Introduction
Endometriosis is a heterogeneous estrogen-dependent
chronic gynecological disease in women of reproductive age
(1–3).
The condition is characterized by endometrial tissues ectopically
implanting outside the uterine cavity, and is subdivided mainly
into ovarian, peritoneal and deep infiltrating endometriosis
according to the different implant locations (4,5). The most
common clinical symptoms of endometriosis include dysmenorrhea,
chronic pelvic pain and diminished fertility potential, which
greatly influence the quality of life of affected individuals
(6). Despite numerous studies being
performed on this research field, the molecular etiology of
endometriosis is not yet fully understood (7,8).
It has long been accepted that endometriosis is
actually an inflammatory disease (9,10), with
cytokine levels that are elevated in the peritoneal fluid, serum
and endometriotic lesion tissues (11–13). Based
on these observations, certain researchers formulated the
inflammation hypothesis that leukocytes are recruited by the
endometrial stromal cells within endometriotic lesions, and that
proinflammatory cytokines are then secreted from these leukocytes,
which will facilitate the progression of endometriosis (14,15).
Furthermore, it has also been proposed that the changed
progesterone responsiveness may be the main reason for the elevated
production of cytokines in endometriosis (16–18).
However, in spite of advances (1,19), there
have not been any therapeutic strategies targeting inflammatory
factors in endometriosis that have been successfully applied to
samples with endometriosis, implying a great underappreciation of
the role of inflammation in endometriosis (20,21).
Caspase recruitment domain family member 11 (CARD11)
belongs to the CARD protein family; it can bind with B-cell
CLL/lymphoma 10 and activate the inflammation-associated nuclear
factor κB (NF-κB) signaling pathway (22). Furthermore, CARD11 also acts as a
scaffold protein to assist the assembly of multiprotein signaling
molecules in the plasma membrane (23). Previous studies showed that CARD11
somatic mutations were detected frequently in multiple human cancer
types, including hematological malignancies, colorectal cancer and
malignant melanoma; the mutations were distributed along the CARD11
coding sequence while being mainly clustered within the coiled-coil
domain (24–28). Most recently, a somatic mutation in
the CARD11 gene (p.R30W, c.C88T) was identified in the
endometriotic lesion of 1 out of 16 samples with ovarian
endometriosis. Considering the fact that endometriosis is a
potential premalignant disorder and the endometriotic lesions
harbored certain genetic alterations in tumor-associated genes
(29–33), and furthermore that mutations in the
paralogous genes usually occurred in the same cancer types
(34,35), we thus hypothesized that mutations in
the paralogous CARD10 and CARD11 may exist in ovarian
endometriosis.
In the present study, a total of 101 patients with
ovarian endometriosis were recruited and analyzed for the presence
of CARD10 and CARD11 mutations, with the aim of exploring the
potential involvement of mutations in these two genes in the
pathogenesis of ovarian endometriosis.
Materials and methods
Samples
The endometriotic lesions of the ovaries and paired
peripheral blood samples were obtained immediately following
surgical resection from a total of 142 patients with ovarian
endometriosis who were treated in the Department of Gynecology,
Jiangxi Provincial Maternal and Child Health Hospital (Nanchang,
Jiangxi, China), between July 2015 and January 2018. All samples
were diagnosed pathologically by two experienced pathologists, and
the 101 cases with an ectopic endometrium purity of >30% were
included in the present study. The study was approved by the
Institutional Review Board of Jiangxi Provincial Maternal and Child
Health Hospital, and the detailed protocol was conducted according
to the Declaration of Helsinki and the Jiangxi Provincial Maternal
and Child Health Hospital. Written informed consent was obtained
from all patients prior to the study.
Mutational analysis of the CARD10 and
CARD11 genes
The genomic DNA was isolated from the endometriotic
lesions and paired peripheral blood samples using the DNeasy Blood
and Tissue kit (catalog no. 69504; Qiagen, Inc., Valencia, CA, USA)
according to the manufacturer's protocols. The entire coding region
of the CARD10 and CARD11 genes in the endometriotic lesions of
patients with ovarian endometriosis were amplified by polymerase
chain reaction (PCR) with sets of primer pairs (Table I). In brief, for each PCR
amplification reaction, ~50 ng total DNA was used in a final volume
of 30 µl, with the following amplification protocols: An initial
pre-denaturation step at 94°C for 3 min, followed by 35 cycles of
denaturation at 94°C for 30 sec, annealing at different
temperatures (52–60°C; Table I) for
30 sec and extension at 72°C for 30 sec, with a final extension at
72°C for 7 min. PCR was performed in a Thermal Cycler 2720 (Applied
Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The
PCR amplification products were then purified and sequenced on an
ABI Prism 3730 DNA sequencer (Applied Biosystems; Thermo Fisher
Scientific, Inc.). An additional independent PCR amplification and
DNA sequencing was performed in the endometriotic lesions for
samples harboring potential CARD10 or CARD11 mutations. The
statuses of the potential somatic mutations were verified by
sequencing the DNA sequences of the paired peripheral blood in the
same patients. The procedure of PCR amplification and DNA
sequencing was performed as described above.
 | Table I.Polymerase chain reaction primers for
the amplification of the CARD10 and CARD11 genes. |
Table I.
Polymerase chain reaction primers for
the amplification of the CARD10 and CARD11 genes.
| Gene and exon | Forward primer
(5′-3′) | Reverse primer
(5′-3′) | Annealing, °C | Amplicon, bp |
|---|
| CARD10 |
|
|
|
|
| 1 |
ACATCTAGCCCTAGGGAGCC |
CCCACTCTACTGATGCGGAG | 52 |
433 |
| 2 |
CCCCTAGACCCTGGGTACAT |
CTTCAGCCTCTCCATGCCTC | 52 |
288 |
| 3 |
GAGCTGCCTATTCTGTCCCC |
CGTCAAAGAGCCAGCAATGG | 52 |
468 |
| 4 |
GCTTCGGCTTCTCTGGGAAT |
CCACAAGCCCCAGTACCTG | 58 |
405 |
|
5–6 |
CTGGTCAGGTGGGTTTGGAG |
GTCCTAACCCAACAGGGAGC | 52 | 1,088 |
| 7 |
TAATTTAGCTCAGGGCCCGG |
CTCCACAATCTTGACCCGCT | 56 |
358 |
|
8–9 |
CACATGGCAGGTGCTCAGTA |
GCTATCCCCAGCCATCTCAC | 52 |
687 |
|
10–11 |
CTTGGGGGCTGTGAGACATT |
TAAAACAGGGGCGAGGCAAT | 60 |
815 |
|
12–13 |
AGGCCTGGGGGTGGGAGTCA |
CAGGAGAAGGGCAATTGG | 55 |
976 |
|
14–15 |
GAAGGTCGGGTCCTGGGAA |
TGGGCACATAGTAGATGACCA | 52 |
756 |
| 16 |
AGGCTCTTCTGGCAAGCCT |
AGCATGACCCCCACTCCTGT | 56 |
372 |
|
17–19 |
TGGGGTATCGACGAGCTG |
AGAGAGGCATCCTCAAGGA | 60 | 1,189 |
| 20 |
ATCTTCTCTCTCTGACTCT |
TGAGGAGAGTGATGGGGAC | 58 | 1,192 |
| CARD11 |
|
|
|
|
| 3 |
GCTGTTCCAGTGAGACTGCT |
GACTGCGGACCCCAGTTTAA | 54 |
394 |
| 4 |
TGAGACCAGCCACAGAGACT |
GTGGTTGACAGACCCCAGTT | 56 |
366 |
| 5 |
CTGCGTCTGGAACCTCCTTT |
CCTGCACCTGCTTTATGGGA | 56 |
482 |
| 6 |
GGGAAGCGTTGCCTTTTCTG |
CAGGTTCATCGTTTCCCCCA | 54 |
326 |
|
7–8 |
CCAGGCACAAAACCTTCTGC |
AAACACTCTGAAGGAGCCGG | 60 | 1,132 |
| 9 |
GATGATGCCTGTCCCTGGG |
CTTCAGGCGTGGGGTCCT | 52 |
314 |
| 10 |
CCATCAGCCCAGCCATCTT |
CCAAGACCCACCCAGAAGC | 52 |
371 |
| 11 |
AAGCCCCAGTGACATGTGTC |
CGCAGGATTGTTCGTTACAG | 52 |
226 |
| 12 |
CTCCCCTCTCTCTCTTCCA |
GCTGGGTCCCTGGATGGCA | 60 |
324 |
| 13 |
TGCCGCCTGAGTAGGAGGC |
GAGGACAGCTGGGTCAGCA | 55 |
305 |
| 14 |
CCTCTCAGAAGCAAGGCCAC |
AGGCTTATCTTTTGTGTTC | 57 |
312 |
| 15 |
GACCAGCCCAGCAGGTCCC |
CCAGGAAGTGATTTCTGACT | 55 |
369 |
|
16–17 |
CACCCAGGCGCCTGATGAC |
GCCTTCCACTGAACAGACGA | 52 |
950 |
|
18–19 |
CTAAGAGCAGCATATTGCACA |
GAATTCATCATCCCAATACGG | 52 | 1,363 |
| 20 |
CTAAGAAAGCGTTAGCAT |
CACTGTGAAGAGTTGCAAGT | 58 |
351 |
| 21 |
TGCCGGGTTGAGGGCAGCA |
GACCGATGTTCCTCCAGGT | 52 |
299 |
|
22–23 |
ACACTGACGGTGGCTTCCA |
TTGTCCCTGGCCCAGGTG | 58 | 1,477 |
| 24 |
CCTGCATCCAGGCCCCGCT |
GGTCAGTCCCGTACTTGGTG | 54 |
336 |
| 25 |
TGGTGCCACACCAGCGCCA |
AGCGTCTGCTGGGGCAGCTC | 52 |
391 |
Evolutionary conservation
analysis
The protein sequences from 21 vertebrate species
from GenBank (https://www.ncbi.nlm.nih.gov/genbank/) were used to
analyze the evolutionary conservation status of the identified
CARD11 mutations, including Homo sapiens (NP_115791),
Macaca mulatta (XP_014988691), Rattus norvegicus
(XP_017454079), Mus musculus (NP_780571), Dipodomys
ordii (XP_012878958), Bos taurus (NP_001103266),
Microcebus murinus (XP_012614839), Equus asinus
(XP_014702187), Pan paniscus (XP_008971413), Cavia
porcellus (XP_013008240), Camelus ferus (XP_014412591),
Ovis aries (XP_014959439), Mustela putorius furo
(XP_0129032), Canis lupus familiaris (XP_005621), Gallus
gallus (NP_001006161), Calidris pugnax (XP_014801661),
Ficedula albicollis (XP_005054487), Lipotes
vexillifer (XP_007467483), Apteryx australis mantelli
(XP_013797455), Stegastes partitus (XP_008283939) and
Ictalurus punctatus (XP_017310081). Multiple sequence
alignment was performed using the ‘ClustalW’ tool of the alignment
function in the Molecular Evolutionary Genetics Analysis software
(MEGA, version 4.0) which was created and developed by Kumar et
al (36).
Bioinformatics programs prediction of
the CARD10 and CARD11 mutations
PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/) (37) and MutationTaster (http://mutationtaster.org/) (38) software were used to analyze the
disease-causing potential for the identified missense mutations,
while the SIFT (http://sift.jcvi.org/) program
(39) was used to predict the
potential pathogenicity of the in-frame deletions. All
bioinformatic analysis was performed on February 6th, 2018.
Together, the software automatically assessed each mutation as
either pathogenic or benign.
Statistical analysis
The frequency difference of CARD11 mutation in the
current study and the previous study (29) was analyzed by two-tailed Fisher's
exact test using SPSS software version 18.0 (SPSS Inc., Chicago,
IL, USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
CARD10 and CARD11 mutations in ovarian
endometriosis
The median age of the sample cohort was 43 years
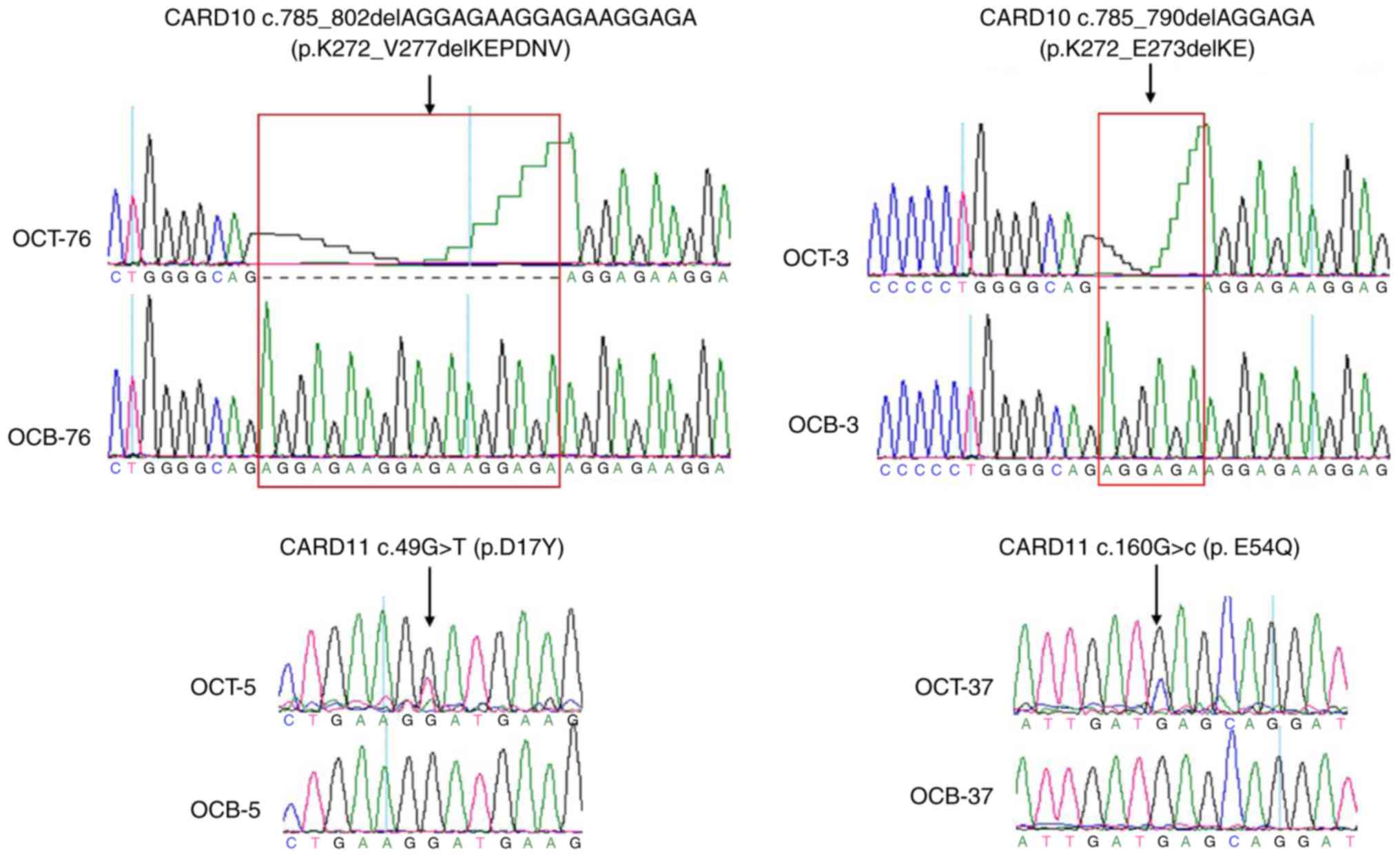
(range, 21–56 years). A total of 4 somatic mutations in CARD10 or
CARD11 genes were identified in 4 out of 101 endometriotic lesions
from the patients with ovarian endometriosis, with the somatic
status confirmed compared with that of the paired peripheral blood
(Fig. 1). The 2 mutations in CARD10
(c.785_790delAGGAGA, p.K272_E273delKE;
c.785_802delAGGAGAAGGAGAAGGAGA, p.K272_V277delKEPDNV) caused
in-frame deletions, while the 2 mutations in CARD11 (c.49G>T,
p.D17Y; c.160G>C, p.E54Q) were missense and heterozygous. It
should be noted that a previous study identified a CARD11 somatic
mutation (p.R30W) in 1 out of 16 (6.3%) patients with ovarian
endometriosis, but no CARD10 mutations (29). In the present study, the sample with
the CARD10 p.K272_E273delKE deletion was from a 47-year-old woman
who was also diagnosed with uterine leiomyoma, while the sample
with the CARD10 p.K272_V277delKEPDNV mutation was from a
43-year-old patient exhibiting a decreased blood eosinophil
granulocyte ratio (0.3%) (normal range: 0.4–8%) and elevated serum
creatine kinase level (314 U/l) (normal range for women: 96–140
U/l). The patient with the CARD11 p.D17Y mutation was 38 years old
and exhibited an increased level of cancer antigen 125 (45.4 U/ml)
(normal range: 0–35 U/ml), while the patient with the CARD11 p.E54Q
mutation was 46 years old and exhibited no other apparent
gynecological conditions.
Evolutionary conservation analysis of
CARD11 missense mutations
The evolutionary conservation analysis of CARD11 in
21 vertebrate species ranging from Homo sapiens to
Ictalurus punctatus showed that the two mutated amino acid
residues (p.D17, p.E54) were highly conserved (Fig. 2).
Causative potential of CARD10 and
CARD11 mutations
The PolyPhen-2 and MutationTaster online prediction
programs were used to predict the disease-causing potentials of the
detected missense mutations. The CARD11 p.D17Y (c.49G>T) and
p.E54Q (c.160G>C) mutations were predicted to be possibly
damaging with a score of >0.77 by Polyphen-2 software, while
predicted to be disease-causing with a score of >29 and a
probability of >0.99999 by MutationTaster. For the two in-frame
deletions in CARD10, p.K272_V277delKEPDNV was predicted to be
disease-causing, while p.K272_E273delKE may be benign, according to
the results predicted by the SIFT program. None of the mutations
were found in the dbSNP (https://www.ncbi.nlm.nih.gov/snp), ExAC (http://exac.broadinstitute.org/) or 1000 Genomes
Project (http://www.internationalgenome.org/) databases.
Discussion
Previous studies revealed that CARD10 and CARD11
served as scaffold proteins to regulate the activation of the NF-κB
signaling pathway under different cellular contexts, by recruiting
and activating the NF-κB repressor IκB kinase (22,40). When
considering that i) endometriosis is an inflammation-related
premalignant disease harboring somatic mutations in multiple genes
(9,10), ii) CARD11 somatic mutations exist in
ovarian endometriosis (29) and iii)
mutations in the paralogous genes occur frequently in multiple
human malignancies (34,35), the present study thus attempted to
investigate the potential mutational spectra of the paralogous
CARD10 and CARD11 genes in patients with ovarian endometriosis.
Frequent CARD11 mutations were detected first in
diffuse large B cell lymphoma (41)
and then also identified in melanoma, colorectal and endometrial
cancer (26–28). Besides being found in these different
cancer types, a previous study also identified a CARD11 somatic
mutation (p.R30W) in 1 out of 16 (6.3%) patients with ovarian
endometriosis (29). In the present
study, 2 novel CARD11 somatic mutations (p.D17Y and p.E54Q) were
detected in 2 out of 101 (2.0%) ovarian endometriosis cases; the
observed frequency in the current study did not reach a
statistically significant level when compared with that in the
prior study (P=0.40) (29). All the
identified mutations in the present study were somatic and not
found in the dbSNP, 1000 Genome Project or ExAC databases. The
evolutionary conservation analysis showed that the mutated amino
acids in CARD11 (p.D17Y and p.E54Q) were highly conserved in 21
vertebrate species. Furthermore, these mutations were predicted to
be disease-causing by Polyphen-2 and MutationTaster. It should be
mentioned that a somatic mutation in the 54th codon of CARD11,
p.E54V (c.161A>T), was previously identified in a patient with
hepatocellular carcinoma (42). Taken
together, these results indicated that these somatic mutations in
CARD11 may serve active roles in the development of ovarian
endometriosis.
Besides CARD11 mutations, 2 somatic in-frame
deletions in CARD10 were also identified in 2 out of 101 (2.0%)
cases of ovarian endometriosis. Since the indel mutations could be
predicted by SIFT, but not by Polyphen-2 and MutationTaster, the
p.K272_V277delKEPDNV deletion was predicted to be disease-causing,
while the p.K272_E273delKE deletion was predicted to be benign by
SIFT only. By contrast, Li et al (29) did not identify any CARD10 mutations in
16 cases with ovarian endometriosis, and we speculate that the
relatively small sample size may be a main reason for this
discrepancy. To the best of our knowledge, the present study is the
first to reveal that CARD10 somatic mutations also exist in ovarian
endometriosis. It should be noted that the p.K272_E273delKE somatic
deletion has also previously been identified in a cancerous tissue
from renal cell carcinoma (43)
(http://cancer.sanger.ac.uk/cosmic).
These results indicated that CARD10 p.K272_V277delKEPDNV deletion
may facilitate the progression of ovarian endometriosis, while the
role of the p.K272_E273delKE deletion requires further
elucidation.
In the present study, the 4 somatic mutations in
CARD10 and CARD11 were identified in 4 different individuals; this
is consistent with previous observations that mutations in the
paralogous genes are usually exhibited in a mutually exclusive
manner (34,35). The present study had several
limitations. Firstly, the sample size was small, which may affect
the mutation frequencies of the CARD10 and CARD11 genes, and the
identified mutations should be verified in a larger sample size.
Secondly, the study failed to obtain the clinical data showing
whether samples with CARD10 or CARD11 mutations will display higher
inflammation levels (such as interleukins) within the local
endometriotic lesions or serum. Finally, further investigation is
required to determine whether mutations would promote ovarian
endometriosis via enhanced inflammation.
In summary, the present study identified 4 novel
somatic mutations in CARD10 and CARD11 in 4.0% (4/101) of patients
with ovarian endometriosis for the first time. The results
indicated that these mutations existed in a mutually exclusive
manner and may serve a positive role in the pathogenesis of ovarian
endometriosis.
Acknowledgements
Not applicable.
Funding
This study was supported by grants from the Natural
Science Foundation of China (no. 81560255) and Jiangxi Province
(nos. 20161ACB21021, 20151BAB205011 and 20143ACG70016).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
YZ performed mutation detection and manuscript
preparation. JYZ and FW performed experiments. ZYZ performed
conservation analysis. FYL and JT performed data analysis. YL
performed mutation analysis. XZ and XDW performed sample
collection. OPH performed study design and manuscript revision.
Ethics approval and consent to
participate
The study was approved by the Institutional Review
Board of Jiangxi Provincial Maternal and Child Health Hospital, and
the detailed protocol was conducted according to the Declaration of
Helsinki and the Jiangxi Provincial Maternal and Child Health
Hospital. Written informed consent was obtained from all patients
prior to the study.
Consent for publication
All the sample donors gave permission for the
publication of their data prior to the study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Han SJ, Jung SY, Wu SP, Hawkins SM, Park
MJ, Kyo S, Qin J, Lydon JP, Tsai SY, Tsai MJ, et al: Estrogen
receptor β modulates apoptosis complexes and the inflammasome to
drive the pathogenesis of endometriosis. Cell. 163:960–974. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ahn SH, Edwards AK, Singh SS, Young SL,
Lessey BA and Tayade C: IL-17A contributes to the pathogenesis of
endometriosis by triggering proinflammatory cytokines and
angiogenic growth factors. J Immunol. 195:2591–2600. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kocbek V, Grandi G, Blank F, Wotzkow C,
Bersinger NA, Mueller MD, Kyo S and McKinnon BD: TNFα-induced IKKβ
complex activation influences epithelial, but not stromal cell
survival in endometriosis. Mol Hum Reprod. 22:768–777. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chan RW, Ng EH and Yeung WS:
Identification of cells with colony-forming activity, self-renewal
capacity, and multipotency in ovarian endometriosis. Am J Pathol.
178:2832–2844. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vouk K, Hevir N, Ribić-Pucelj M,
Haarpaintner G, Scherb H, Osredkar J, Möller G, Prehn C, Rižner TL
and Adamski J: Discovery of phosphatidylcholines and sphingomyelins
as biomarkers for ovarian endometriosis. Hum Reprod. 27:2955–2965.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Barbieri M, Somigliana E, Oneda S, Ossola
MW, Acaia B and Fedele L: Decidualized ovarian endometriosis in
pregnancy: A challenging diagnostic entity. Hum Reprod.
24:1818–1824. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kim TH, Yoo JY, Wang Z, Lydon JP, Khatri
S, Hawkins SM, Leach RE, Fazleabas AT, Young SL, Lessey BA, et al:
ARID1A is essential for endometrial function during early
pregnancy. PLoS Genet. 11:e10055372015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Borrelli GM, Abrão MS, Taube ET,
Darb-Esfahani S, Köhler C, Chiantera V and Mechsner S: (Partial)
Loss of BAF250a (ARID1A) in rectovaginal deep-infiltrating
endometriosis, endometriomas and involved pelvic sentinel lymph
nodes. Mol Hum Reprod. 22:329–337. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Raible MD: Pathophysiology and treatment
of endometriosis. Am J Hosp Pharm. 38:1696–1701. 1981.PubMed/NCBI
|
|
10
|
Gazvani R and Templeton A: Peritoneal
environment, cytokines and angiogenesis in the pathophysiology of
endometriosis. Reproduction. 123:217–226. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kang YJ, Jeung IC, Park A, Park YJ, Jung
H, Kim TD, Lee HG, Choi I and Yoon SR: An increased level of IL-6
suppresses NK cell activity in peritoneal fluid of patients with
endometriosis via regulation of SHP-2 expression. Hum Reprod.
29:2176–2189. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Suen JL, Chang Y, Chiu PR, Hsieh TH, His
E, Chen YC, Chen YF and Tsai EM: Serum level of IL-10 is increased
in patients with endometriosis, and IL-10 promotes the growth of
lesions in a murine model. Am J Pathol. 184:464–471. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jiang JF, Deng Y, Xue W, Zheng TP and Sun
AJ: Increased expression of Interleukin 37 in the eutopic and
ectopic endometrium of patients with ovarian endometriosis. Reprod
Sci. 23:244–248. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Taylor RN, Kane MA and Sidell N:
Pathogenesis of endometriosis: Roles of retinoids and inflammatory
pathways. Semin Reprod Med. 33:246–256. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Taylor RN, Yu J, Torres PB, Schickedanz
AC, Park JK, Mueller MD and Sidell N: Mechanistic and therapeutic
implications of angiogenesis in endometriosis. Reprod Sci.
16:140–146. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Attia GR, Zeitoun K, Edwards D, Johns A,
Carr BR and Bulun SE: Progesterone receptor isoform A but not B is
expressed in endometriosis. J Clin Endocrinol Metab. 85:2897–2902.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Osteen KG, Bruner-Tran KL and Eisenberg E:
Reduced progesterone action during endometrial maturation: A
potential risk factor for the development of endometriosis. Fertil
Steril. 83:529–537. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Luk J, Seval Y, Kayisli UA, Ulukus M,
Ulukus CE and Arici A: Regulation of interleukin-8 expression in
human endometrial endothelial cells: A potential mechanism for the
pathogenesis of endometriosis. J Clin Endocrinol Metab.
90:1805–1811. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nothnick W and Alali Z: Recent advances in
the understanding of endometriosis: The role of inflammatory
mediators in disease pathogenesis and treatment. F1000Res. 5:pii:
F1000 Faculty Rev-186. 2016.PubMed/NCBI
|
|
20
|
Platteeuw L and D'Hooghe T: Novel agents
for the medical treatment of endometriosis. Curr Opin Obstet
Gynecol. 26:243–252. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bruner-Tran KL, Herington JL, Duleba AJ,
Taylor HS and Osteen KG: Medical management of endometriosis:
Emerging evidence linking inflammation to disease pathophysiology.
Minerva Ginecol. 65:199–213. 2013.PubMed/NCBI
|
|
22
|
Yang YK, Yang C, Chan W, Wang Z, Deibel KE
and Pomerantz JL: Molecular determinants of scaffold-induced linear
ubiquitinylation of B cell lymphoma/leukemia 10 (Bcl10) during T
cell receptor and oncogenic caspase recruitment domain-containing
protein 11 (CARD11) signaling. J Biol Chem. 291:25921–25936. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Thome M and Tschopp J: TCR-induced
NF-kappaB activation: A crucial role for Carma1, Bcl10 and MALT1.
Trends Immunol. 24:419–424. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Davis RE, Ngo VN, Lenz G, Tolar P, Young
RM, Romesser PB, Kohlhammer H, Lamy L, Zhao H, Yang Y, et al:
Chronic active B-cell-receptor signalling in diffuse large B-cell
lymphoma. Nature. 463:88–92. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
da Silva Almeida AC, Abate F, Khiabanian
H, Martinez-Escala E, Guitart J, Tensen CP, Vermeer MH, Rabadan R,
Ferrando A and Palomero T: The mutational landscape of cutaneous T
cell lymphoma and Sézary syndrome. Nat Genet. 47:1465–1470. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Giannakis M, Hodis E, Mu Jasmine X,
Yamauchi M, Rosenbluh J, Cibulskis K, Saksena G, Lawrence MS, Qian
ZR, Nishihara R, et al: RNF43 is frequently mutated in colorectal
and endometrial cancers. Nat Genet. 46:1264–1266. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mouradov D, Sloggett C, Jorissen RN, Love
CG, Li S, Burgess AW, Arango D, Strausberg RL, Buchanan D, Wormald
S, et al: Colorectal cancer cell lines are representative models of
the main molecular subtypes of primary cancer. Cancer Res.
74:3238–3247. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shain AH, Garrido M, Botton T, Talevich E,
Yeh I, Sanborn JZ, Chung J, Wang NJ, Kakavand H, Mann GJ, et al:
Exome sequencing of desmoplastic melanoma identifies recurrent
NFKBIE promoter mutations and diverse activating mutations in the
MAPK pathway. Nat Genet. 47:1194–1199. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li X, Zhang Y, Zhao L, Wang L, Wu Z, Mei
Q, Nie J, Li X, Li Y, Fu X, et al: Whole-exome sequencing of
endometriosis identifies frequent alterations in genes involved in
cell adhesion and chromatin-remodeling complexes. Hum Mol Genet.
23:6008–6021. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lee AW, Templeman C, Stram DA, Beesley J,
Tyrer J, Berchuck A, Pharoah PP, Chenevix-Trench G and Pearce CL:
Ovarian Cancer Association Consortium: Evidence of a genetic link
between endometriosis and ovarian cancer. Fertil Steril. 105:35–43.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Matsumoto T, Yamazaki M, Takahashi H,
Kajita S, Suzuki E, Tsuruta T and Saegusa M: Distinct β-catenin and
PIK3CA mutation profiles in endometriosis-associated ovarian
endometrioid and clear cell carcinomas. Am J Clin Pathol.
144:452–463. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Worley MJ Jr, Liu S, Hua Y, Kwok JS,
Samuel A, Hou L, Shoni M, Lu S, Sandberg EM, Keryan A, et al:
Molecular changes in endometriosis-associated ovarian clear cell
carcinoma. Eur J Cancer. 51:1831–1842. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Anglesio MS, Papadopoulos N, Ayhan A,
Nazeran TM, Noë M, Horlings HM, Lum A, Jones S, Senz J, Seckin T,
et al: Cancer-associated mutations in endometriosis without cancer.
N Engl J Med. 376:1835–1848. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yan H, Parsons DW, Jin G, McLendon R,
Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ,
et al: IDH1 and IDH2 mutations in gliomas. N Engl J Med.
360:765–773. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zou Y, Zeng Y, Zhang DF, Zou SH, Cheng YF
and Yao YG: IDH1 and IDH2 mutations are frequent in Chinese
patients with acute myeloid leukemia but rare in other types of
hematological disorders. Biochem Biophys Res Commun. 402:378–383.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kumar S, Stecher G and Tamura K: MEGA7:
Molecular evolutionary genetics analysis version 7.0 for bigger
datasets. Mol Biol Evol. 33:1870–1874. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Adzhubei IA, Schmidt S, Peshkin L,
Ramensky VE, Gerasimova A, Bork P, Kondrashov AS and Sunyaev SR: A
method and server for predicting damaging missense mutations. Nat
Methods. 7:248–289. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Schwarz JM, Cooper DN, Schuelke M and
Seelow D: MutationTaster2: Mutation prediction for the
deep-sequencing age. Nat Methods. 11:361–362. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kumar P, Henikoff S and Ng PC: Predicting
the effects of coding non-synonymous variants on protein function
using the SIFT algorithm. Nat Protoc. 4:1073–1081. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Blonska M and Lin X: NF-κB signaling
pathways regulated by CARMA family of scaffold proteins. Cell Res.
21:55–70. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lenz G, Davis RE, Ngo VN, Lam L, George
TC, Wright GW, Dave SS, Zhao H, Xu W, Rosenwald A, et al: Oncogenic
CARD11 mutations in human diffuse large B cell lymphoma. Science.
319:1676–1679. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kan Z, Zheng H, Liu X, Li S, Barber TD,
Gong Z, Gao H, Hao K, Willard MD, Xu J, et al: Whole-genome
sequencing identifies recurrent mutations in hepatocellular
carcinoma. Genome Res. 23:1422–1433. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Arai E, Sakamoto H, Ichikawa H, Totsuka H,
Chiku S, Gotoh M, Mori T, Nakatani T, Ohnami S, Nakagawa T, et al:
Multilayer-omics analysis of renal cell carcinoma, including the
whole exome, methylome and transcriptome. Int J Cancer.
135:1330–1342. 2014. View Article : Google Scholar : PubMed/NCBI
|