Introduction
Renal cell carcinoma (RCC) is a common cancer of the
genitourinary tract. Previous research suggests that RCC is
responsible for ~3% of all adult tumors, and its frequency and
associated mortality rates are increasing worldwide over the past 8
years (1,2). Typically, patients present with
advanced-stage RCC at the time of diagnosis due to the insidious
onset of RCC, which accounts for the associated poor prognosis, and
the high morbidity and mortality rates of RCC (3). Increasing resistance to targeted
therapies is a common occurrence in advanced RCC (4). Resistance to targeted therapy may be
mediated by several mechanisms including dysregulation of genes,
angiogenesis and decreased intake of TKI inhibitors by cancer cells
(5). Thus, there is an urgent
requirement to identify effective biomolecular markers to assist in
early diagnosis and provide prognosistic information for RCC, as
these do not currently exist in clinical practice.
High-throughput sequencing has been widely used to
generate mass data, particularly in cancer. For example, the
cBioPortal database (6,7), which includes vast quantities of
clinical samples and sequence data, provides visible, free-access
and clear genomics datasets. In the present study, a series of long
non-coding (lnc)RNAs associated with poor prognosis for patients
with breast cancer was elucidated. Following the selection of one
lncRNA for further study, it was identified that a specific
knockout significantly inhibited cell proliferation in breast
cancer (8). Previous bioinformatic
analyses suggested certain genes involved in tumorigenesis; for
example, by using a bioinformatic approach to study ovarian cancer,
researchers identified key genes, including CCNB1, CENPF, KIF11,
and ZWINT, and, therefore, were able to suggest potential
therapeutic targets (9).
The present study compared two sets of microarray
data to identify the various common genes. These genes were
subsequently investigated using a bioinformatics approach, and a
portable network graphic was established. A significant module of
11 hub genes was obtained from this network, prior to further
investigation using the cBioPortal dataset. Finally, among these
hub genes, COPS7B was identified to serve an oncogenic function and
to be associated with the prognosis of RCC.
Materials and methods
Gene expression ombnibus (GEO)
database and data processing
PubMed (https://www.ncbi.nlm.nih.gov/pubmed) and the GEO
database (https://www.ncbi.nlm.nih.gov/geo) were used in the
present study. The datasets GSE40435 (10) and GSE53757 (11) were the most abundant sources of
relevant data. The GSE40435 and GSE53757 gene expression profile
datasets were downloaded from the GEO database. The GSE40435 gene
expression profiles included 101 RCC tissues and 101 normal
tissues. The GSE53757 gene expression profiles consisted of 60 RCC
tissues and 60 normal tissues. Comparisons of the two GSE datasets
were performed and the common genes were identified; the expression
profiles of these common genes in GSE40435 were copied to another
file. A heat map was constructed using Morpheus (https://software.broadinstitute.org/morpheus), and the
metric in the heat map was set as the signal to noise (STN), where
altered STN suggested significant expression alteration. The values
(>3 or ≤3) suggest significant fold change of expressions. Class
A and B were set as tumor and normal, respectively. The present
study ran the Gene Ontology (GO) and Kyoto Encyclopedia of Genes
and Genomes (KEGG) analyses in DAVID version 6.8 (http://david.abcc.ncifcrf.gov).
Gene network building and hub genes
analysis
The present study included all common genes
identified in the two GEO Serious (GSE) datasets (GSE40435 and
GSE53757) located within the STRING database (http://string.embl.de/). A protein-protein interaction
(PPI) network was constructed using Cytoscape version 3.6
(http://www.cytoscape.org). All datasets were
identical to those previously reported (9).
Database search of cBioPortal
All hub genes were input and recognized in the
cBioPortal (http://www.cbioportal.org/). The dataset for RCC
carcinoma (TCGA, provisional) was selected as it has numerous
cases, compared with other datasets. Alterations in the mutations,
copy number and expression of various hub genes were analyzed.
Clinical feathers, including stage, metastasis, the overall
survival and disease free survival, sex, age, laterality and volume
were analyzed based on the clinical data from cBioPortal. The
cBioPortal also generated the Kaplan-Meier curve following input of
the gene name for disease/progression-free-survival. P<0.05 was
considered to indicate a statistically significant difference.
Cell culture, regents and siRNA
transfection
The Human kidney cortex proximal tubule cell line
(HK-2) and 786-O, 769-P and A498 cell lines were obtained from the
American Type Culture Collection (Manassas, VA, USA) and cultured
in RPMI-1640 medium (Hyclone; GE Healthcare Life Sciences, Logan,
UT, USA) supplemented with 10% fetal bovine serum (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA). and 2 mM glutamine
(Sangon Biotech Co., Ltd., Shanghai, China). Polymerase chain
reaction (PCR) primers (Table I) were
purchased from Sangon Biotech Co., Ltd. (Shanghai, China). The
small interfering (si)RNA negative control and COPS7B siRNA at the
concentration of 40 nM were purchased from Shanghai GenePharma Co.,
Ltd., Shanghai, China. The siRNAs were transfected into cells using
Lipofectamine®2000 (Invitrogen, Thermo Fisher
Scientific, Inc., Waltham, MA, USA) for 48 h prior to subsequent
experimentation. A random sequence (Sense:
5′-UUCUCCGAACGUGUCACGUTT-3′ Antisense: 5′-ACGUGACACGUUCGGAGAATT-3′)
from Shanghai GenePharma Co., Ltd. was used as the negative
control.
 | Table I.Primers in the present study. |
Table I.
Primers in the present study.
| Gene | Sense | Antisense |
|---|
| CA9 |
AGGTGAATGCAGAGGTGACA |
AAGCTGGAGAGAGAGGGAGA |
| NDUFA4L2 |
AAAAGTTGGCATGGCAGGAG |
GCTCCGGGTTGTTCTTTCTG |
| HRG |
CTTCTACCACCAAGCCTCCA |
GCAGGACATGGGCAATAGTG |
| KCNJ1 |
TCTTCGGAAATGGGTCGTCA |
CTGCATACCACAGGAGACCA |
| KNG1 |
GGTTGGCTCTGACACGTTTT |
TGGGTAGCCACGGAGAATTT |
| UMOD |
GTTTTCATCCCGCAGAGCAA |
ATCGCCCGGTTTAAATGTCG |
| COPS7B |
AAAGACCTGGAGATGCGGAA |
AGCCATCACACCATTCATGC |
Reverse transcription-quantitative
(RT-q)PCR
Total RNA of RCC tissue and cell lines were
extracted with TRIzol® reagent (Thermo Fisher
Scientific, Waltham, MA, USA.) prior to concentration determination
with a Nanodrop reader (Thermo Fisher Scientific, Inc.). RNA was
reverse transcribed to cDNA with ReverTra Ace-α-®
(Toyobo, Osaka, Japan). The RT-qPCR was performed with Brilliant
SYBR Green Master mix (Bio-Rad Laboratories, Hercules, CA, USA) on
a Roche LightCycler® 480 Instrument II (Roche Applied
Science, Mannheim, Germany) according to the study of Liu et
al (8). All primers are listed in
Table I. The thermocycling conditions
were as follows: 95°C for 5 min, 95°C for 10 sec and 60°C for 30
sec (40 cycles), 95°C for 15 sec, 60°C for 60 sec and 95°C for 15
sec.
Cell survival assay
The 769-P cells (2×104) were seeded in
12-well plates and cultured overnight following transfection with
siRNA-COP9 signalosome subunit 7B (COPS7B) or siRNA control for 48
h. In total, 10 µl Cell Counting Kit-8 (Dojindo, Rockville, MD,
USA) was added after 24, 48 and 72 h. Optical density (OD) values
were determined based on the absorbance at 450 nm with Nanodrop
reader. Cell survival was calculated according to the following
formula: [OD(time)-OD(blank)]/[OD(0
h)-OD(blank)].
Colony formation
Following transfection with the siRNA control or
si-COPS7B at 37°C for 48 h, 2,000 cells/well were seeded in 6-well
plates. At 2 weeks after seeding, colonies were fixed and stained
with 0.1% crystal violet at room temperature for 10 min. Finally,
all the colonies were counted with the naked eye, and then
colony-forming efficiency was calculated as described previously
(8).
Cell migration assay
Following transfection with the siRNA control or
si-COPS7B at 37°C for 48 h, 769-P cells were starved for 3 h prior
to cell counting. In total, 1×104 cells in serum-free
medium were seeded into the upper chambers of a BioCoat invasion
system from BD Biosciences (San Jose, CA, USA) and completed
RPMI-1640 was placed in the lower chamber. Then it was incubated at
37°C and 5% CO2 overnight. After 24 h, cells in the
upper membrane were wiped away with a cotton pad. The cells on the
lower surface of the membrane were stained with crystal violet at a
concentration of 0.1% at room temperature for 30 min. The stained
769-P cells were counted and the mean cell numbers were calculated
in five fields with light microscope at ×40 magnification (12).
Statistical analysis
Statistical analysis was performed using SPSS
software (version 17; SPSS Inc., Chicago, IL, USA). One-way
analysis of variance was conducted, with the Tamhane's T2 post hoc
test to consider heterogeneity of variance. Data are expressed as
the mean ± standard error of the mean. Data were analyzed using the
Student's t-test. The log rank test and Kaplan-Meier were conducted
to determinate overall survival and recurrence. P-values were
two-sided, and P<0.05 was considered to indicate a statistically
significant difference.
Results
Identification of genetic alterations
and analysis of functions in GEO datasets
To acquire sufficient data from the gene
microarrays, two GEO chip files, GSE53757 and GSE40435 were
downloaded. The present study identified 174 and 149 genes that
have significant STN ratios in the GSE53757 and GSE40435 datasets,
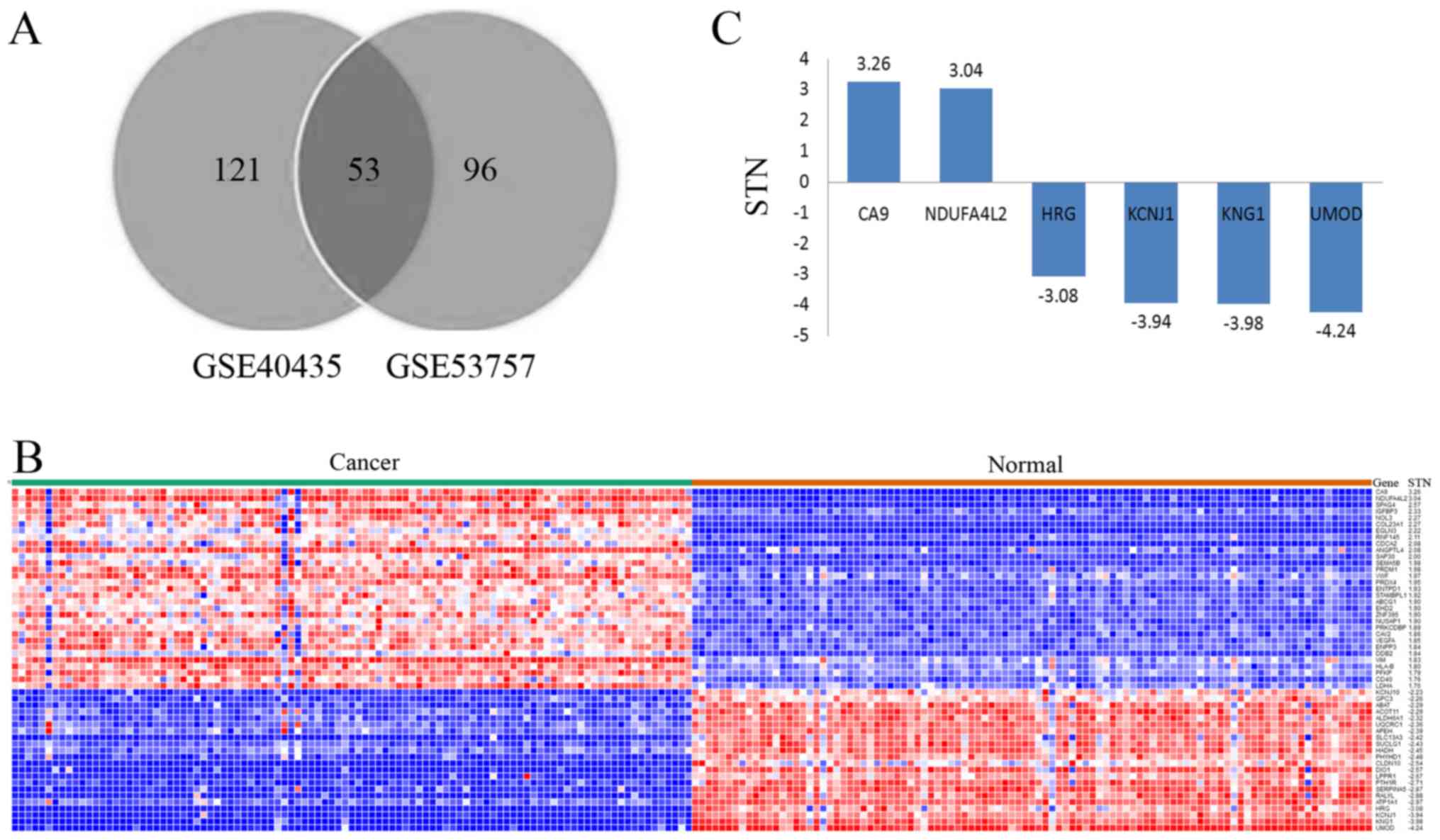
respectively. Following comparisons between these two datasets, 53
of the genes were selected as they featured in both sets (Fig. 1A). All data associated with these 53
genes (available upon request) were input to Morpheus (https://software.broadinstitute.org) and a new
heat map was constructed (Fig. 1B).
Among these genes, carbonic anhydrase 9 (CA9) and NDUFA4L2
possessed an STN ratio of >3, whereas HRG, KCNJ1, KNG1 and UMOD
had an STN ratio of <-3 (Fig. 1C).
Furthermore, to investigate the gene function(s) and the potential
pathways involved, the present study analyzed the data using DAVID
(13,14), in which GO and KEGG analysis were
conducted. All 53 genes were recognized by DAVID. GO analysis
indicated that PRKCDBP, EHD2KCNJ10, ATP1A1, KCNJ1 and EHD2 may be
involved in the process of cortical actin cytoskeleton
organization, potassium-ion import, positive regulation of
endocytic recycling and membrane tabulation, respectively. In
addition, ALDH6A1, LDHA, SUCLG1 and ABAT may be involved in the
propanoate metabolism pathway.
Analysis of hub genes in RCC
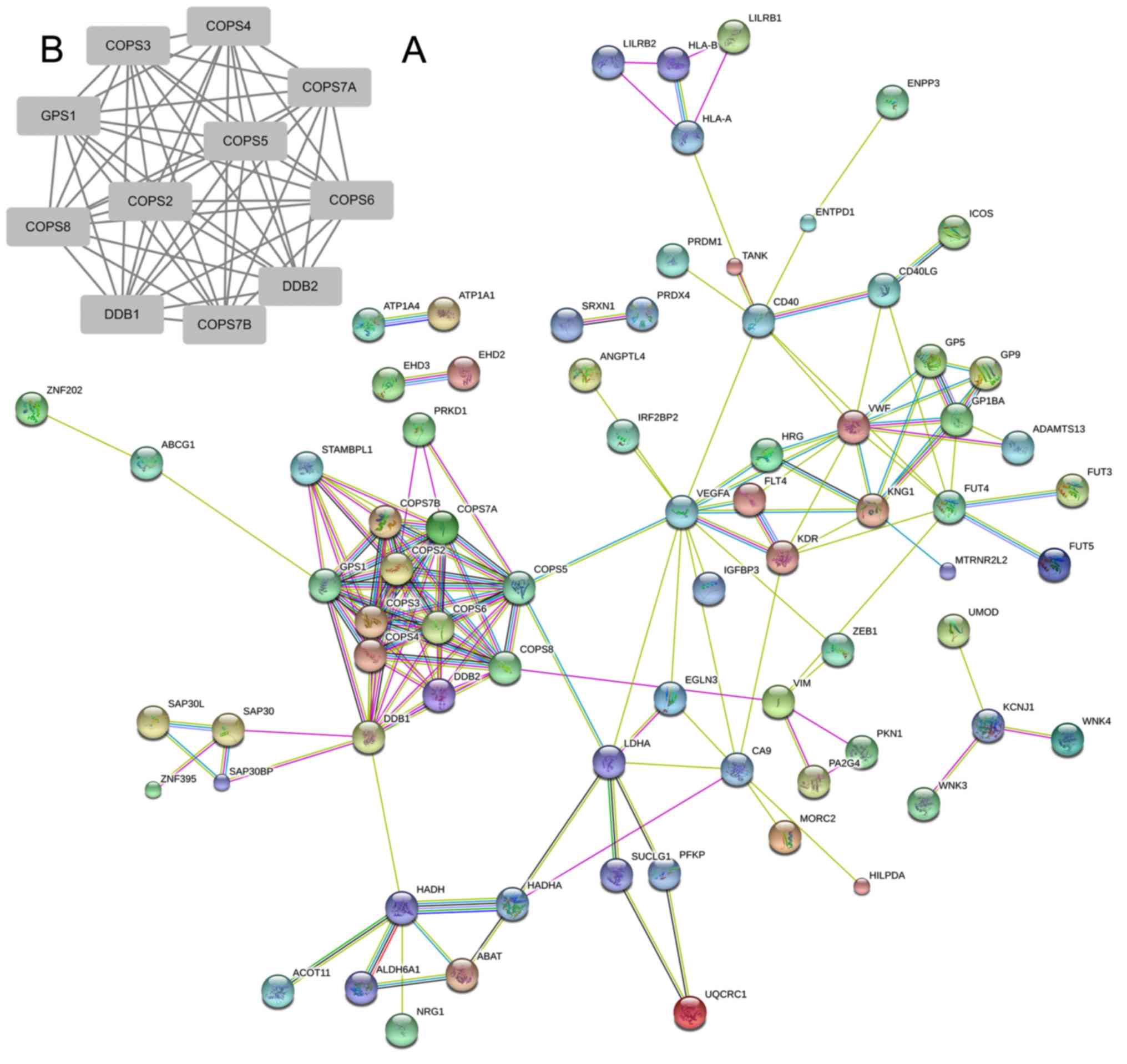
To elucidate the critical hub genes in RCC, the
STRING database was used to build a portable network graphic which
consisted of 106 genes. As presented in Fig. 2A, a cluster of genes was identified to
be markedly associated with other genes. It included DDB2, COPS6,
DDB1, COPS5, COPS7A, COPS8, COPS4, GPS1, COPS3 COPS7B and COPS2.
Among these genes, eight are members of the COP9 family; the
remaining two are members of the DDB family. These results suggest
that both COP9 and DDB may be crucial in the detection of RCC. To
further validate the hub genes in RCC, Cytoscape was used to
construct a PPI network (15). A
significant module was obtained from the network using MCODE in
Cytoscape, which included 11 nodes (Fig.
2B). These results revealed the crucial role of the COP9 family
in this module; these were also similar to the results derived from
STRING analysis. Notably, GPS1 was associated with all other genes
in the module, which may provide direction for further study in
RCC.
Hub genes are significantly associated
with overall survival and recurrence in RCC
The 11 genes specified in Fig. 2B were analyzed using the cBioPortal
database, which contained 538 clinical cases of RCC and sequenced
data. As presented in the cBioPortal database, genetic alterations
are comprised of three parameters: Mutation, Copy Number Alteration
(CNA) and expression. The present study focused on the expression
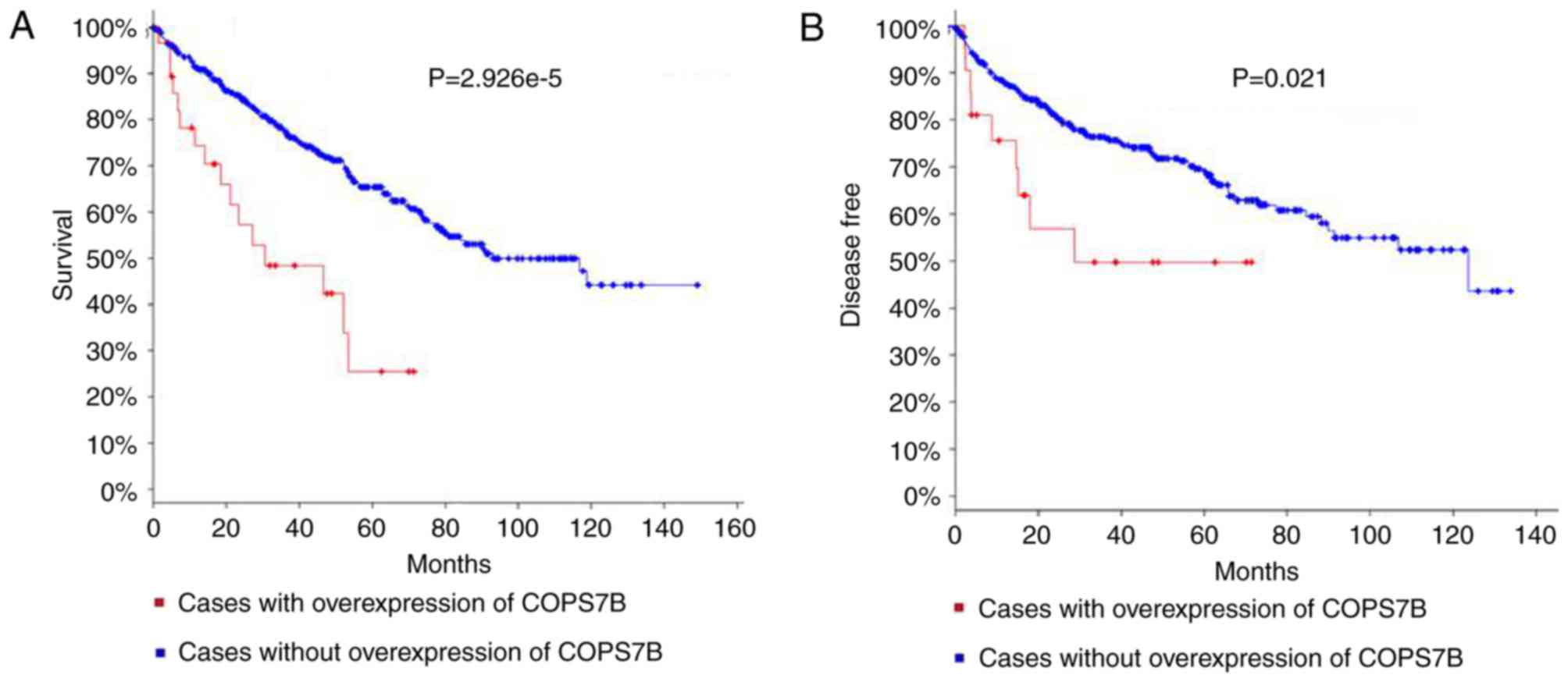
of the hub genes. The significant associations of COPS7B with
overall survival and recurrence were observed using a Kaplan-Meier
curve. The log rank test P-values for these are
2.926×10−5 and 0.021, respectively, (Fig. 3A and B). No other hub genes were
associated with overall survival and disease free survival. These
results indicate that the COPS7B may be important in monitoring the
development of RCC and also serve as a valuable biological
marker.
Clinical features of COPS7B in
cBioPortal
To reveal the clinical significance of COPS7B in
RCC, genetic alterations in COPS7B, as well as alterations to other
clinical features (information available upon request), were
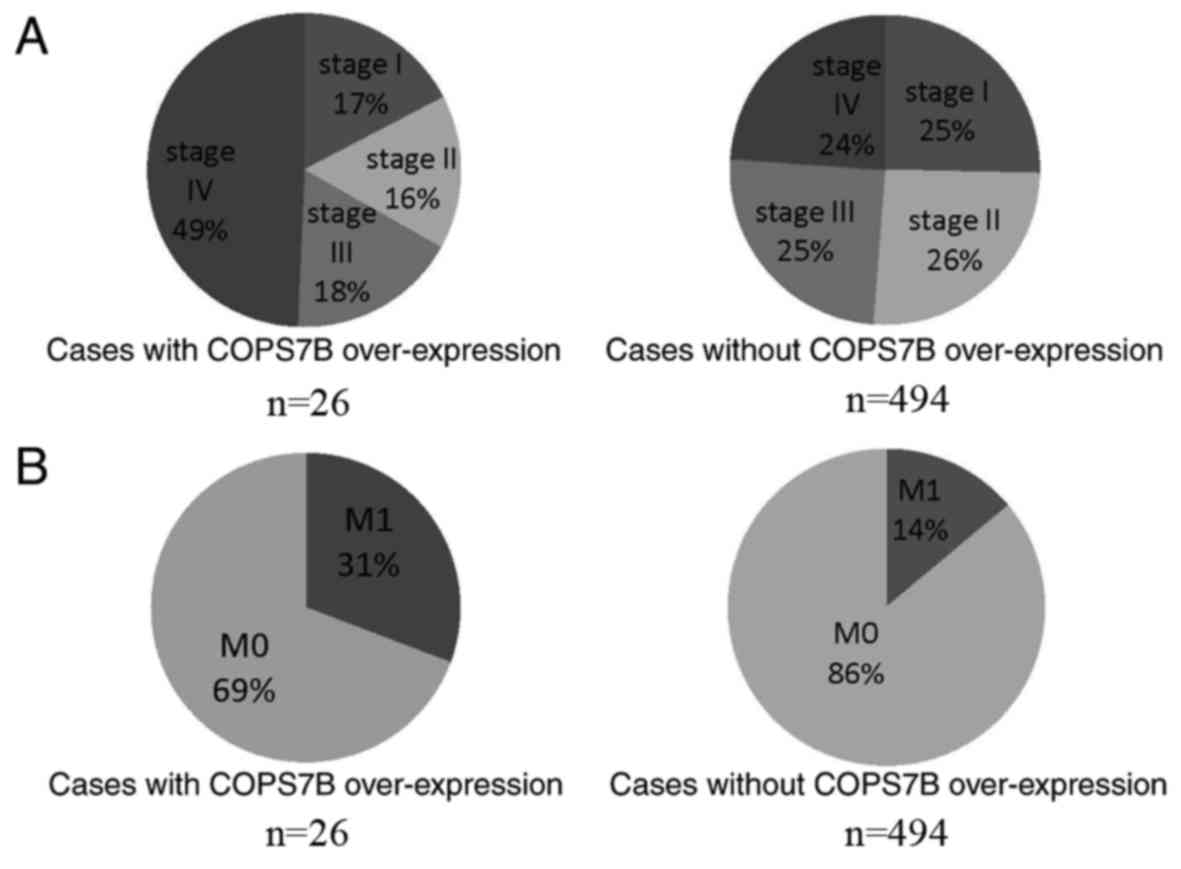
investigated. Notably, COPS7B was identified to be associated with
clinical stage and metastasis. As presented in Fig. 4A, 49% of cases with COPS7B
overexpression were diagnosed with stage IV RCC. On the other hand,
24% of cases with normal expression of COPS7B were stage IV,
suggesting that COPS7B may be associated with advanced-stage IV RCC
(Fig. 4A). Furthermore, metastasis
was identified in 31% of cases in which COPS7B was overexpressed,
while 14% presented with metastasis without concomitant COPS7B
overexpression (Fig. 4B). Other
clinical features, including sex, age, laterality and volume, were
evaluated separately, and no significant associations with COPS7B
were observed.
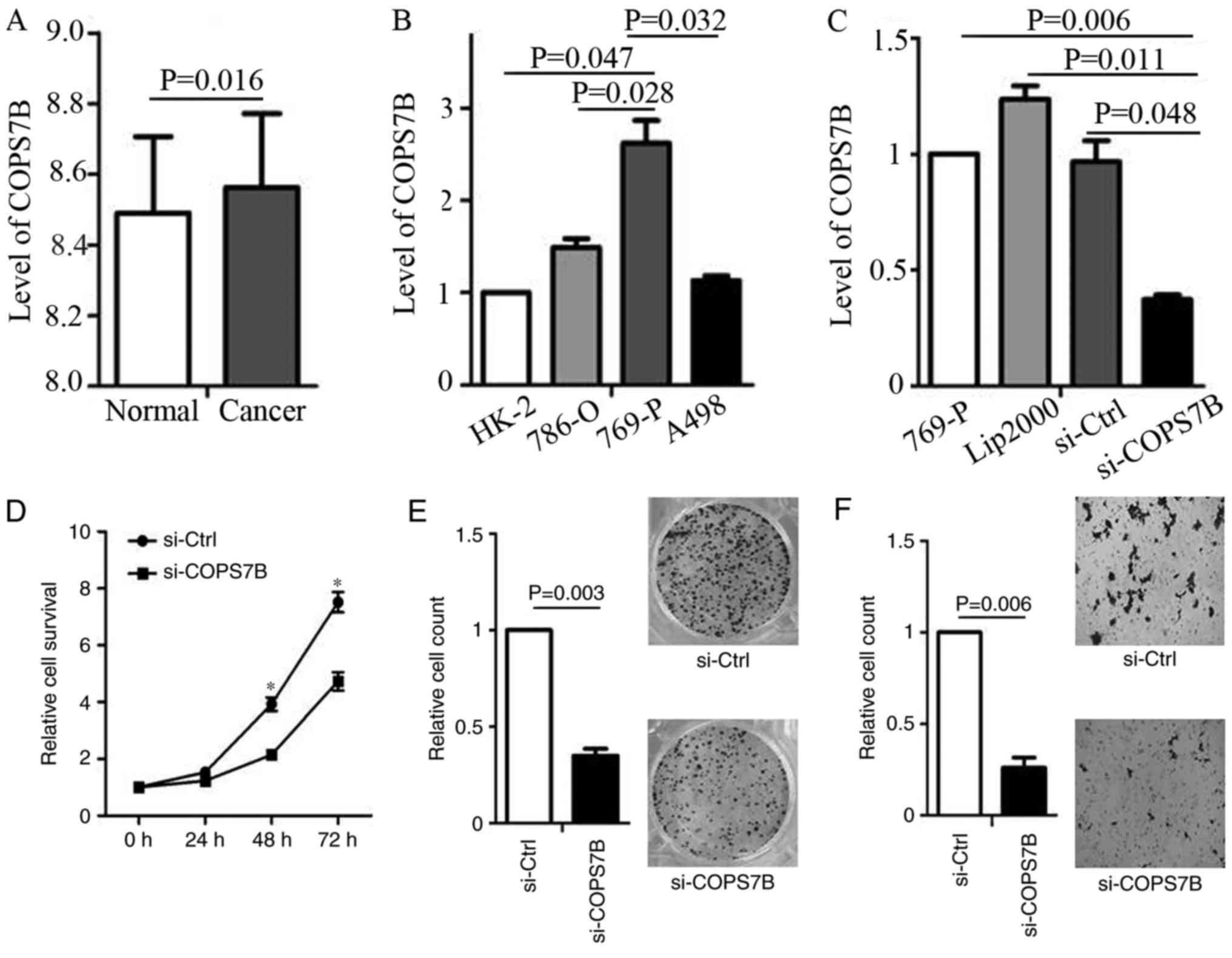
Knockdown of COPS7B by siRNA in RCC
cell lines
The present study demonstrated the overexpression of
COPS7B is associated with poor overall survival, recurrence,
advanced stage and metastasis. Based on the data provided by the
GEO chip file GSE40435 (203 tissues), the expression of COPS7B was
determined. It indicated that the COPS7B was overexpressed in RCC
compared with in adjacent normal tissues (Fig. 5A). Furthermore, these results
indicated that COPS7B may function as an oncogene in RCC. To
confirm this function, we focused on the level of COPS7B expression
in various RCC cell lines (786-O, 769-P and A498) as well as in
normal renal tubular epithelial HK-2 cells. The results
demonstrated that COPS7B was overexpressed in 769-P cells, compared
with in HK-2 cells (Fig. 5B).
Furthermore, we then used siRNA to knock-down COPS7B in 769-P cells
(Fig. 5C).
Oncogenic role of COPS7B in RCC
Cell survival assays suggested that the knock-down
of COPS7B inhibited cell proliferation 48 h following transfection,
as compared with the siRNA control (Fig.
5D). To confirm that knock-down of COPS7B inhibits cell
proliferation, colony formation was assessed following
transfection, with effective siRNA knockdown (Fig. 5E). Considering that COPS7B is
associated with metastasis, as determined based on the data from
cBioPortal, an invasion assay was performed following COPS7B
knockdown. The invasion assay demonstrated that knockdown of COPS7B
inhibited the invasion ability of 769-P cells, indicating the
oncogenic function of COPS7B in RCC (Fig.
5F).
Discussion
A previous study investigated the molecular markers
of RCC; however, little is known regarding the clinical
significance of any specific marker (16). The present study focused on two
microarray datasets and used a series of biological software
investigations to identify hub genes. The present study also
screened the hub genes in cBioPortal, a database containing a large
amount of sequencing data, as well as clinical features that may be
associated with RCC. Thus, these hub genes were characterized as
follows: 1) Hub genes were generated based on expression
alterations in 161 tumors and 161 tumor adjacent tissues; 2) hub
genes were screened in the cBioPortal with 538 RCC cases, and
COPS7B was associated with prognosis; 3) The hub gene COPS7B is
predictive of overall survival and recurrence for patients with
RCC. These results suggest that COPS7B is a potential prognostic
marker in RCC and warrants further investigation.
A previous study investigating molecular markers to
determine prognosis in RCC focused on VHL, hypoxia inducible factor
1-α (HIF1-α), VEGF, CA9 and Ki-67 (16). The results of a previous study
demonstrated that VHL is a tumor suppressor gene in RCC. Its
product, phosphorylated-VHL, is relevant to numerous cell-signaling
pathways including the HIF1-α pathway, and regulates oxidative
phosphorylation. The inactivation of VHL leads to aberrant
VHL-HIF-VEGF pathway activation associated with oncogenesis and
tumor development in RCC (17). In
the GSE40435 dataset, the present study surveyed VHL expression in
202 samples. It was identified that VHL was overexpressed in
tumors, a result consistent with those of other studies. However,
VHL does not meet the criteria for a hub gene. This is may be due
to several reasons, including variation in the detection methods of
VHL between datasets. Furthermore, VHL did not meet the
requirements when constructing the visual molecular interaction
networks (Cytoscape). Therefore, COPS7B may be more important as a
hub gene, compared with VHL.
Additionally, previous study suggests that several
genes in the serum were indicated to be non-invasive and associated
with metastasis, recurrence and prognosis. CA9 was over-expressed
in RCC compared with in normal renal samples (18). CA9 expression, which is detectable in
serum, has previously been reported to be associated with prognosis
due to early-activated HIF. However, the present study observed
that COPS7B is a novel molecular marker, based on analysis of a
large number of clinical cases, suggesting that COPS7B may serve as
a clinical biomarker for RCC. Currently, little is known about
biomarkers in the urine. The expression of NMP22 in the urine of
patients with RCC was significantly increased, compared with the
controls. A previous study reported that urinary NMP22 may,
therefore, allow the early detection of RCC (19). However, due to the low positive test
rate, the clinical significance of NMP22 requires further
evaluation. The potential oncogenic gene COPS7B may offer an
alternative and promising diagnosis strategy for RCC by analyzing
its expression levels in urine.
This bioinformatic study provides new perspectives
to identify novel biomarkers in RCC. Future investigations
analyzing additional online datasets will allow screening for other
potential biomarkers for RCC. For example, data from the metastasis
dataset provides a number of genes associated with metastasis
(20). In addition, there are various
clinical stage-associated genes, including ephA2, in the datasets
that warrant further study (21).
Thus, biomarkers for different biological functions may be
uncovered based on these microarray results. A previous study on
ovarian cancer screened out 345 genes and 36 differentially
expressed miRNAs, suggesting that a number of genes and miRNAs may
serve crucial functions in ovarian carcinoma (9). Compared with the findings of this
previous study, the present study focused on the hub genes that
were identified in the cBioPortal database (538 clinical cases) to
reveal the role of the hub genes in overall survival and disease
recurrence. Furthermore, the present study validated the oncogenic
function in various RCC cell lines. Collectively, these studies
suggest that online data analysis is an effective tool in the
evaluation of cancer prognosis and identifying potential
therapeutic targets. Furthermore, it is possible to identify the
underlying molecular mechanisms using online software. For example,
COPS7B was analyzed using the cBioPortal, identifying the
co-expression of COPS7B and ATG4B, with a Pearson correlation of up
to 0.79, therefore indicating that COPS7B is closely associated
with ATG4B. Thus, ATG4B may be upstream or downstream of COPS7B.
Similarly, other genes associated with COPS7B must be identified
prior to elucidating the mechanisms.
CSN7 is an important subunit of COP9 signalosome
(CSN), which is a conserved protein complex that includes 8 CSNs.
CSN7 includes the COPS7A and COPS7B complexes, and is one of the
most prominent phosphorylated subunits formed in response to DNA
damage (22,23). CSN has notable effects on multiple
cell signaling functions, including DNA repair, cell cycle control,
angiogenesis and tumor development (24). An increase in the CSN holo-complex
under cancerous conditions may lead to the development of drug
resistance (25).
Furthermore, COPS7A may be considered a biomarker
predicting recurrence and prognosis of RCC. Considering the
important roles of CSN in cancer, it is likely that COPS7B may also
have important roles in other solid tumors. Additionally, the
knockdown of COPS7B inhibited HIF1-α (information available upon
request). Thus, the inhibition of HIF1-α following COPS7B knockdown
may be a mechanism of COPS7B in RCC. The mechanisms associated with
this phenotype in RCC have not been clearly demonstrated in the
present study; however, it is possible that HIF1-α may be involved
downstream, thus warranting further investigation.
It is also reported that COPS7A and COPS7B complexes
co-exist in human red blood cells (26). The results of the present study
suggest that COPS7A and COPS7B may also coexist and exert
synergistic effects in RCC. These synergistic effects may be based
on the spatial structure of the two proteins, which are similar to
the miRNA previously reported (27).
Further investigation of the COPS7A and COPS7B complexes may
explain the role of COPS7B suggested by the present study and
assist in identifying novel targets for RCC therapy. Although the
role and underlying mechanism of COPS7B in RCC remains to be
determined, its association with overall survival and disease
recurrence is supported by the results of the present study, which
indicate that COPS7B has an oncogenic role. Considering that COPS7B
is regarded as a hub gene, it is hypothesized that COPS7B may be a
driver gene and serve as a biological marker as well as a
therapeutic target in RCC.
The present study provided an example of an
integrated bioinformatics analysis approach to identify potential
biomarkers, hub genes for diagnosis and for the prediction of
overall survival, disease recurrence and therapeutic targets. Due
to the extensive clinical chip data now available online, the
present study demonstrated how researchers may utilize a
bioinformatics approach to discover genes that serve key functions
in cancer. Specifically, the cBioPortal encompasses genomic data
from 105 types of cancer, and its expansion may provide new
research opportunities as well as save time and resources.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81402100 and
31600952), the Foundation of Health and Family Planning Commission
of Jiangsu Province (grant no. Q201408), the Social Development
Foundation of Zhenjiang (grant nos. SH2016031 and SH2014026), the
foundation from the Foundation for Jiangsu Provincial Medical Youth
Talent (grant no. QNRC2016840), and the Six Talent Peaks Project of
Jiangsu Province (grant no. WSW-007).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
BC took responsibility for the majority of the
present study, including PCR experimentation, data analysis and
manuscript preparation. ZJ, XY, ZQ, JG and HS participated in study
design, helped to draft the manuscript and contributed
intellectually. JG provided specific academic guidance on
experimental design, assistance in data analysis and review of the
manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
the Affiliated Hospital of Jiangsu University.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
RCC
|
renal cell carcinoma
|
|
GEO
|
gene expression omnibus
|
|
GO
|
gene ontology
|
|
KEGG
|
Kyoto encyclopedia of genes and
genomes
|
|
CSN
|
COP9 signalosome
|
References
|
1
|
Chow WH, Dong LM and Devesa SS:
Epidemiology and risk factors for kidney cancer. Nat Rev Urol.
7:245–257. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Patard JJ, Pignot G, Escudier B, Eisen T,
Bex A, Sternberg C, Rini B, Roigas J, Choueiri T, Bukowski R, et
al: ICUD-EAU international consultation on kidney cancer 2010:
Treatment of metastatic disease. Eur Urol. 60:684–690. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Arabsalmani M, Mohammadian-Hafshejani A,
Ghoncheh M, Hadadian F, Towhidi F, Vafaee K and Salehiniya H:
Incidence and mortality of kidney cancers, and human development
index in Asia; a matter of concern. J Nephropathol. 6:30–42. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Waalkes S, Kramer M, Herrmann TR, Schrader
AJ, Kuczyk MA and Merseburger AS: Present state of target therapy
for disseminated renal cell carcinoma. Immunotherapy. 2:393–398.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Duran I, Lambea J, Maroto P,
González-Larriba JL, Flores L, Granados-Principal S, Graupera M,
Sáez B, Vivancos A and Casanovas O: Resistance to targeted
therapies in renal cancer: The importance of changing the mechanism
of action. Target Oncol. 12:19–35. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu H, Li J, Koirala P, Ding X, Chen B,
Wang Y, Wang Z, Wang C, Zhang X and Mo YY: Long non-coding RNAs as
prognostic markers in human breast cancer. Oncotarget.
7:20584–20596. 2016.PubMed/NCBI
|
|
9
|
Xu Z, Zhou Y, Cao Y, Dinh TL, Wan J and
Zhao M: Identification of candidate biomarkers and analysis of
prognostic values in ovarian cancer by integrated bioinformatics
analysis. Med Oncol. 33:1302016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
von Roemeling CA, Radisky DC, Marlow LA,
Cooper SJ, Grebe SK, Anastasiadis PZ, Tun HW and Copland JA:
Neuronal pentraxin 2 supports clear cell renal cell carcinoma by
activating the AMPA-selective glutamate receptor-4. Cancer Res.
74:4796–4810. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wozniak MB, Le Calvez-Kelm F,
Abedi-Ardekani B, Byrnes G, Durand G, Carreira C, Michelon J,
Janout V, Holcatova I, Foretova L, et al: Integrative genome-wide
gene expression profiling of clear cell renal cell carcinoma in
Czech Republic and in the United States. PLoS One. 8:e578862013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen B, Liu J, Ho TT, Ding X and Mo YY:
ERK-mediated NF-kB activation through ASIC1 in response to
acidosis. Oncogenesis. 5:e2792016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
da Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
da Huang W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Barata PC and Rini BI: Treatment of renal
cell carcinoma: Current status and future directions. CA Cancer J
Clin. 67:507–524. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Schonenberger D, Rajski M, Harlander S and
Frew IJ: Vhl deletion in renal epithelia causes HIF-1α-dependent,
HIF-2α-independent angiogenesis and constitutive diuresis.
Oncotarget. 7:60971–60985. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tostain J, Li G, Gentil-Perret A and
Gigante M: Carbonic anhydrase 9 in clear cell renal cell carcinoma:
A marker for diagnosis, prognosis and treatment. Eur J Cancer.
46:3141–3148. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kaya K, Ayan S, Gokce G, Kilicarslan H,
Yildiz E and Gultekin EY: Urinary nuclear matrix protein 22 for
diagnosis of renal cell carcinoma. Scand J Urol Nephrol. 39:25–29.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jones J, Otu H, Spentzos D, Kolia S, Inan
M, Beecken WD, Fellbaum C, Gu X, Joseph M, Pantuck AJ, et al: Gene
signatures of progression and metastasis in renal cell cancer. Clin
Cancer Res. 11:5730–5739. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Herrem CJ, Tatsumi T, Olson KS, Shirai K,
Finke JH, Bukowski RM, Zhou M, Richmond AL, Derweesh I, Kinch MS
and Storkus WJ: Expression of EphA2 is prognostic of disease-free
interval and overall survival in surgically treated patients with
renal cell carcinoma. Clin Cancer Res. 11:226–231. 2005.PubMed/NCBI
|
|
22
|
Dessau M, Halimi Y, Erez T, Chomsky-Hecht
O, Chamovitz DA and Hirsch JA: The Arabidopsis COP9 signalosome
subunit 7 is a model PCI domain protein with subdomains involved in
COP9 signalosome assembly. Plant Cell. 20:2815–2834. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Beli P, Lukashchuk N, Wagner SA, Weinert
BT, Olsen JV, Baskcomb L, Mann M, Jackson SP and Choudhary C:
Proteomic investigations reveal a role for RNA processing factor
THRAP3 in the DNA damage response. Mol Cell. 46:212–225. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gummlich L, Rabien A, Jung K and Dubiel W:
Deregulation of the COP9 signalosome-cullin-RING ubiquitin-ligase
pathway: Mechanisms and roles in urological cancers. Int J Biochem
Cell Biol. 45:1327–1337. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Feist M, Huang X, Müller JM, Rau B and
Dubiel W: Can hyperthermic intraperitoneal chemotherapy efficiency
be improved by blocking the DNA repair factor COP9 signalosome? Int
J Colorectal Dis. 29:673–680. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rozen S, Tieri A, Ridner G, Stark AK,
Schmaler T, Ben-Nissan G, Dubiel W and Sharon M: Exposing the
subunit diversity within protein complexes: A mass spectrometry
approach. Methods. 59:270–277. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen B, Duan L, Yin G, Tan J and Jiang X:
Simultaneously expressed miR-424 and miR-381 synergistically
suppress the proliferation and survival of renal cancer cells-Cdc2
activity is up-regulated by targeting WEE1. Clinics (Sao Paulo).
68:825–833. 2013. View Article : Google Scholar : PubMed/NCBI
|