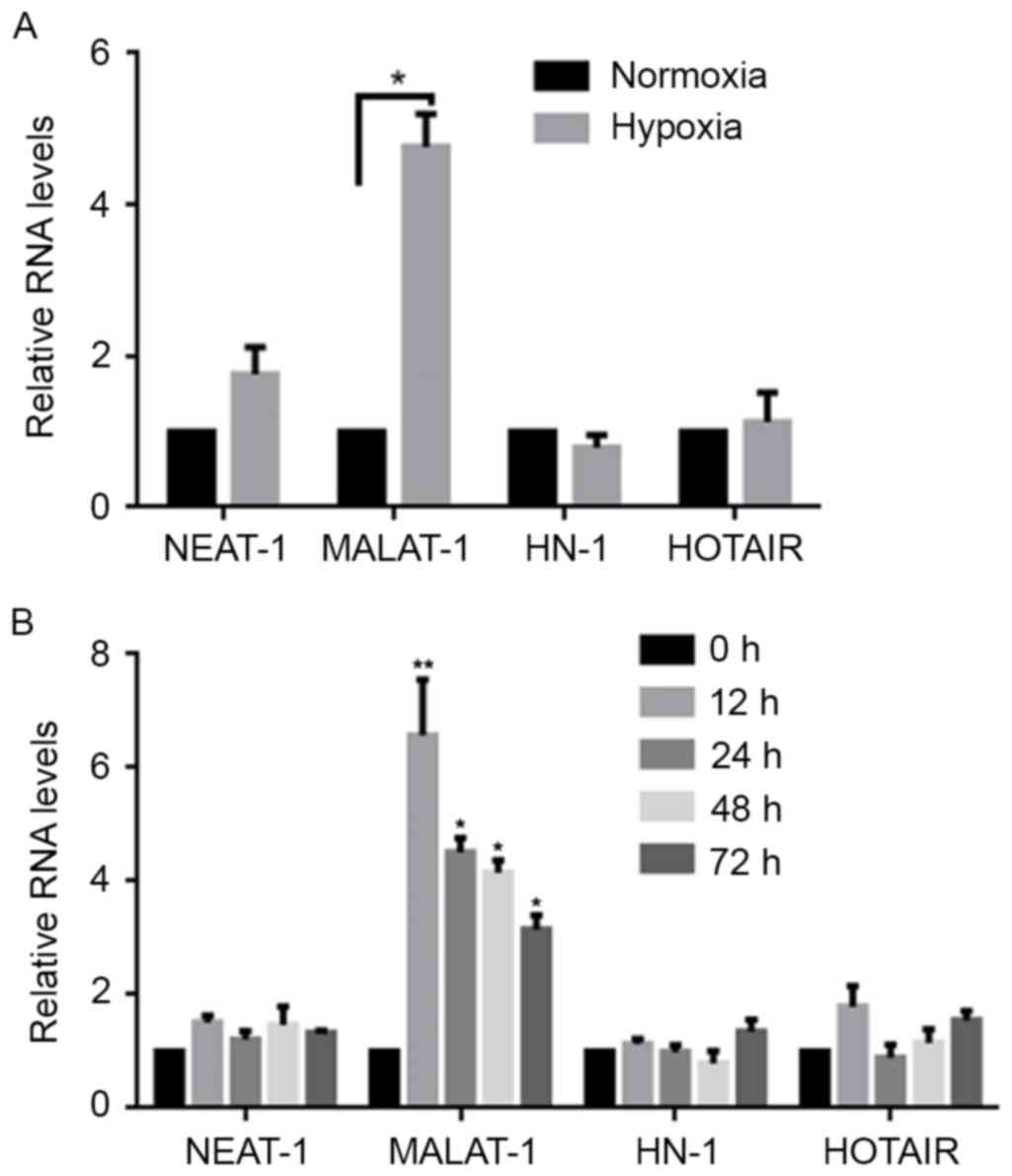 | Figure 1.Hypoxia exposure induces the
expression of MALAT-1. (A) Using a normoxia treatment group for
comparison, the expression levels of NEAT-1, MALAT-1, HN and HOTAIR
in hypoxia-treated A549 cells were detected by RT-qPCR. Data were
normalized to expression of β-actin mRNA. *P<0.05. (B) Following
hypoxia exposure, the expression levels of NEAT-1, MALAT-1, HN and
HOTAIR were detected by RT-qPCR at 0, 12, 24, 48 and 72 h after
hypoxia exposure. *P<0.05 and **P<0.01 vs. 0 h. MALAT-1,
metastasis-associated lung adenocarcinoma transcript-1; NEAT-1,
nuclear paraspeckle assembly transcript 1; HN; HOTAIR, HOX
transcript antisense RNA; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction. |
Upregulated MALAT-1 lncRNA binds to
PSF and releases it from the GAGE6 promoter region
MALAT-1 has been reported to be a key regulator in
controlling the malignancy of lung cancer cells (13,14).
Therefore, the present study was designed to investigate the
regulatory roles of MALAT-1 and PSF in the effects of hypoxia in
A549 cells. MALAT-1/PSF binding was detected by RIP in
hypoxia-exposed A549 cells. Following confirmation of the
upregulation of MALAT-1 and consistent PSF expression levels after
hypoxia exposure for 24 h (Fig. 2A),
it was evident that the amount of MALAT-1 bound to PSF increased
significantly after exposure to hypoxic conditions (Fig. 2B). As Li et al (13) showed that the binding of MALAT-1 to
PSF dissociates the PSF/GAGE6 promoter complex, we chose to detect
whether hypoxia may be responsible for the dissociation of the
PSF/GAGE6 promoter region. Hypoxia significantly released the
binding of PSF to the GAGE6 promoter region, as shown by ChIP assay
(Fig. 2C). In order to further
confirm that upregulated MALAT-1 caused this dissociation,
siMALAT-1 was introduced into hypoxia-exposed A549 cells and, 24 h
later, the efficiency of MALAT-1 knockdown was detected by RT-qPCR
(Fig. 2D). A ChIP assay identified
increased binding of PSF to the GAGE6 promoter region (Fig. 2E).
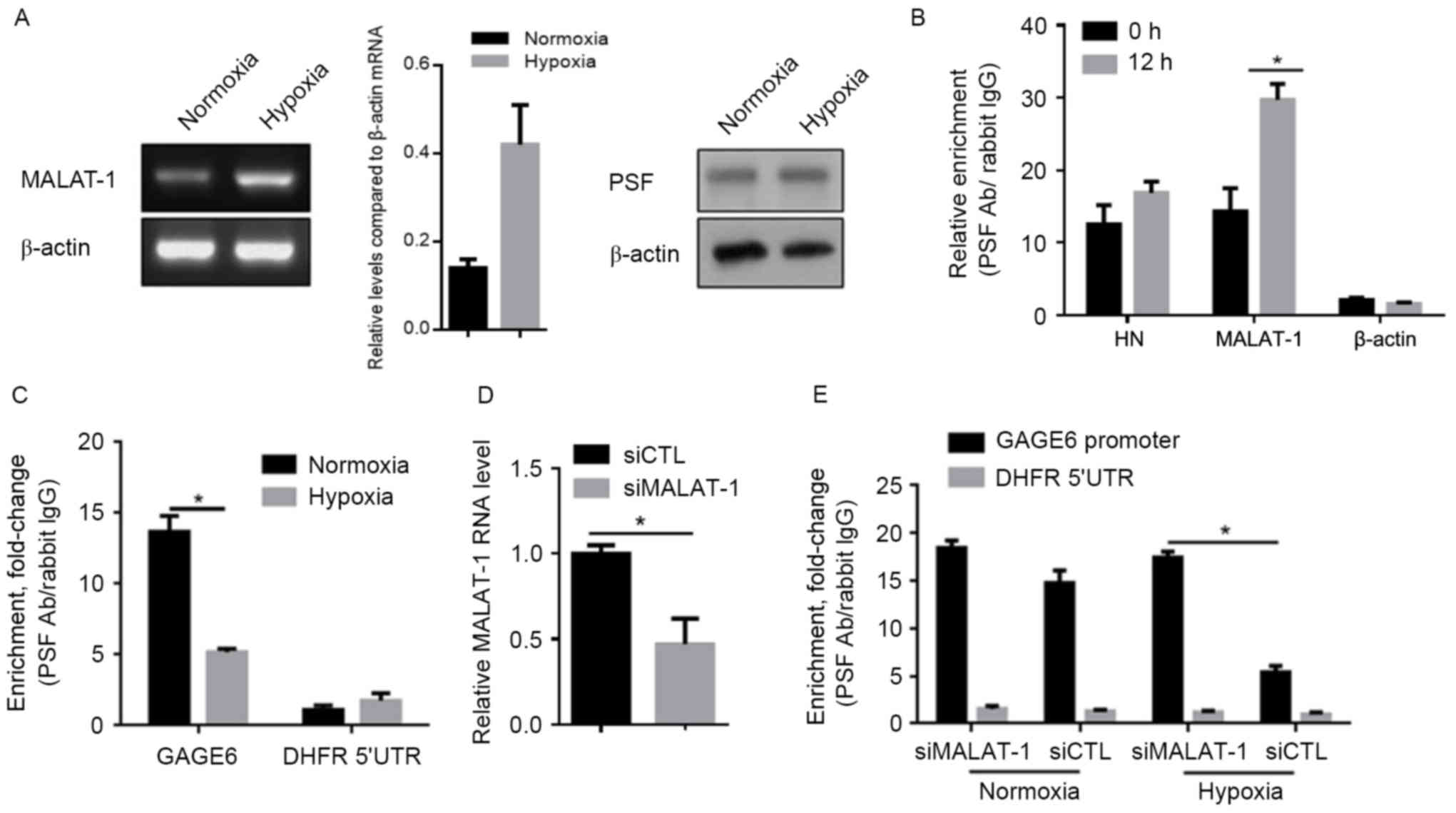 | Figure 2.Hypoxia exposure promotes the
interaction between MALAT-1 and PSF, and thus leads to the
dissociation of PSF from the GAGE6 promoter. (A) Semi-quantitative
PCR (left panel) and semi-quantitative western blot analysis (right
panel) were performed to confirm the changes in the expression
levels of MALAT-1 and PSF, respectively, after hypoxia exposure.
(B) A RIP assay was performed to detect the binding of MALAT-1 to
PSF protein after 24 h exposure. (C) After hypoxic exposure, siRNA
targeted to MALAT-1 was introduced for further RIP assay. (D)
Detection of MALAT-1 knockdown efficiency by RT-qPCR after siRNA
transfection for 24 h. (E) A ChIP assay was performed to detect the
effects of hypoxia-induced MALAT-1 expression on the binding of PSF
to its target gene, GAGE6. *P<0.05. MALAT-1,
metastasis-associated lung adenocarcinoma transcript-1; PSF,
PTB-associated splicing factor; RIP, RNA immunoprecipitation;
RT-qPCR, reverse transcription-quantitative polymerase chain
reaction; ChIP, chromatin immunoprecipitation; Ab, antibody; GAGE6,
G antigen 6; DHFR 5′UTR, Dihydrofolate reductase 5′-untranslated
region; siCTL, control siRNA; siMALAT-1, siRNA targeting
MALAT-1. |
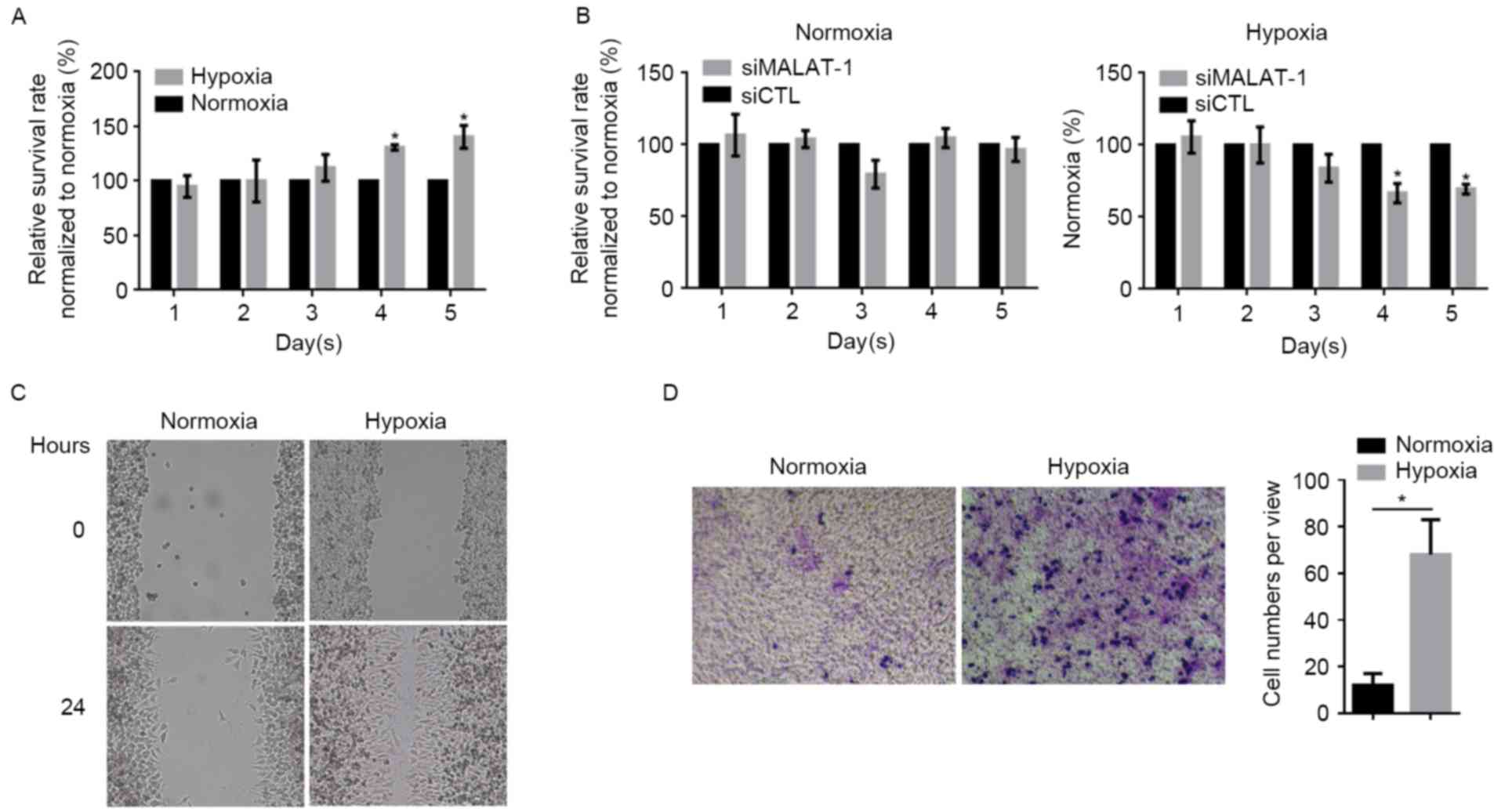
Hypoxia promotes proliferation,
migration and invasion partially via upregulating MALAT-1
The effects of hypoxia exposure on the proliferation
in A549 cells were detected by CCK-8 assay, which demonstrated
that, following hypoxia, the proliferation rate markedly increased
from day 4 onwards compared with the normoxia group at the same
time-point (Fig. 3A). In order to
establish whether the upregulation of MALAT-1 was involved,
siMALAT-1 was introduced into A549 cells after hypoxia treatment.
When the level of MALAT-1 lncRNA was downregulated by the siRNA,
the proliferation rate of hypoxia-treated A549 cells decreased
compared with the control siRNA-transected group (Fig. 3B); however, in normoxia-treated A549
cells, the knockdown of MALAT-1 led to only a marginal difference
(P>0.05). Due to the involvement of MALAT-1 in regulating cell
migration and invasion, the potential effects of hypoxia on
migration and invasion were investigated. Wound healing and
Transwell assays demonstrated that hypoxia exposure strongly
promoted migration and invasion in A549 cells (Fig. 3C and D).
Discussion
It has been reported that hypoxia serves critical
roles in various malignant behaviors of lung cancer (16–18). The
present study identified that the expression level of lncRNA
MALAT-1 was upregulated after hypoxia exposure in lung
adenocarcinoma A549 cells, while the levels of several other
lncRNAs were unaltered. Following the removal of hypoxic
conditions, upregulated MALAT-1 lncRNA persisted for >72 h.
Notably, MALAT-1 epigenetically binds to the RBD of PSF, which is a
tightly regulated region due to its DNA-binding activity.
Consequently, the present study investigated whether the
upregulated MALAT-1 RNA affects the interaction between PSF and the
GAGE6 promoter region. The ChIP assay results revealed that hypoxia
exposure dramatically inhibited the binding of PSF to the GAGE6
promoter region and resulted in the upregulation of GAGE6 mRNA and
protein. In accordance with the initial hypothesis, hypoxia
promoted proliferation, migration and invasion in A549 cells. These
results indicate that hypoxia regulates certain malignant
characteristics of lung cancer cells through transcriptionally
regulating the lncRNA MALAT-1. Taken together, the results
indicated a potential molecular mechanism underlying the effects of
hypoxia on the malignancy of lung cancer cells.
Previous studies have used various oxygen
concentrations for hypoxia exposure, ranging from 0.5 to 5%
O2 (19–21). To define an effective concentration of
oxygen for tumor proliferation and migration, A549 cells were
exposed to a range of oxygen concentrations for 24 h, and
proliferation was detected after 48 h. The results of the present
study demonstrated that, with the exception of 0.5%, any oxygen
concentration <5% significantly promoted cell proliferation.
Instead of promoting cell proliferation, 0.5% oxygen exerted a
fatal effect on A549 cells (data not shown).
Hypoxia exposure has been found to promote tumor
progression in vivo in numerous studies involving different
types of cancer. However the results have varied. In D12 and R-18
melanoma, hypoxia exposure was reported to increase tumor
progression (19,22). Lung cancer cells pre-treated with
hypoxia presented higher metastasis in human fibrosarcoma in mice.
Contrarily, Büchler et al (23) demonstrated that hypoxia exposure
influenced the number of metastatic lesions, but not the
proliferation or tumor formation processes. Cairns et al
(21) reported that acute hypoxia
exposure of the cervical carcinoma cell line ME-180 significantly
promoted lymph node but not lung metastasis (24). In the present study, to demonstrate
the effects of hypoxia exposure on proliferation, migration and
invasion, the lung adenocarcinoma cell line A549 was exposed to 1%
oxygen for 24 h. Significant effects on proliferation, migration
and invasion in A549 cells were discovered and thus enabled the
conclusion that hypoxia was a stimulating factor for tumor
progression.
Previous studies have suggested that the expression
levels of many lncRNAs are altered in various human cancers and
play crucial roles in tumor progression (25). lncRNA-LET, which is closely associated
with hepatocellular carcinoma metastasis, was demonstrated to be
attenuated in a hypoxic microenvironment (26). In an independent study, lncRNA-p21 was
identified to be involved in the regulation of the physiological
processes following hypoxic exposure by modulating hypoxia-enhanced
glycolysis (27). However, the
mechanism of lncRNAs in hypoxia in cancer has not been thoroughly
elucidated.
MALAT-1 RNA is overexpressed in numerous human
cancer types, such as non-small cell lung cancer (28), hepatocellular carcinoma (29) and bladder cancer (30). MALAT-1 is localized in nuclear
speckles (31). Its abundance and
aberrant expression in various cancers indicate its critical roles
in tumor progression, including metastasis. Tian et al
(32) transfected melanoma A-375
cells with MALAT-1 siRNA and analyzed the migration and invasion by
Transwell assay with or without Matrigel. Cells with impaired
expression of MALAT-1 migrated and invaded less effectively. Han
et al (33) observed the
upregulation of MALAT-1 in bladder urothelial carcinoma compared
with the matched normal urothelial tissues, indicating that
increased expression of MALAT-1 was associated with high-grade and
high-stage bladder urothelial carcinoma. Furthermore, silencing
MALAT-1 inhibited proliferation, migration and invasion in bladder
urothelial carcinoma T24 and 5,637 cells (33).
In conclusion, the results of the present study
indicate that the expression of the lncRNA MALAT-1 is significantly
increased in hypoxia-exposed cells and promotes hypoxia-induced
proliferation, migration and invasion in A549 cells. The
transcriptional regulation of GAGE6, a proto-oncogene, by PSF is
inhibited by the upregulation of MALAT-1 in an epigenetic manner,
and thus promotes proliferation, migration and invasion. However,
more convincing evidence from animal and clinical studies is
required to support these findings. Further study will explore the
exact mechanism of the upregulation of MALAT-1 by hypoxia exposure
in A549 cells, and employ animal studies for testing the expression
of MALAT-1 in vivo. In summary, these findings suggest that
the hypoxia/MALAT-1/PSF pathway may contribute to the development
of novel anti-cancer therapeutics directed against hypoxic tumor
targets.
Acknowledgements
The authors would like to thank Dr Tao Hong of
Sichuan University (Chengdu, China) for English language
editing.
Funding
No funding received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
LG contributed to the design of the study. LH was
responsible for project design, organization and manuscript
writing. JT, XH and TZ conducted the molecular experiments. XF
contributed to the project design and organisation and the
supervision of experimental progress and final approval of the
version to be published.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ginsberg RJ and Rubinstein LV: Randomized
trial of lobectomy versus limited resection for T1 N0 non-small
cell lung cancer. Lung Cancer Study Group. Ann Thorac Surg.
60:615–622. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Timmerman R, Paulus R, Galvin J, Michalski
J, Straube W, Bradley J, Fakiris A, Bezjak A, Videtic G, Johnstone
D, et al: Stereotactic body radiation therapy for inoperable early
stage lung cancer. JAMA. 303:1070–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chi A, Liao Z, Nguyen NP, Xu J, Stea B and
Komaki R: Systemic review of the patterns of failure following
stereotactic body radiation therapy in early-stage non-small-cell
lung cancer: Clinical implications. Radiother Oncol. 94:1–11. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Martini N, Bains MS, Burt ME, Zakowski MF,
McCormack P, Rusch VW and Ginsberg RJ: Incidence of local
recurrence and second primary tumors in resected stage I lung
cancer. J Thorac Cardiovasc Surg. 109:120–129. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sarkar SP and Adshead G: Whose DNA is it
anyway? European court, junk DNA and the problem with prediction. J
Am Acad Psychiatry Law. 38:247–250. 2010.PubMed/NCBI
|
|
7
|
Li G, Zhang H, Wan X, Yang X, Zhu C, Wang
A, He L, Miao R, Chen S and Zhao H: Long noncoding RNA plays a key
role in metastasis and prognosis of hepatocellular carcinoma.
Biomed Res Int. 2014:7805212014.PubMed/NCBI
|
|
8
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lelli A, Nolan KA, Santambrogio S,
Gonçalves AF, Schönenberger MJ, Guinot A, Frew IJ, Marti HH,
Hoogewijs D and Wenger RH: Induction of long noncoding RNA MALAT1
in hypoxic mice. Hypoxia (Auckl). 3:45–52. 2015.PubMed/NCBI
|
|
10
|
Song X, Wang B, Bromberg M, Hu Z,
Konigsberg W and Garen A: Retroviral-mediated transmission of a
mouse VL30 RNA to human melanoma cells promotes metastasis in an
immunodeficient mouse model. Proc Natl Acad Sci USA. 99:6269–6273.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Song X, Sui A and Garen A: Binding of
mouse VL30 retrotransposon RNA to PSF protein induces genes
repressed by PSF: Effects on steroidogenesis and oncogenesis. Proc
Natl Acad Sci USA. 101:621–626. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
French NS and Norton JD: Structure and
functional properties of mouse VL30 retrotransposons. Biochim
Biophys Acta. 1352:33–47. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li L1, Feng T, Lian Y, Zhang G, Garen A
and Song X: Role of human noncoding RNAs in the control of
tumorigenesis. Proc Natl Acad Sci USA. 106:12956–12961. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Semenza GL: HIF-1 and tumor progression:
Pathophysiology and therapeutics. Trends Mol Med. 8 4
Suppl:S62–S67. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhong H, De Marzo AM, Laughner E, Lim M,
Hilton DA, Zagzag D, Buechler P, Isaacs WB, Semenza GL and Simons
JW: Overexpression of hypoxia-inducible factor 1alpha in common
human cancers and their metastases. Cancer Res. 59:5830–5835.
1999.PubMed/NCBI
|
|
18
|
Semenza GL: Oxygen sensing, homeostasis,
and disease. N Engl J Med. 365:537–547. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rofstad EK, Rasmussen H, Galappathi K,
Mathiesen B, Nilsen K and Graff BA: Hypoxia promotes lymph node
metastasis in human melanoma xenografts by up-regulating the
urokinase-type plasminogen activator receptor. Cancer Res.
62:1847–1853. 2002.PubMed/NCBI
|
|
20
|
Zhang L and Hill RP: Hypoxia enhances
metastatic efficiency in HT1080 fibrosarcoma cells by increasing
cell survival in lungs, not cell adhesion and invasion. Cancer Res.
67:7789–7797. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cairns RA, Kalliomaki T and Hill RP: Acute
(cyclic) hypoxia enhances spontaneous metastasis of KHT murine
tumors. Cancer Res. 61:8903–8908. 2001.PubMed/NCBI
|
|
22
|
Rofstad EK, Galappathi K, Mathiesen B and
Ruud EB: Fluctuating and diffusionlimited hypoxia in
hypoxia-induced metastasis. Clin Cancer Res. 13:1971–1978. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Büchler P, Reber HA, Lavey RS, Tomlinson
J, Büchler MW, Friess H and Hines OJ: Tumor hypoxia correlates with
metastatic tumor growth of pancreatic cancer in an orthotopic
murine model. J Surg Res. 120:295–303. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kalliomäki TM, McCallum G, Lunt SJ, Wells
PG and Hill RP: Analysis of the effects of exposure to acute
hypoxia on oxidative lesions and tumour progression in a transgenic
mouse breast cancer model. BMC Cancer. 8:1512008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Reis EM and Verjovski-Almeida S:
Perspectives of long non-coding RNAs in cancer diagnostics. Front
Genet. 3:322012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP,
Wang F and Sun SH: Repression of the long noncoding RNA-LET by
histone deacetylase 3 contribuetes to hypoxia-mediated metastasis.
Mol Cell. 49:1083–1096. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang F, Zhang H, Mei Y and Wu M:
Reciprocal regulation of HIF-1α and lincRNA-p21 modulates the
Warburg effect. Mol Cell. 53:88–100. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ji Q, Liu X, Fu X, Zhang L, Sui H, Zhou L,
Sun J, Cai J, Qin J, Ren J and Li Q: Resveratrol inhibits invasion
and metastasis of colorectal cancer cells via MALAT1 mediated
Wnt/b-catenin signal pathway. PLoS One. 8:e787002013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY,
Zhang F, Wu LM, Chen LM and Zheng SS: Long non-coding RNA MALAT-1
overexpression predicts tumor recurrence of hepatocellular
carcinoma after liver transplantation. Med Oncol. 29:1810–1816.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xu C, Yang M, Tian J, Wang X and Li Z:
MALAT-1: A long non-coding RNA and its important 30 end functional
motif in colorectal cancer metastasis. Int J Oncol. 39:169–175.
2011.PubMed/NCBI
|
|
31
|
Tripathi V, Ellis JD, Shen Z, Song DY, Pan
Q, Watt AT, Freier SM, Bennett CF, Sharma A and Bubulya PA: The
nuclear-retained noncoding RNA MALAT1 regulates alternative
splicing by modulating SR splicing factor phosphorylation. Mol
Cell. 39:925–938. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tian Y, Zhang X, Hao Y, Fang Z and He Y:
Potential roles of abnormally expressed long noncoding RNA UCA1 and
Malat-1 in metastasis of melanoma. Melanoma Res. 24:335–341. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Han Y, Liu Y, Nie L, Gui Y and Cai Z:
Inducing cell proliferation inhibition, apoptosis and motility
reduction by silencing long noncoding ribonucleic acid
metastasis-associated lung adenocarcinoma transcript 1 in
urothelial carcinoma of the bladder. Urology. 81:209.e1–e7. 2013.
View Article : Google Scholar
|