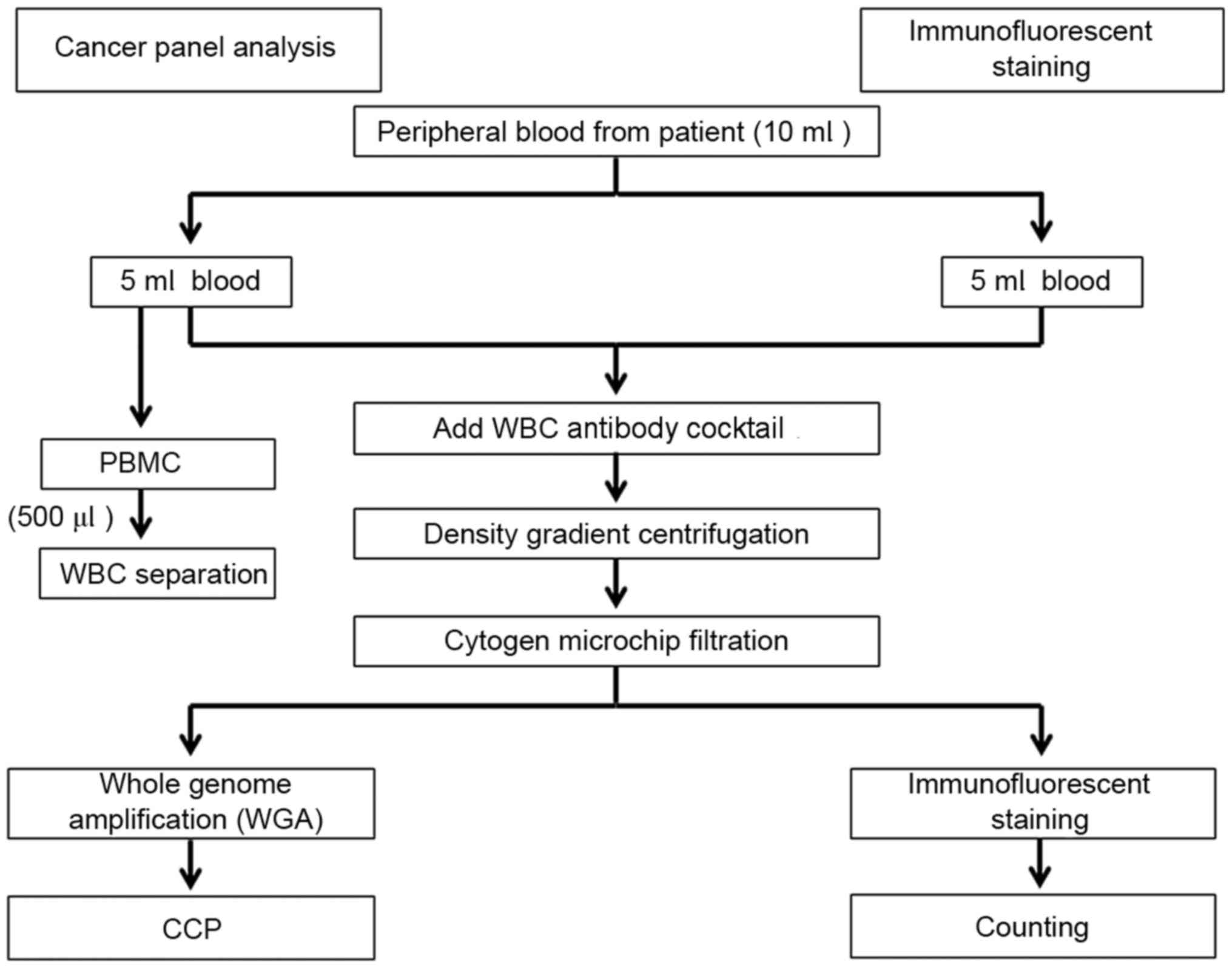
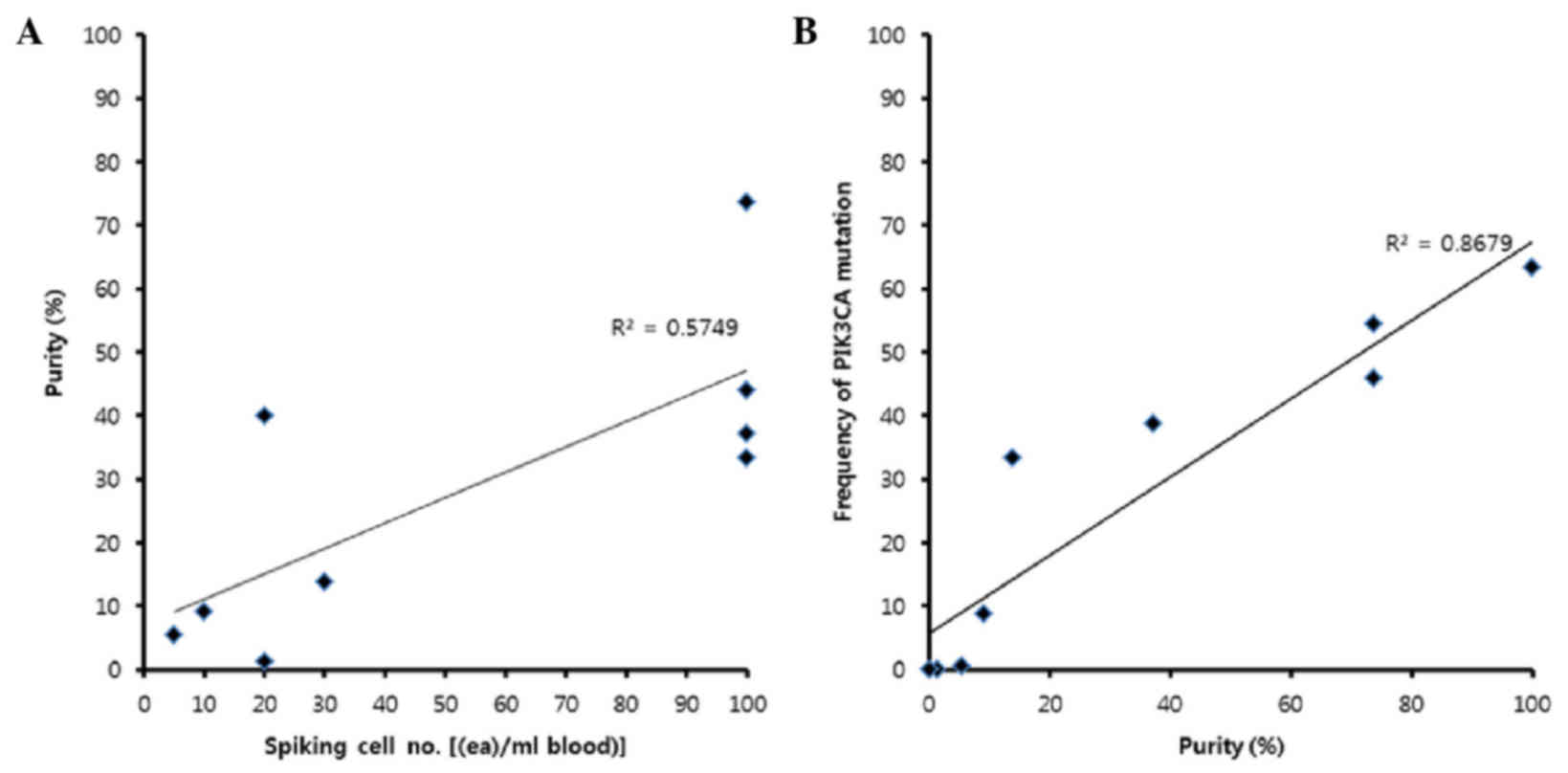
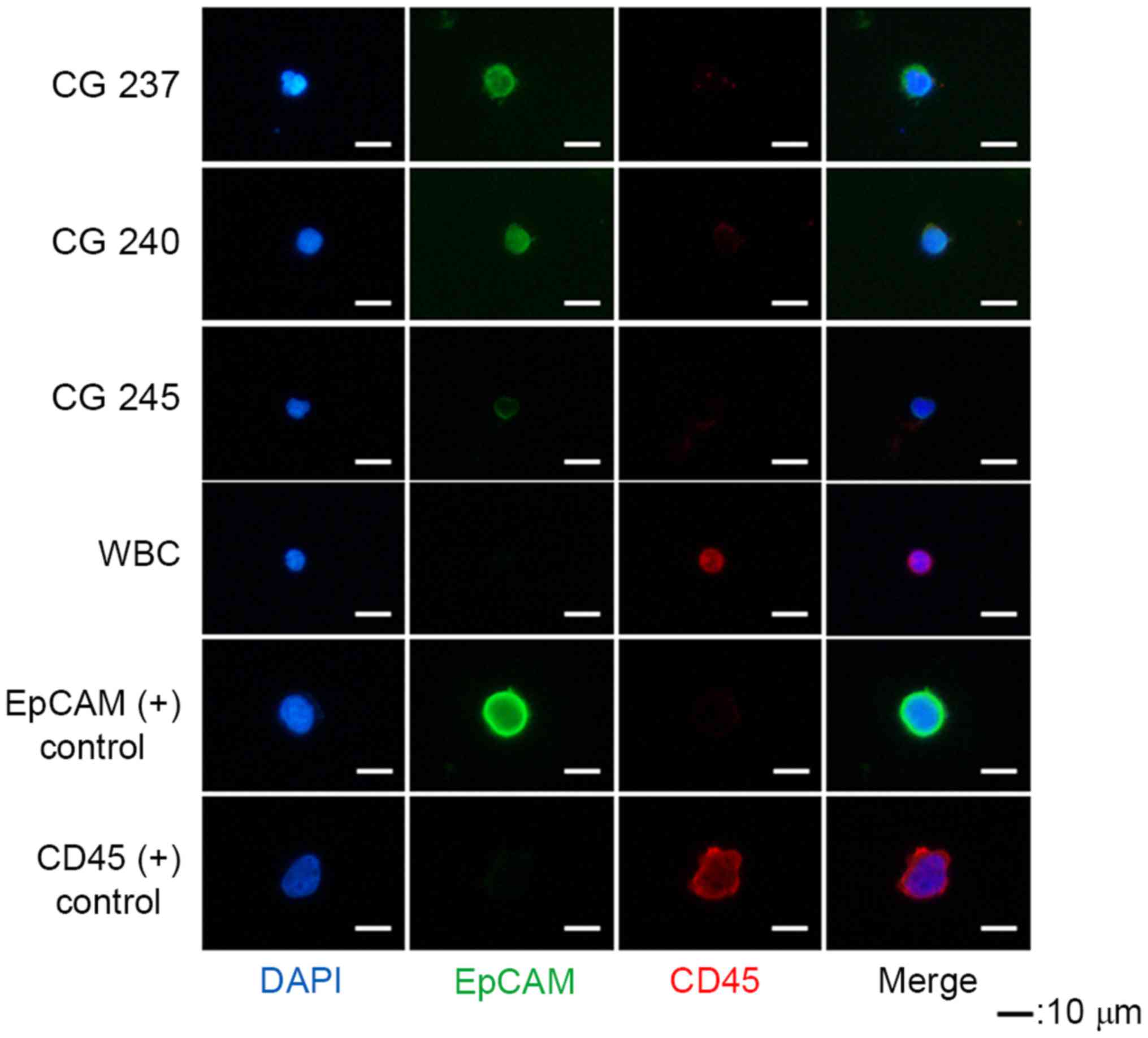
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jung KW, Won YJ, Kong HJ, Oh CM, Cho H,
Lee DH and Lee KH: Cancer statistics in Korea: Incidence,
mortality, survival, and prevalence in 2012. Cancer Res Treat.
47:127–141. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lorusso G and Rüegg C: New insights into
the mechanisms of organ-specific breast cancer metastasis. Semin
Cancer Boil. 22:226–233. 2012. View Article : Google Scholar
|
|
4
|
Suzuki M and Tarin D: Gene expression
profiling of human lymph node metastases and matched primary breast
carcinomas: Clinical implications. Mol Oncol. 1:172–180. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Von Minckwitz G, du Bois A, Schmidt M,
Maass N, Cufer T, de Jongh FE, Maartense E, Zielinski C, Kaufmann
M, Bauer W, et al: Trastuzumab beyond progression in human
epidermal growth factor receptor 2-positive advanced breast cancer:
A german breast group 26/breast international group 03–05 study. J
Clin Oncol. 27:1999–2006. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Peake BF and Nahta R: Resistance to
HER2-targeted therapies: A potential role for FOXM1. Breast Cancer
Manag. 3:423–431. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Blackwell KL, Burstein HJ, Storniolo AM,
Rugo H, Sledge G, Koehler M, Ellis C, Casey M, Vukelja S, Bischoff
J, et al: Randomized study of Lapatinib alone or in combination
with trastuzumab in women with ErbB2-positive,
trastuzumab-refractory metastatic breast cancer. J Clin Oncol.
28:1124–1130. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Modi S, Stopeck A, Linden H, Solit D,
Chandarlapaty S, Rosen N, D'Andrea G, Dickler M, Moynahan ME,
Sugarman S, et al: HSP90 inhibition is effective in breast cancer:
A phase II trial of tanespimycin (17-AAG) plus trastuzumab in
patients with HER2-positive metastatic breast cancer progressing on
trastuzumab. Clin Cancer Res. 17:5132–5139. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Franken B, de Groot MR, Mastboom WJ,
Vermes I, van der Palen J, Tibbe AG and Terstappen LW: Circulating
tumor cells, disease recurrence and survival in newly diagnosed
breast cancer. Breast Cancer Res. 14:R1332012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cohen SJ, Punt CJ, Iannotti N, Saidman BH,
Sabbath KD, Gabrail NY, Picus J, Morse M, Mitchell E, Miller MC, et
al: Relationship of circulating tumor cells to tumor response,
progression-free survival, and overall survival in patients with
metastatic colorectal cancer. J Clin Oncol. 26:3213–3221. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Van de Stolpe A, Pantel K, Sleijfer S,
Terstappen LW and den Toonder JM: Circulating tumor cell isolation
and diagnostics: Toward routine clinical use. Cancer Res.
71:5955–5960. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Giuliano M, Giordano A, Jackson S, De
Giorgi U, Mego M, Cohen EN, Gao H, Anfossi S, Handy BC, Ueno NT, et
al: Circulating tumor cells as early predictors of metastatic
spread in breast cancer patients with limited metastatic
dissemination. Breast Cancer Res. 16:4402014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim EH, Lee JK, Kim BC, Rhim SH, Kim JW,
Kim KH, Jung SM, Park PS, Park HC, Lee J, et al: Enrichment of
cancer cells from whole blood using a microfabricated porous
filter. Anal Biochem. 440:114–116. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
American Joint Committee on Cancer: AJCC
Cancer Staging Manual. 7th edition. Springer; 2010
|
|
15
|
Forbes SA, Bindal N, Bamford S, Cole C,
Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, et al:
COSMIC: Mining complete cancer genomes in the catalogue of somatic
mutations in cancer. Nucleic Acids Res. 39:D945–D950. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lang JM, Casavant BP and Beebe DJ:
Circulating tumor cells: Getting more from less. Sci Transl Med.
4:141ps132012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mostert B, Sleijfer S, Foekens JA and
Gratama JW: Circulating tumor cells (CTCs): Detection methods and
their clinical relevance in breast cancer. Cancer Treat Rev.
35:463–474. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Riethdorf S, Fritsche H, Müller V, Rau T,
Schindlbeck C, Rack B, Janni W, Coith C, Beck K, Jänicke F, et al:
Detection of circulating tumor cells in peripheral blood of
patients with metastatic breast cancer: A validation study of the
cellsearch system. Clin Cancer Res. 13:920–928. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Casavant BP, Mosher R, Warrick JW, Maccoux
LJ, Berry SM, Becker JT, Chen V, Lang JM, McNeel DG and Beebe DJ: A
negative selection methodology using a microfluidic platform for
the isolation and enumeration of circulating tumor cells. Methods.
64:137–143. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Giordano A, Gao H, Anfossi S, Cohen E,
Mego M, Lee BN, Tin S, De Laurentiis M, Parker CA, Alvarez RH, et
al: Epithelial-mesenchymal transition and stem cell markers in
patients with HER2-positive metastatic breast cancer. Mol Cancer
Ther. 11:2526–2534. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wu J and Crowe DL: The histone
methyltransferase EZH2 promotes mammary stem and luminal progenitor
cell expansion, metastasis and inhibits estrogen receptor-positive
cellular differentiation in a model of basal breast cancer. Oncol
Rep. 34:455–460. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shao S and Zhao X, Zhang X, Luo M, Zuo X,
Huang S, Wang Y, Gu S and Zhao X: Notch1 signaling regulates the
epithelial-mesenchymal transition and invasion of breast cancer in
a Slug-dependent manner. Mol Cancer. 14:282015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yuan X, Zhang M, Wu H, Xu H, Han N, Chu Q,
Yu S, Chen Y and Wu K: Expression of Notch1 correlates with breast
cancer progression and prognosis. PLoS One. 10:e01316892015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen Z, Trotman LC, Shaffer D, Lin HK,
Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, et al:
Crucial role of p53-dependent cellular senescence in suppression of
PTEN-deficient tumorigenesis. Nature. 436:725–730. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fernandez SV, Bingham C, Fittipaldi P,
Austin L, Palazzo J, Palmer G, Alpaugh K and Cristofanilli M: TP53
mutations detected in circulating tumor cells present in the blood
of metastatic triple negative breast cancer patients. Breast Cancer
Res. 16:4452014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu S, Wang H, Zhang L, Tang C, Jones L,
Ye H, Ban L, Wang A, Liu Z, Lou F, et al: Rapid detection of
genetic mutations in individual breast cancer patients by
next-generation DNA sequencing. Hum Genomics. 9:22015. View Article : Google Scholar : PubMed/NCBI
|