Introduction
Cancer is one of the leading causes of mortality and
a major public health issue worldwide. In 2015, there were 17.5
million cancer cases globally and 8.7 million cases resulting in
mortality. Between 2005 and 2015, cancer cases increased by 33%
(1). By 2030, the annual incidence of
new cancer cases and cancer-associated mortalities is projected to
increase to, 26.4 million and 17.0 million, respectively (2). The incidence and mortality of malignant
tumor has exhibited a sustained growth trend in China (3). The primary cause of mortality in
patients with cancer is tumor cell invasion and metastasis
(4).
Talin was discovered ~3 decades ago as a highly
abundant cytosolic protein important for cytoskeleton organization
and cell-extracellular matrix (ECM) adhesion (5). Extensive genetic and cell biological
studies have established that Talin is essential for the regulation
of a variety of integrin-mediated cell adhesion dependent
processes, including cell-shape changes, growth, differentiation
and migration (6–8). In vertebrates, there are two Talin
genes, Talin1 and Talin2 (9,10), which encode similar proteins (74%
amino acid sequence identity) (11,12).
Talin1 is expressed in nearly all tissues, and Talin2 is expressed
primarily in the heart, brain, testis and muscle (11–14).
Talin1 is also known to regulate focal adhesion dynamics, cell
migration and cell invasion (15–17). In a
recent study, Talin2 knockdown was demonstrated to inhibit the
migration of hepatocellular carcinoma cells (18). However, the role of the Talin2
isoforms in cell invasion and metastasis remains unclear.
In the present study, Talin2 expression was
disrupted in MDA-MB-231 cells using short hairpin-RNA
(shRNA)-mediated interference, to establish Talin2 knockdown stable
cell lines and Talin2-specific functions were examined. The effects
of Talin2 were further studied on MDA-MB-231 cell migration and
invasion. In addition, the role of Talin2 in the occurrence and
development of breast cancer was analyzed, and the underlying
mechanisms were examined. The present study provides a novel method
and basis for the diagnosis and treatment of breast cancer.
Materials and methods
Patients and samples
A total of 32 breast cancer tissue specimens from
tumor and adjacent tissues were obtained from previously untreated
female patients aged 36–73 with a mean age of 53 years who
underwent surgical treatment at the Department of General Surgery,
The First Affiliated Hospital of Wenzhou Medical University
(Wenzhou, China) between January 2016 and June 2017. All samples
were cut into two pieces. One piece was embedded in paraffin and
processed for routine histopathological examination, while the
other piece of tissue was frozen immediately in liquid nitrogen and
stored at −80°C for further studies. For all patients, the
diagnosis of breast cancer was based on clinical and
histopathological examination. Tumor stage was defined according to
Union Internationale Contre le Cancer. Tumor pathological grade was
defined based on the World Health Organization classification. The
present study was approved by the Board and Ethical Committee of
The First Affiliated Hospital of Wenzhou Medical University. All
study participants provided written informed consent in accordance
with the Declaration of Helsinki.
Reagents
Anti-Talin1 (1:1,000 dilution; cat. no. MCA4770) and
anti-Talin2 [1:1,000 dilution in the western blot analysis and 1:25
dilution in the immunohistochemical (IHC) assay; cat. no.
MCA4771GA] antibodies were obtained from Bio-Rad Laboratories, Inc.
(Hercules, CA, USA); anti-poly ADP-ribose polymerase (PARP; 1:1,000
dilution; cat. no. AY3523) and anti-cleaved PARP (1:1,000 dilution;
cat. no. CY3838) were obtained from Shanghai Abways Biotechnology
Co., Ltd. (Shanghai, China); anti-cleaved Caspase-8 (1:1,000
dilution; cat. no. RLC0011) and anti-cleaved Caspase-3 (1:1,000
dilution; cat. no. RLC0004) were obtained from Suzhou Rui Sheng
Biological Technology Co., Ltd. (Suzhou, China); anti-Caspase-8
(1:1,000 dilution, cat. no. 9746S), anti-Caspase-3 (1:1,000
dilution, cat. no. 9661S), anti-Tubulin (1:1,000 dilution, cat. no.
2148S) and anti-GAPDH (1:1,000 dilution, cat. no. 2188S) antibodies
were obtained from Cell Signaling Technology, Inc. (Danvers, MA,
USA). Control shRNA TRC1/1.5
(5′-CCGGGCGCGATAGCGCTAATAATTTCTCGAGAAATTATTAGCGCTATCGCGCTTTTT-3′),
Talin2 shRNA clones TRCN0000122990 (Talin2 90,
5′-CCGGCCATGTTAGAAGAGTCCGTTTCTCGAGAAACGGACTCTTCTAACATGGTTTTTG-3′)
and TRCN0000122992 (Talin2 92,
5′-CCGGCCATGCAGTTTGAACCATCTACTCGAGTAGATGGTTCAAACTGCGTTTTTG-3′) were
obtained from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany).
Horseradish peroxidase (HRP)-conjugated goat anti-rabbit secondary
antibody (1:1,000; cat no. A0208) and DAB (0.05–0.03%) horseradish
peroxidase color development kits (cat no. ST033) were obtained
from Beyotime Institute of Biotechnology (Haimen, China).
Lipofectamine™ 2000, Dulbecco's modified Eagle's medium (DMEM) and
fetal bovine serum (FBS) were purchased from Invitrogen (Thermo
Fisher Scientific, Inc., Waltham, MA, USA). SuperSignal™ West Pico
PLUS Chemiluminescent substrate was purchased from Thermo Fisher
Scientific, Inc. Penicillin G and streptomycin were purchased from
Sangon Biotechnology Co., Ltd. (Shanghai, China).
Cell culture
MDA-MB-231 human breast cancer and 293T cells were
obtained from the American Type Culture Collection (Manassas, VA,
USA), and were maintained in DMEM containing 10% FBS, penicillin
(100 U/ml) and streptomycin (100 mg/ml). Cells were cultured at
37°C in a humidified 5% CO2 atmosphere incubator.
Preparation of viruses and cell
infection
To produce the lentivirus, VSV-G-pseudotyped
lentiviral vectors were produced by co-transfecting
6×106 293T cells with 2 pmol of the respective control
shRNA, Talin2 90 (TRCN0000122990) and Talin2 92 (TRCN0000122992)
along with 1 pmol pMDLg.RRE, 0.5 pmol pMD2.G and 0.5 pmol pRSV.REV.
using Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. After 8
h, the original medium [DMEM containing 10% FBS, penicillin (100
U/ml) and streptomycin (100 mg/ml)] was replaced with fresh medium,
and the lentiviral supernatant was collected at 48 and 72 h
post-transfection. After each collection, the supernatant was
filtered through a cellulose acetate membrane (0.45 µm pore).
A total of 1×105 MDA-MB-231 cells were
plated into each well of a 6 well plate and infected with a total
volume of 100 µl lentiviruses for 24 h. Cells that stably expressed
lentiviral shRNAs were obtained by selecting the infected cells
with 1 mg/ml puromycin for a period of 3–4 weeks at 37°C in a
humidified 5% CO2 atmosphere incubator.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from MDA-MB-231 and
MDA-MB-231 Talin2 knockdown cells using the PureLink RNA kit
(Ambion; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. cDNA was synthesized at 42°C for 50 min
using the SuperScript First Strand Synthesis kit (Invitrogen;
Thermo Fisher Scientific, Inc.) using 0.5–1.0 µg RNA samples
according to the manufacturer's protocol. qPCR reactions were
performed using SYBR Green PCR master mix reagents (cat no.
4309155; Invitrogen; Thermo Fisher Scientific, Inc.) on an ABI 7500
Fast Real-Time PCR System (Applied Biosystems). The relative
quantification of gene expression for each sample was analyzed
using the 2−ΔΔCq method (19). The following primers were used: Talin1
forward, 5′-TGTTCCCCAGAGCCACCTGCC-3′ and reverse,
5′-GAAGCCGCACATCAGGGGC-3′; Talin2 forward,
5′-GGGGAATGTGTGGGGATTGCATCC-3′ and reverse,
5′-GATGAGGCGATGCGGCAGGCA-3′; 18S rRNA forward,
5′-ACCTGGTTGATCCTGCCAGT-3′ and reverse, 5′-CTGACCGGGTTGGTTTTGAT-3′.
The thermocycling conditions were as follows: 20 sec at 95°C;
followed by 40 cycles of 5 sec at 95°C and 30 sec at 60°C. Each
experiment was repeated three times in duplicate.
Colony formation assay
MDA-MB-231 cells were trypsinized, counted by an
automatic cell counter (Invitrogen Countess; Invitrogen; Thermo
Fisher Scientific, Inc.) and seeded for the colony formation assay
in six-well plates at a density of 1×103 cells/well,
then incubated at 37°C in a humidified 5% CO2 atmosphere
incubator. During colony growth, the culture medium was replaced
every 3 days. A colony was only acknowledged if it contained
>50% cells. After 2 weeks, cells were fixed for 15 min with 3.7%
formaldehyde at room temperature and stained using 0.1% crystal
violet in 10% ethanol for 30 min at room temperature. The number of
invaded cells per field was counted under a light microscope at a
magnification of ×400. The colony formation rate was calculated
using the formula: (Number of colonies/number of seeded cells)
×100%. Three independent experiments were performed for each
group.
Cell migration assay
After cells had grown to 100% confluence in six-well
culture plates, an artificial wound was created by scratching the
cell monolayer with the tip of a 10-µl pipette. The wound area was
inspected after 24 and 48 h using an inverted phase-contrast
microscope with a digital camera (magnification, ×400). The wound
healing speed was calculated as the percentage of the initial wound
at different time points (24, 36 and 48 h) until total wound
closure.
Invasion assays
To examine the effect of Talin2 on cell invasion,
100 µl of Matrigel (0.35 mg/ml; 1:30 dilution in serum-free DMEM)
was added to each Transwell polycarbonate filter (6-mm diameter;
8-µm pore size) and incubated with the filters at 37°C for 6 h.
Breast cancer cells MDA-MB-231 were trypsinized and washed three
times with DMEM containing 1% FBS. The cells were resuspended in
DMEM containing 1% FBS at a density of 5×105 cells/ml.
The cell suspensions (100 µl) were seeded into the upper chambers
and 600 µl of DMEM medium containing 10% FBS was added to the lower
chambers. The cells were allowed to invade for 12 h in a
CO2 incubator, then fixed, stained and quantified as
described previously (20).
IHC staining
Representative tumor and adjacent regions were
marked on the paraffin-embedded blocks. Paired cancer and normal
cores were punched and transferred to a recipient block to make the
tissue microarray (TMA) block. For each patient, one tumor core and
one normal core were taken from the donor block. IHC staining was
performed as described previously (21). Briefly, 4-µm thick TMA slides were
baked for 1 h at 60°C, deparaffinized, dehydrated and treated in
citrate buffer (pH 6.0). The TMA slides were blocked using equine
serum (Beijing Solarbio Science & Technology Co., Ltd.,
Beijing, China), and then incubated with 3% hydrogen peroxide at
room temperature for 1 h, followed by overnight incubation with
anti-Talin2 (1:25 dilution) at 4°C. On the second day, the slides
were incubated with secondary antibodies (goat anti-rabbit
immunoglobulin G conjugated with horseradish peroxidase antibody
(1:2,000; cat no. PV-6001; OriGene Technologies, Inc., Rockville,
MD, USA) at 37°C for 1 h. then stained using hematoxylin (cat no.
H8070; Beijing Solarbio Science & Technology Co., Ltd.),
dehydrated, cleared and mounted. The distribution area of Talin2 in
different tissues was analyzed with ImagePro Plus software 6.0
(Media Cybernetics, Inc., Rockville, MD, USA). The positive rate of
stained tumor cells and the corresponding score were assigned as
follows: 0 (0%), 1 (1–25%), 2 (26–50%), 3 (51–75%) and 4 (76–100%).
The intensity of CDK5 staining was scored from 0 to 3, and the
detailed standard was as follows: 0, no staining; 1, weak staining;
2, moderate staining; and 3, strong staining, and observed under an
Eclipse 50i/55i Microscope (Optical microscope) at a magnification
of ×400. Positive staining intensity: colorless, 0; light yellow,
1; pale brown, 2; and brown, 3.
Hoechst 33342 staining
Hoechst 33342 staining was performed to detect
altered nuclear morphology in MDA-MB-231 and MDA-MB-231 Talin2
knockdown cells. Cells were plated on coverslips overnight;
subsequently, the cells were stained with 10 µM Hoechst 33342
solution for 15 min at 37°C. After staining, cells were washed
three times with PBS, then viewed and counted by eye under a
fluorescence microscope (magnification, ×200) with standard
excitation filters (Nikon Corporation, Tokyo, Japan). The
excitation wavelength used was 346 nm and the emission wavelength
was 460 nm.
Annexin V/fluorescein isothiocyanate
(FITC) flow cytometric assay
The flow cytometric assay was performed with the
Annexin V kit (BD Pharmingen; BD Biosciences, San Jose, CA, USA).
Cells were seeded in six-well plates at a density of
1×105 cells/well and incubated at 37°C in a humidified
5% CO2 atmosphere incubator. After 12 h, cells were
harvested and pelleted via centrifugation (1,000 × g for 5 min) at
4°C, immediately resuspended in binding buffer and subsequently
stained with 5 µl FITC Annexin V and 5 µl propidium iodide (PI),
according to the kit protocol. The mixture was placed on ice (4°C)
in the dark and analyzed using a FACS system with BD Accuri C6
Software (BD 1.0.264.21; BD Biosciences).
Western blot analysis
A total of 2×106 cells (MDA-MB-231 or
MDA-MB-231 Talin2 knockdown cells) were seeded overnight in
six-well plates, cells were washed with ice-cold PBS and harvested
using RIPA buffer (cat no. P0013C; Beyotime Institute of
Biotechnology). The protein concentration was determined using the
BCA method (cat no. P0011; Beyotime Institute of Biotechnology) and
20 µg of total protein was separated using 12% SDS-PAGE prior to
western blot analysis. The proteins were transferred to a
nitrocellulose membrane that was blocked in 5% milk for 1 h at room
temperature. The expression levels of Talin1, Talin2, Caspase-3,
cleaved Caspase-3, GAPDH and Tubulin were determined by incubating
the membrane with the specific aforementioned primary antibodies
(1:1,000 dilution in 5% milk) overnight at 4°C. This was followed
by incubation with a HRP-conjugated secondary antibody (Beyotime
Institute of Biotechnology) for 1 h at room temperature prior to
development using SuperSignal West Pico PLUS Chemiluminescent
substrate. Tubulin or GAPDH was used as the loading control. All
protein expressions levels were quantified using ImageJ software
(ImageJ 1.43u/Java 1.6.0–10; National Institutes of Health,
Bethesda, MD, USA).
Statistical analysis
All data were presented as the mean ± standard
deviation and analyzed using single-factor analysis of variance
(one-way ANOVA) for comparison between groups. Multiple comparisons
were performed among groups using ANOVA followed by the least
significant difference test. The software package SPSS 19.0 (SPSS,
Inc., Chicago, IL, USA) was used for all statistical analyses. The
results are representative of three independent experiments.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Construction of stable Talin2
knockdown cells
To establish the stable Talin2 90 or Talin2 92 cell
lines, Talin2 recombinant lentivirus infected MDA-MB-231 cells were
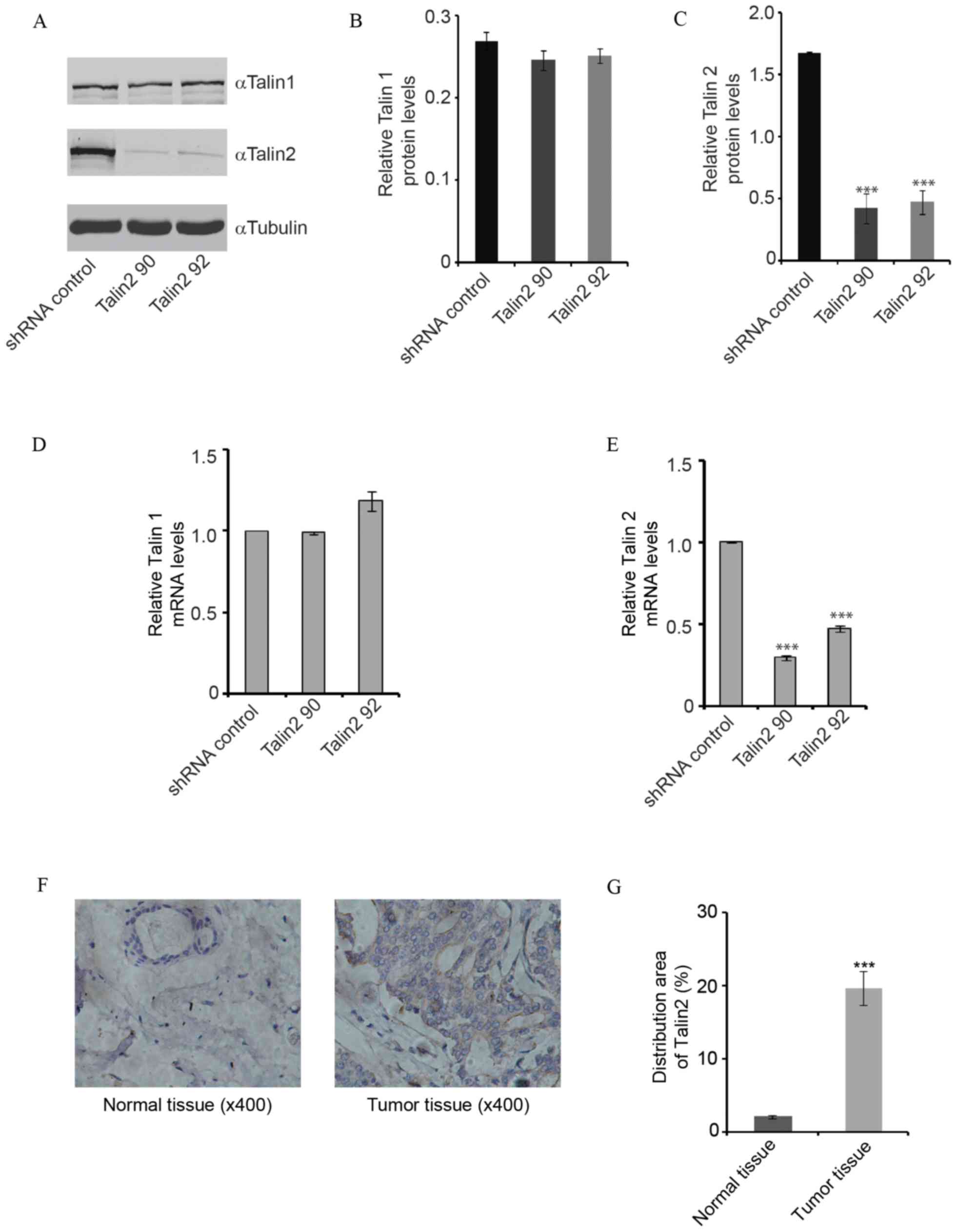
screened with puromycin for 3–4 weeks and then amplified (Fig. 1). The protein and mRNA expression
levels of Talin2 in the knockdown cell lines were significantly
lower compared with that in the Talin2 knockdown groups (Talin2 90
and Talin2 92) and shRNA control group, with ~2-fold decreases in
mRNA and protein levels (all P<0.001). However, no significant
differences in the mRNA or protein expression levels of Talin1 were
observed among the three groups (all P>0.05; Fig. 1A-F). The results revealed successful
construction of stable Talin2 knockdown cell lines, which laid the
foundation for the subsequent experiments.
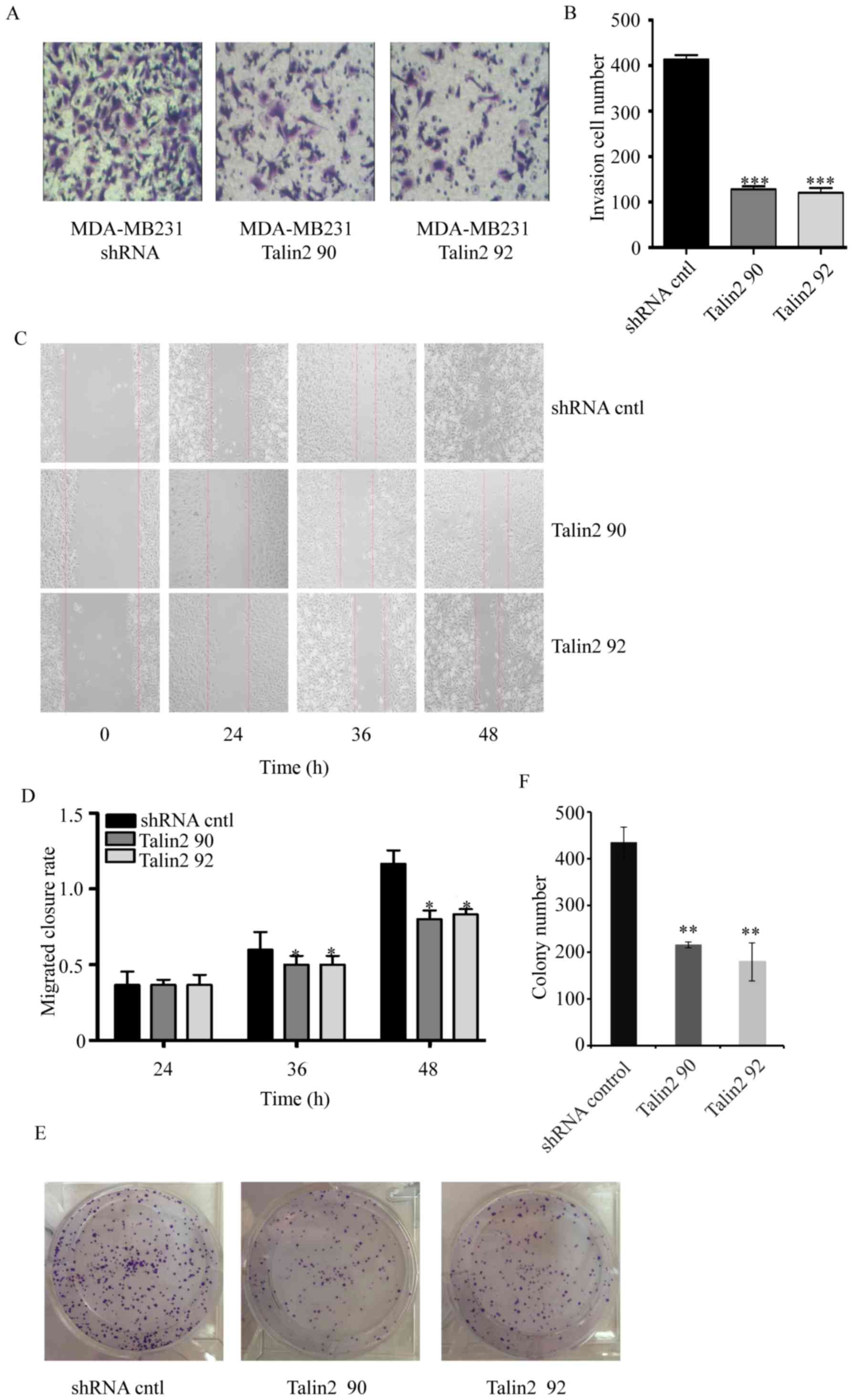
Talin2 regulates cancer cell invasion
and migration
To examine the role of Talin2 in breast cancer cell
invasion, MDA-MB-231 cells were infected with lentiviruses that
expressed Talin2 shRNA or shRNA control. The invasive growth
potential of these cells was measured by examining the functional
capacity of the cells to penetrate through Transwell membranes
coated with 0.35 mg/ml Matrigel (Fig.
2). The results indicated that the invasiveness of MDA-MB-231
cells was significantly reduced following Talin2 depletion (Talin2
90 and Talin2 92) as compared with that in the shRNA control group,
whereby the rates were reduced by 70 in Talin2 92 and 58% in Talin2
90 (Fig. 2A and B).
Talin2 expression levels were also analyzed in
different breast cancer tissues by IHC. As shown in Fig. 1F and G, the distribution area of
Talin2 in tumorous tissue was 8.4 times greater compared with that
in normal tissue (P<0.001). The results suggested that Talin2
expression is upregulated in breast cancer. To determine whether
Talin2 regulates the migration of breast cancer cells, an
artificial wound was created by scratching the cell monolayer of
MDA-MB-231 cells with the tip of a 10-µl pipette. The wound area
was examined after 24 and 48 h using an inverted phase-contrast
microscope with a digital camera. The effect of Talin2 on the
migratory ability of cells is shown in Fig. 2C and D. At 36 h, the wound healing
rate of Talin2 knockdown (Talin2 90 and Talin2 92) cells was
significantly reduced compared with that of cells in the blank
control group (P<0.05). At 48 h, the migratory ability of Talin2
knockdown cells was 0.625 times (Talin2 90) and 0.7 times (Talin2
92) that of the control cells. These results indicated that Talin2
had a significant suppressive effect on the migratory ability of
MDA-MB-231 cells. The colony formation assay also demonstrated that
the proliferative ability of the Talin2 knockdown (Talin2 90 and
Talin2 92) cells was significantly lower compared with that of
cells in the control group (Fig. 2E and
F).
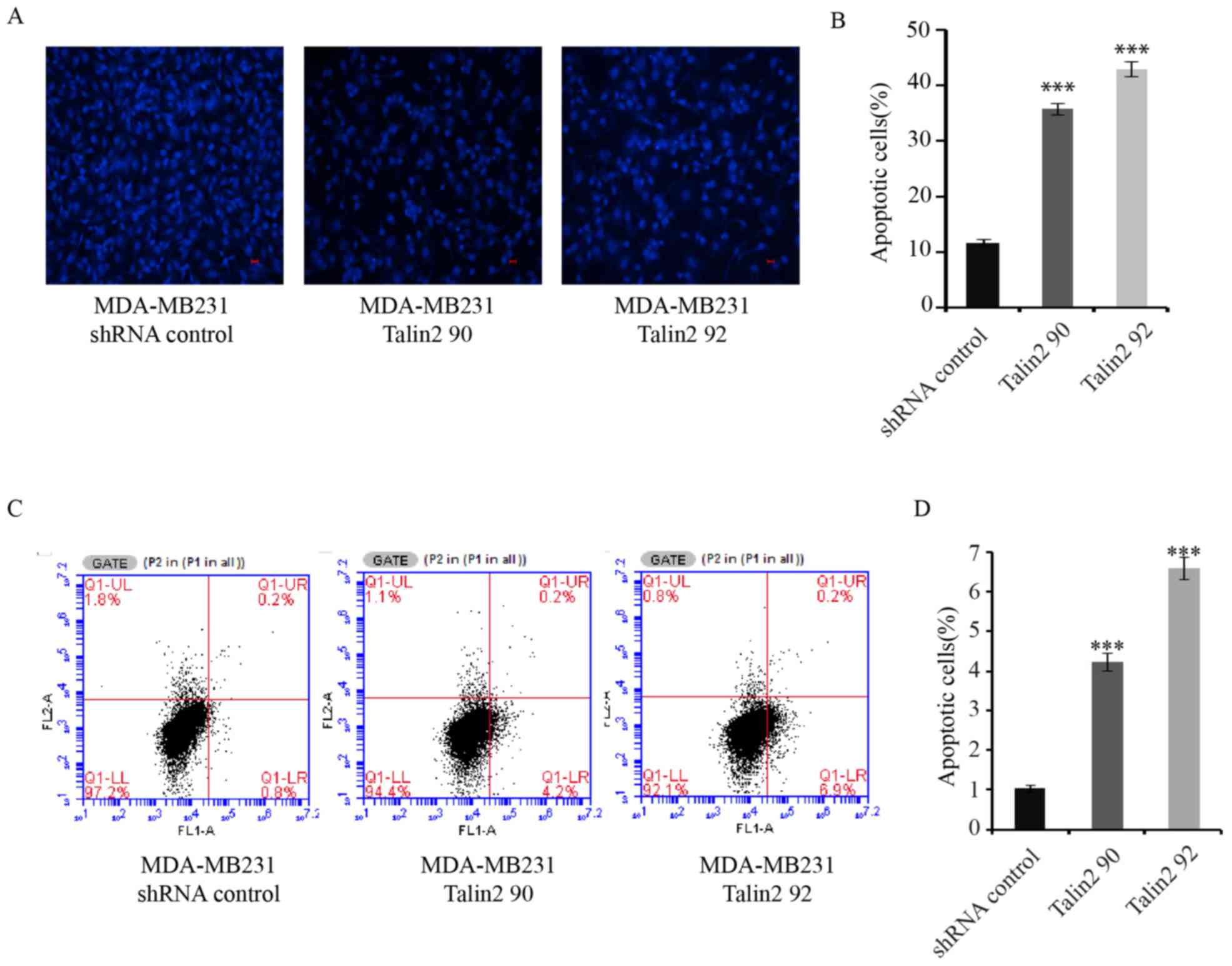
Talin2 knockdown promotes apoptosis of
MDA-MB-231 cells
The development of tumors is associated with the
disruption of homeostasis between cell proliferation and apoptosis
(22). The morphological changes in
MDA-MB-231 cells with Talin2 knockdown were examined with Hoechst
33342 staining. In Talin2-depleted (Talin2 90 and Talin2 92) cells,
the cell nuclei became agglutinated, were smaller in size and the
chromatin was partially condensed into small spherical or crescent
shapes, indicating that cells underwent apoptosis (Fig. 3A and B). In order to analyze the
effect of Talin2 on the apoptosis of MDA-MB-231 cells, the cell
apoptotic rate was detected by FACS analysis with AnnexinV-FITC and
PI double labeling. As shown in Fig. 3C
and D, the apoptotic rate of MDA-MB-231 cells was 1.03±0.088%,
while the apoptotic rates of Talin2 knockdown cells were 4.23±0218
(Talin2 90) and 6.6±0.265% (Talin2 92). The difference between the
Talin2 knockdown groups and the normal control group were
statistically significant (both P<0.001). These data suggest
that Talin2 knockdown promotes apoptosis of breast cancer
MDA-MB-231 cells.
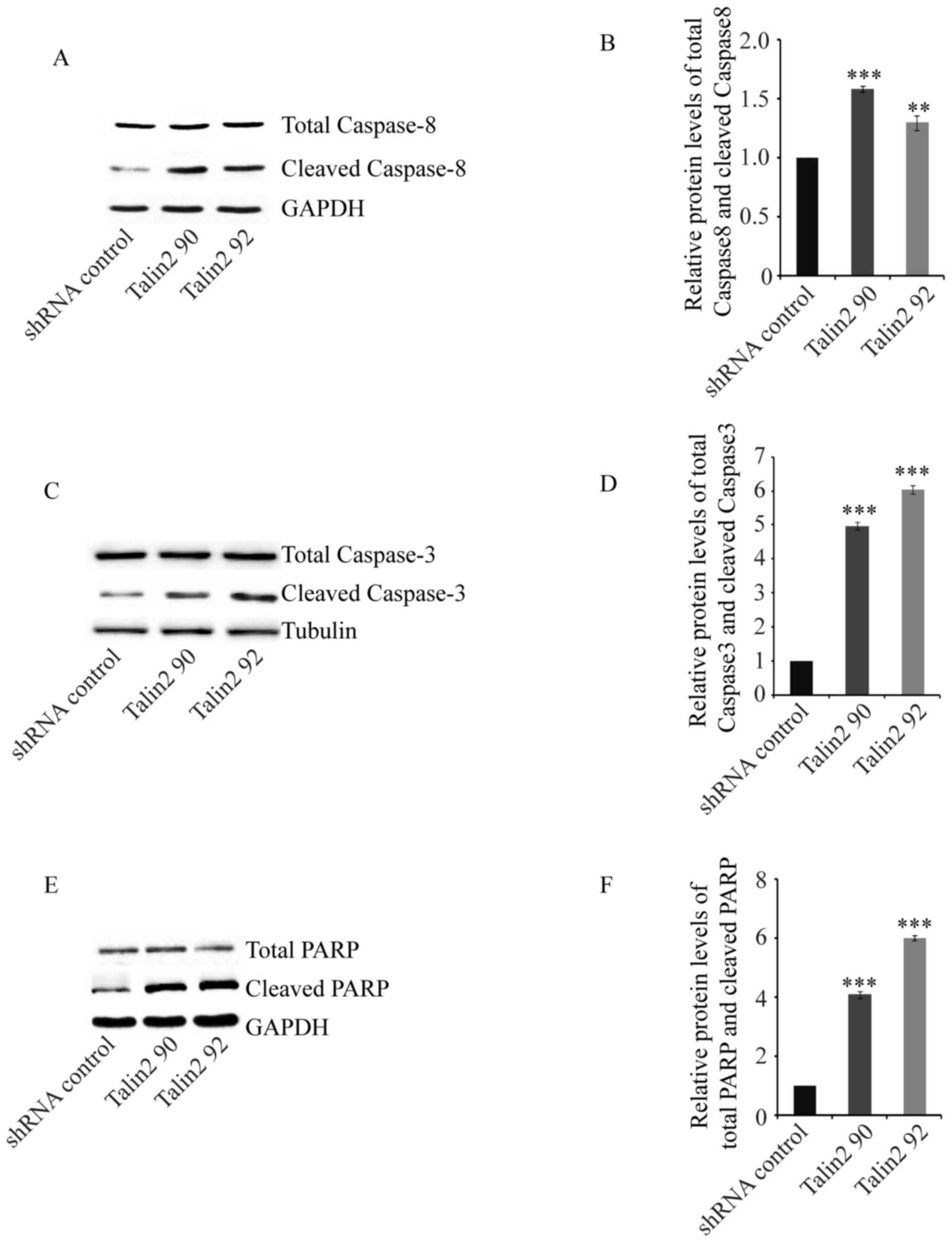
To further analyze the cause of Talin2-induced
apoptosis, the effect of Talin2 on the tumor apoptosis signaling
pathway was determined using western blot analysis. The protein
expression levels of Caspase-8, Caspase-3 and PARP (a substrate of
Caspase-3) were detected. The expression level of Caspase-8 is
shown in Fig. 4A and B. The total
expression levels of Caspase-8 in MDA-MB-231 cell and Talin2
knockdown (Talin2 90 and Talin2 92) cell were comparable; however,
the level of cleaved Caspase-8 in Talin2 knockdown cells was
significantly higher compared with that in shRNA
control-transfected MDA-MB-231 cells (P<0.001). Similarly, the
total expression levels of Caspase-3 in MDA-MB-231 cells and Talin2
knockdown (Talin2 90 and Talin2 92) cells were comparable; however,
the expression level of cleaved Caspase-3 in Talin2 knockdown cells
was significantly higher compared with that in the shRNA control
group (Fig. 4C and D; P<0.001).
PARP, the cleaved substrate of Caspase, is considered to be an
important indicator of cell apoptosis; it is also considered as an
indicator of Caspase-3 activation. The results of western blot
analysis revealed that the total expression of PARP was comparable
between MDA-MB-231 cells and Talin2 knockdown (Talin2 90 and Talin2
92) cells; however, PARP in Talin2 knockdown cells was
significantly activated (Fig. 4E and
F). This indicated that Caspase-8 induced by Fas cell surface
death receptor or tumor necrosis factor during apoptosis, directly
or indirectly activated the downstream effector molecules
Caspase-3, and that the activated Caspase-3, in turn, cleaved PARP
which reactivated the endonuclease, resulting in DNA fragmentation
and the promotion of apoptosis of tumor cells.
Discussion
The incidence of malignant tumors and the associated
mortality rate has steadily increased in China from 2000 to 2011
(2). The invasive growth of cancer
cells and metastasis are key impediments to the treatment of cancer
and a major contributor to cancer-associated mortality (4). The tumor microenvironment serves an
essential role in tumor invasion and metastasis. The ECM is the
primary component of the tumor microenvironment, and contains
various growth factors and cytokines, that modulate tumor
properties (23). Talin is a key
molecule among the ECM, integrins and cytoskeleton (24). Vertebrates express two closely
associated Talins encoded by separate genes. While it is well
established that Talin1 serves an important role in cell adhesion
and dissemination, little is known regarding the role of Talin2
(25).
To investigate the function of Talin2 in breast
cancer cells, Talin2 knockdown stable expression cell lines were
constructed and the functions of Talin2 were examined. The colony
formation assay demonstrated that Talin2 knockdown significantly
inhibited the proliferative ability of MDA-MB-231 cells. Wound
healing and invasion assays were used to investigate the effects of
Talin12 on MDA-MB-231 cell migration and invasion. The results of
the wound healing assay revealed that the migratory rate of Talin2
knockdown cells was significantly slower compared with that of
cells in the control group. At 48 h, the migratory ability of
Talin2 knockdown cells was ~0.7 times that of cells in the control
group. These results indicated that Talin2 regulates the migration
of MDA-MB-231 cells. Furthermore, Talin2 knockdown reduced the
invasive ability to ~60% of that observed in the control group,
indicating that Talin2 knockdown significantly inhibited the
invasiveness of MDA-MB-231 cells. In a recent study by Le et
al (26), increased miR-194
expression was revealed to markedly reduce the expression levels of
the cytoskeletal protein Talin2 and specifically inhibit the
migration of breast cancer cells. The present study did not
investigate the direct association between Talin2 and breast cancer
metastasis, which is a limitation and a potential focus for our
future research.
Apoptosis is the process of programmed cell death
that is regulated by multiple genes and is required for the
stability of the internal environment and the development of
multiple systems (27). Evasion of
apoptosis by tumor cells is a key challenge in the clinical
treatment of tumors. In the current study, Hoechst 33342 staining
demonstrated that the nuclei of Talin2 knockdown cells were
agglutinated and smaller, and the chromatin was partially condensed
into small spherical or crescent shapes. Furthermore, the results
of FACS Annexin V/PI assay demonstrated that the apoptosis rate in
the Talin2 knockdown group was significantly higher compared with
that in the control group. The aforementioned results suggest that
Talin2 knockdown promotes the apoptosis of MDA-MB-231 cells.
The apoptotic process is divided into two
categories: Caspase dependent and non-Caspase dependent. The
Caspase family is a mediator and executor of programmed cell death
in mammals. Proapoptotic signals culminate in activation of
different initiator Caspases that, in turn, activate effector
Caspases through enzyme cascade pathways. Activated effectors
cleave a set of substrates resulting in cellular disassembly.
Caspase is synthesized and stored in the form of an inactive
precursor under normal conditions, whereby apoptotic signaling then
activates the Caspase cascade reaction. Caspase-3 is known as the
‘executor’, and Caspase-8 and Capase-9 are referred to as the
‘initiators’ (28). Caspase-3 serves
a key role in the process of apoptosis. Upon activation, it
triggers a cascade of activation reactions, which lead to cell
apoptosis. Detection of Caspase-3 reflects cell apoptosis rate and
is also a marker of initiation of programmed apoptosis in the
cells. The western blot analysis results in the present study
revealed that the protein levels of cleaved Caspase-8 and Caspase-3
in the experimental groups were significantly higher compared with
that in the control group. PARP is a Caspase substrate and is
considered to be an important indicator of cell apoptosis. The
western blotting results demonstrated that PARP phosphorylation
levels in the Talin2 knockdown groups were higher compared with
those in the control group. Hence, we hypothesize that Talin2
knockdown activates Caspase-3, which then initiates a cascade
reaction, leading to cell apoptosis.
In conclusion, the results of the present study
suggest that Talin2 knockdown regulates MDA-MB-231 cell migration
and invasion through apoptosis. Thus, these results may provide a
novel target and basis for the diagnosis and treatment of breast
cancer.
Acknowledgements
Not applicable.
Funding
This work was supported in part by grants from the
Natural Science Foundation of Zhejiang Province (grant nos.
LY15C070003 and LY13H100003), Zhejiang University Student Science
and Technology Innovation Activity Plan and Xinmiao Talents Program
(grant nos. 2017R413056 and 2017R413087).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XL and YLL conceived and designed the experiments.
YFL, HWC, LJ and XFX performed the experiments. XL, YFL, JFD, HWC
analyzed the data. XL and YLL wrote the manuscript. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Board and Ethical
Committee of the First Affiliated Hospital of Wenzhou Medical
University. All study participants provided written informed
consent in accordance with the Declaration of Helsinki.
Consent for publication
All patients provided consent for the publication of
their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Fitzmaurice C, Allen C, Barber RM,
Barregard L, Bhutta ZA, Brenner H, Dicker DJ, Chimed-Orchir O,
Dandona R, Dandona L, et al: Global, regional, and national cancer
incidence, mortality, years of life lost, years lived with
disability, and disability-adjusted life-years for 32 cancer
groups, 1990 to 2015: A systematic analysis for the global burden
of disease study. JAMA Oncol. 3:524–548. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: Globocan 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Christofori G: New signals from the
invasive front. Nature. 441:444–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Burridge K and Connell L: A new protein of
adhesion plaques and ruffling membranes. J Cell Biol. 97:359–367.
1983. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Critchley DR: Biochemical and structural
properties of the integrin-associated cytoskeletal protein talin.
Annu Rev Biophys. 38:235–254. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Moser M, Legate KR, Zent R and Fassler R:
The tail of integrins, talin, and kindlins. Science. 324:895–899.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wehrle-Haller B: Structure and function of
focal adhesions. Curr Opin Cell Biol. 24:116–124. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Monkley SJ, Pritchard CA and Critchley DR:
Analysis of the mammalian talin2 gene TLN2. Biochem Biophys Res
Commun. 286:880–885. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Debrand E, El Jai Y, Spence L, Bate N,
Praekelt U, Pritchard CA, Monkley SJ and Critchley DR: Talin 2 is a
large and complex gene encoding multiple transcripts and protein
isoforms. FEBS J. 276:1610–1628. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Senetar MA, Moncman CL and McCann RO:
Talin2 is induced during striated muscle differentiation and is
targeted to stable adhesion complexes in mature muscle. Cell Motil
Cytoskeleton. 64:157–173. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tsujioka M, Yoshida K and Inouye K: Talin
B is required for force transmission in morphogenesis of
Dictyostelium. EMBO J. 23:2216–2225. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Praekelt U, Kopp PM, Rehm K, Linder S,
Bate N, Patel B, Debrand E, Manso AM, Ross RS, Conti F, et al: New
isoform-specific monoclonal antibodies reveal different
sub-cellular localisations for Talin1 and Talin2. Eur J Cell Biol.
91:180–191. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Senetar MA and McCann RO: Gene duplication
and functional divergence during evolution of the cytoskeletal
linker protein Talin. Gene. 362:141–152. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang C, Rajfur Z, Yousefi N, Chen Z,
Jacobson K and Ginsberg MH: Talin phosphorylation by Cdk5 regulates
Smurf1-mediated Talin head ubiquitylation and cell migration. Nat
Cell Biol. 11:624–630. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jin JK, Tien PC, Cheng CJ, Song JH, Huang
C, Lin SH and Gallick GE: Talin1 phosphorylation activates β1
integrins: A novel mechanism to promote prostate cancer bone
metastasis. Oncogene. 34:1811–1821. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sakamoto S, McCann RO, Dhir R and
Kyprianou N: Talin1 promotes tumor invasion and metastasis via
focal adhesion signaling and anoikis resistance. Cancer Res.
70:1885–1895. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fang KP, Dai W, Ren YH, Xu YC, Zhang SM
and Qian YB: Both Talin-1 and Talin-2 correlate with malignancy
potential of the human hepatocellular carcinoma MHCC-97 L cell. BMC
Cancer. 16:452016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wu Z, Li X, Sunkara M, Spearman H, Morris
AJ and Huang C: PIPKIgamma regulates focal adhesion dynamics and
colon cancer cell invasion. PLoS One. 6:e247752011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu B, Pang B, Hou X, Fan H, Liang N,
Zheng S, Feng B, Liu W, Guo H, Xu S and Pang Q: Expression of
high-mobility group AT-hook protein 2 and its prognostic
significance in malignant gliomas. Hum Pathol. 45:1752–1758. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li Na, Gao Junyan and Liu Min: Research on
the relationship between apoptosis and tumor. Contemporary
Medicine. 15:13–14. 2009.
|
|
23
|
Boggs AE, Vitolo MI, Whipple RA,
Charpentier MS, Goloubeva OG, Ioffe OB, Tuttle KC, Slovic J, Lu Y,
Mills GB and Martin SS: α-Tubulin acetylation elevated in
metastatic and basal-like breast cancer cells promotes
microtentacle formation, adhesion, and invasive migration. Cancer
Res. 75:203–215. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Haining AW, Lieberthal TJ and Del Rio
Hernandez A: Talin: A mechanosensitive molecule in health and
disease. FASEB J. 30:2073–2085. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li L, Li X, Qi L, Rychahou P, Jafari N and
Huang C: The role of talin2 in breast cancer tumorigenesis and
metastasis. Oncotarget. 8:106876–106887. 2017.PubMed/NCBI
|
|
26
|
Le XF, Almeida MI, Mao W, Spizzo R, Rossi
S, Nicoloso MS, Zhang S, Wu Y, Calin GA and Bast RC Jr: Modulation
of MicroRNA-194 and cell migration by HER2-targeting trastuzumab in
breast cancer. PLoS One. 7:e411702012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Krysko DV, Vanden Berghe T, D'Herde K and
Vandenabeele P: Apoptosis and necrosis: Detection, discrimination
and phagocytosis. Methods. 44:205–221. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li JY, Luo H and Yu L: FMNL2 regulates
cell migration and Src and Talin expressionin colorectaI cancer
cell. Shijie Huaren Xiao Hua Za Zhi. 20:289–295. 2012.
|