Introduction
Glypican 3 (GPC3) is a 60 kDa cell-surface protein
that belongs to the glypican family, and its gene is located at
chromosome Xq26 (1). Glypicans are
cell surface proteoglycans that are associated with the outer
leaflet of the plasma membrane by a glycosyl phosphatidyl inositol
anchor (2,3). This gene is expressed in a
tissue-specific manner, exhibits its peak expression during
embryonic tissue development and is downregulated in mature tissues
(4). Glypicans are expressed
predominantly during development, suggesting that they serve a role
in morphogenesis (5,6).
Glypican 3 mutations have been identified as the
genetic defects associated with Simpson Golabi-Behmel syndrome
(SGBS), which is characterized by overgrowth, dysplasia and
multiple congenital anomalies, and by an increased prevalence of
Wilm's tumors, nephroblastoma and hepatoblastoma (7,8), malignant
melanoma (9), ovarian cancer
(10) and testicular germ cell tumors
(11). The role of this protein in
different types of cancer has not yet been well-defined. The
Antigen Ki-67 protein (Ki-67) is an antigen associated with active
cell proliferation (12). Higher
levels of Ki-67 expression in tumor tissues were identified to be
associated with a higher mitotic activity (12).
By the end of 2012, liver cancer is the second most
frequently diagnosed cancer among the men in less development
countries (13). China has observed
an increasing incidence of liver cancer, and this type of malignant
tumor has become the estimated second most common cause of
cancer-associated mortality in China (14). GPC3 was first identified as a
potential biomarker of liver cancer since its level increased
significantly in the serum of patients with liver cancer in
comparison with healthy donors and patients with benign liver
tumors (15). The role GPC3 serves in
liver cancer is controversial and its precise mechanism of action
and clinical significance remains uncharacterized.
In the present study, the correlation between GPC3
expression levels in 114 patients with liver cancer and the levels
of cancer cell differentiation and proliferation were
retrospectively analyzed. The in vitro effects of GPC3 on
liver cancer cells growth were observed, to verify the assumption
that elevated GPC3 is a significant risk factor in liver cancer
mortality due to its capability of promoting growth in tumor
cells.
Materials and methods
Subjects
A total of 114 patients with
histopathologically-confirmed liver cancer were recruited for the
present study from Beijing You'An Hospital Affiliated to Capital
Medical University (Beijing, China). All the specimens were
obtained via surgical resection or liver transplantation
procedures. The patient ages ranged from 19–67 years (mean age,
51.2 years), and including 97 meals and 17 females. The diagnosis
of liver cancer was performed following the WHO Classification of
Tumors of the Digestive System (16).
All the tissue samples were reviewed by 2 independent experienced
pathologists (Department of Pathology, Beijing You'An Hospital
Affiliated to Capital Medical University, Beijing, China). The
study protocol was approved by the Ethical Committee of Beijing
You'An Hospital. Written informed consent was obtained from all
patients.
Immunohistochemical (IHC)
staining
Liver cancer specimens were fixed in 10%
neutral-buffered formalin for >24 h at room temperature.
Paraffin-embedded tissue blocks were cut into 4-µm sections for
immunohistochemical staining following the tissues being handled by
dehydrating, clearing, dipping wax and embedding. The expression
levels of GPC3 and Ki-67 in tissues were assessed by IHC staining
with monoclonal anti-GPC3 and anti-Ki-67 antibodies (catalog nos.,
sc-390587 and sc-23900; dilution, 1:100 and 1:200; both Santa Cruz
Biotechnology, Inc., Dallas, TX, USA). A non-immune mouse IgG
antibody (catalog no. ZM-0491; dilution, 1:50; OriGene
Technologies, Inc., Beijing, China) was used as negative control
reagent for each specimen. In brief, tissue sections were
de-paraffinized in xylene at room temperature for 15 min and
rehydrated in 100, 95, 85, 70 and 50% ethyl alcohol for 5 min at
each concentration at room temperature, and heat-induced epitope
retrieval was performed in a 10 mmol/l citrate buffer (pH 6.0) at
92°C for 15 min. Then, endogenous peroxidase was blocked with 3%
H2O2 followed by incubation with primary
antibodies overnight at 4°C. The tissues were then incubated for
additional 60 min at room temperature on the second day with a
biotin-free horseradish peroxidase-labeled sheep anti-mouse IgG
secondary antibody (catalog no., ZDR-5307; dilution, 1:500; OriGene
Technologies, Inc.). Staining was performed with
3,3′-diaminobenzidine (ZLI-9019, dilution concentrations: 1:20;
OriGene Technologies, Inc.) at room temperature for 5 min.
GPC3 staining was considered positive when brown
granules were located in the cytoplasm, membrane or canaliculi
(17). The results were evaluated
according to the proportion and staining intensity of positive
cells. The proportions of positive cells were scored as: 0, <5;
1, 5–25; 2, 26–50; and 3, >50%. The staining intensities were
scored as: 0, no staining; 1, weak; 2, intermediate; and 3, strong.
Next, the results of multiplying the proportion scores by the
intensity scores were classified into 4 grades as follow: -, 0–1;
+, 2–4; ++, 5–7; and +++, >8. Positive Ki-67 staining was
indicated by brown granules located in nucleus. In order to
determine the proliferation index, 500 tumor cells and the number
of Ki-67-positive cells were counted at ×400 magnification in
several dense positive cell areas. The cell proliferation indexes
were evaluated according to the percentage of the number of
positive cells/500 tumor cells, as follows: -, <5; +, 5–25; ++,
26–50; and +++, >50%. All results were reviewed by light
microscopy (×200 magnification) by two blinded, experienced
pathologists, as previously stated.
Cell lines and treatment
The GPC3-producing HepG2 cell line, demonstrated to
be a hepatoblastoma cell line (18)
and the non-GPC3-producing hepatocellular carcinoma HLE cell line
were cultured in Dulbecco's modified Eagle's medium (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% w/v fetal
bovine serum (Thermo Fisher Scientific, Inc.) at 37°C in a
humidified atmosphere of 5% CO2. HepG2 and HLE cells
were gifts from the Department of Biochemistry and Molecular
Biology, Peking University Health Science Center (Beijing,
China).
GPC3 (10088-H08H; Sino Biological, Inc. Beijing,
China) was dissolved in PBS to final concentrations of 0.01, 0.1, 1
and 10 mg/ml, which were then used to treat the 2 cultured cell
lines. PBS, which served as the solvent of GPC3, was used as a
control.
Western blotting
Western blotting was performed for the analysis of
expression of GPC3 in HepG2 and HLE cells. Briefly, cells were
lysed in RIPA lysis buffer (Beyotime Institute of Biotechnology,
Beijing, China) and 40 µg protein was utilized for each western
blot. BCA assay was used to determinate the protein concentrations
(Beyotime Institute of Biotechnology). Electrophoretic transfer of
proteins was performed from 10% SDS-PAGE gels onto nitrocellulose
membranes. Membranes were blocked by immersion in 5% non-fat milk
(w/v)/PBS for 1 h at room temperature, and then incubated at 4°C
overnight with anti-GPC3 (dilution, 1:500; catalog no., sc-390587)
and β-actin monoclonal antibodies (dilution, 1:1,000; catalog no.,
sc-130656; both Santa Cruz Biotechnology, Inc., Dallas, TX, USA).
Subsequent to rinsing with PBST for three times, membranes were
incubated at 37°C with a horseradish peroxidase-conjugated IgG
secondary antibody for 1 h. Immunocomplexes were visualized by
incubation of the membranes with the Enhanced Chemiluminescence kit
(catalog no., 32109, Thermo Fisher Scientific, Inc.) at room
temperature for 1 min according to the manufacturer's protocol.
Immunofluorescence
Localization of GPC3 in HepG2 and HLE cells was
observed at ×1,000 magnification. Immunofluorescence staining was
performed by a routine staining method (19). Mouse anti-human GPC3 and secondary
goat anti-mouse antibody conjugated with fluorescence rhodamine
(TRITC) (sc-362277, dilution 1:200) against GPC3 were purchased
from Santa Cruz Biotechnology, Inc. Nuclei were counterstained with
DAPI (100 µg/ml) at room temperature for 1 min. Intracellular
localization of proteins in each group were observed, and images
were captured with a fluorescence microscope (magnification,
×1,000).
MTT and Cell Counting Kit-8 (CCK-8)
assays
To assess the effect of GPC3 on cell proliferation,
MTT colorimetric assays and analyses were performed with the CCK-8
kit (Dojindo Molecular Technologies, Inc., Kumamoto, Japan). HepG2
and HLE cells were seeded into 96-well plates at a density of
5×103 cells/well in 200 µl medium, and then treated with
various concentrations of GPC3 (0, 0.01, 0.1, 1 and 10 mg/ml).
Following 24 h incubation at 37°C, MTT solution was added to the
wells, followed by incubation at 37°C for 1 h. The medium was
removed carefully, and dimethyl sulfoxide was added to dissolve the
blue formazan in the living cells. Absorbance was measured at 570
nm for MTT assay and 450 nm for CCK-8 with an ELISA reader. The
percentage of cell proliferation for each treatment was calculated
as [(A570 sample-background)/(A570 control-background)] × 100.
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR)
The RNeasy Mini kit (Qiagen GmbH, Hilden, Germany)
was used to isolate total RNA from cultured HepG2 and HLE cells.
cDNA was then synthesized by reverse transcribing 1 mg extracted
RNA using a SuperScript II First-stand Synthesis System
(Invitrogen; Thermo Fisher Scientific, Inc.). SYBR Green (QPK-201;
Toyobo Life Science, Osaka, Japan) was used to detect the
double-stranded DNA products during the qPCR assay. The mRNA
content was normalized to the housekeeping gene GAPDH. All primer
sequences for RT-qPCR are summarized in Table I. The reaction conditions were: 95°C
for 5 min, then followed by 40 cycles at 95°C for 10 sec and 55°C
for 40 sec with ABI 9500 sequence detection system. The RT-qPCR
results were normalized to GADPH according to the 2−ΔΔCq
method (20).
 | Table I.Sequences of oligonucleotides used as
primers for reverse transcription quantitative polymerase chain
reaction. |
Table I.
Sequences of oligonucleotides used as
primers for reverse transcription quantitative polymerase chain
reaction.
| Gene name | Primer sequences |
|---|
| SHH |
|
|
Sense |
5′-ACAGCGACTTCCTCACTTTCCT-3′ |
|
Antisense |
5′-CCGCGTCTCGATCACGTAGA-3′ |
| GLI1 |
|
|
Sense |
5′-TTCCTACCAGAGTCCCAAGT-3′ |
|
Antisense |
5′-CCCTATGTGAAGCCCTATTT-3′ |
| PTCH |
|
|
Sense |
5′-GGCAGGAGGAGTTGATTG-3′ |
|
Antisense |
5′-CGTACATTTGCTTGGGAGT-3′ |
| SMO |
|
|
Sense |
5′-AGCTTCCGGGACTATGTGC-3′ |
|
Antisense | 5′-GCTCGGGCGATTCTTGAT
−3′ |
| GAPDH |
|
|
Sense |
5′-TGAAGGTCGGAGTCAACGGA-3′ |
|
Antisense |
5′-CCTGGAAGATGGTGATGGGAT-3′ |
Statistical analysis
All data were statistically analyzed using SPSS
software version 17.0 (SPSS, Inc., Chicago, IL, USA). The
χ2 test was performed to analyze the association between
GPC3 and cell differentiation or cell proliferation. Correlation
analysis was performed using Spearman's rank test. The in
vitro results of multiple observations were presented as the
mean ± standard deviation of at least three separate experiments,
and analyzed using a one-way analysis of variance followed by a
Least Significant Difference test to compare the treatment and
control groups. P≤0.05 was considered to indicate a statistically
significant difference.
Results
Association between GPC3 expression
and the differentiation of liver cancer
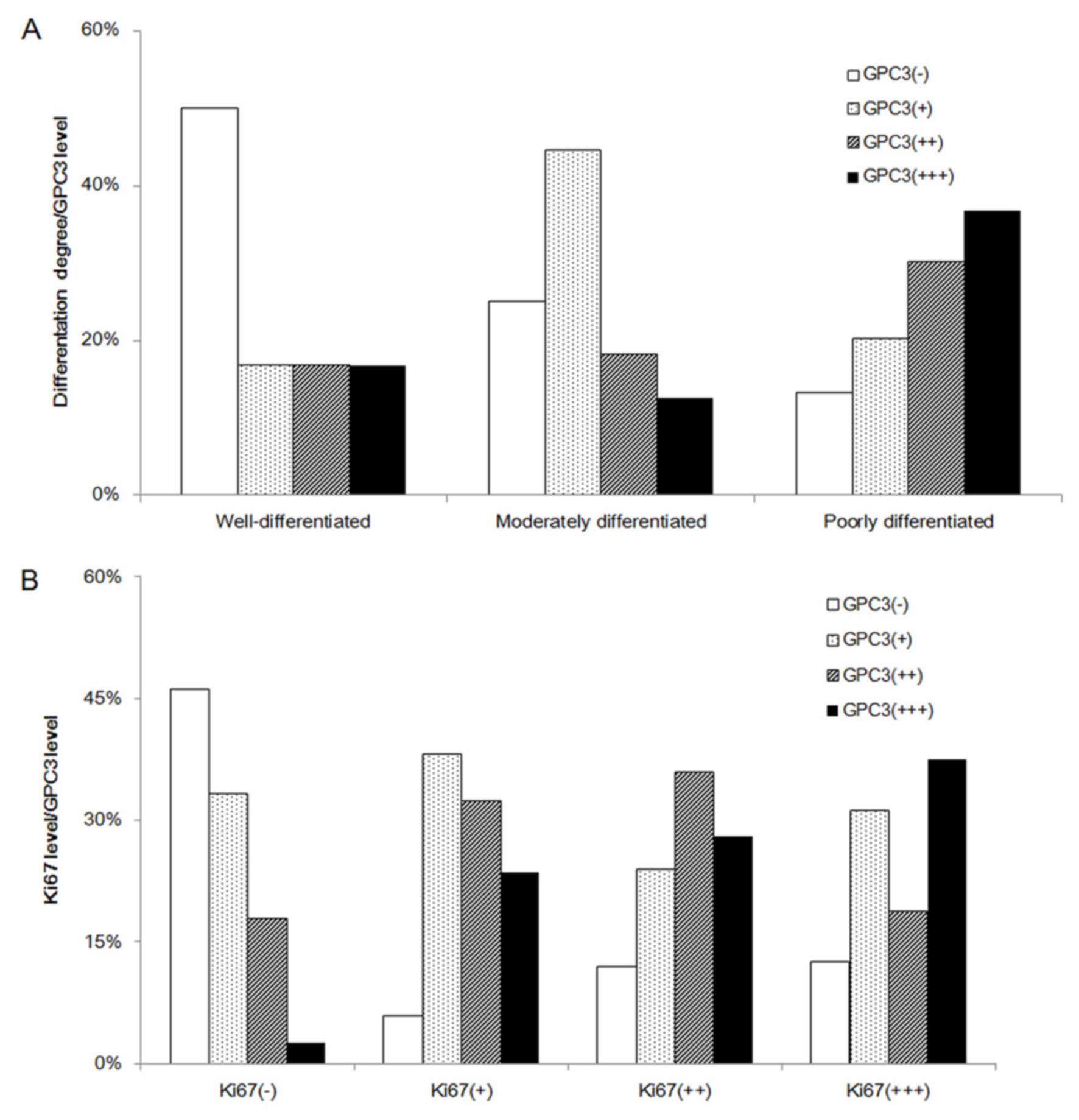
Among the 114 patients with liver cancer, 12 cases
were well-differentiated. Of these, 50% (6/12) expressed GPC3, with
1+ expression in 2 cases, 2+ expression in 2 cases and 3+
expression in 2 cases. A total of 72 patients with liver cancer
were moderately differentiated, and 75% (54/72) expressed GPC3. Of
these, there were 32 cases with + expression (44.44%), 13 cases
with ++ expression (18.06%) and 9 cases with +++ expression
(12.50%). Of the other 30 patients with liver cancer with poor
differentiation, 6 cases exhibited + expression (20.00%), 9 cases
exhibited ++ expression (30.00%) and 11 cases exhibited +++
expression (36.67%). These results demonstrated a statistical
significance between GPC3 expression and liver cancer
differentiation (χ2=16.306, P=0.008), and there was a
significant correlation between GPC3 expression and liver cancer
differentiation (r=0.302, P=0.01): The poorer the differentiation
stage, the higher the level of the GPC3 expression (Table II; Figs.
1 and 2A).
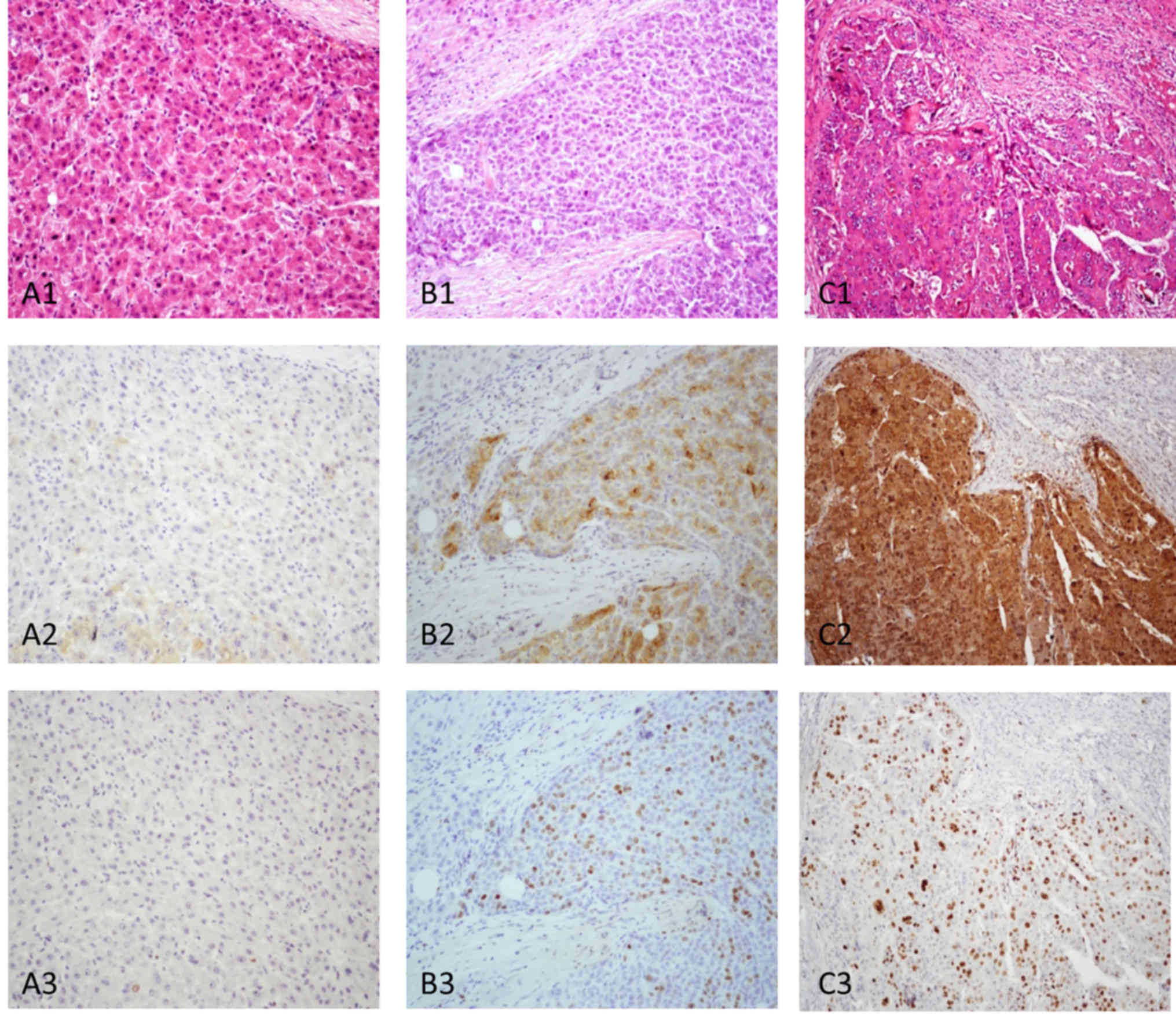 | Figure 1.Association between GPC3 expression
and the differentiation of liver cancer. (A) Well-differentiated
liver cancer tissue and the same field. (A1). The tumor cells
demonstrated a thin trabecular pattern with mild atypical cells (HE
at magnification, ×200). (A2) GPC3 was absent (GPC3 immunostaining
at magnification, ×200). (A3) Positive cells were <5% (Ki-67
immunostaining at magnification, ×200). (B)
Moderately-differentiated liver cancer. (B1) Tumor cells arranged
in trabeculae measuring ≥3 cells-thick (HE at magnification, ×200).
(B2) Tumor tissue indicated moderate staining with GPC3 (GPC3
immunostaining at magnification, ×200). (B3) Positive cells were
~30% (Ki-67 immunostaining, ×200). (C) Poorly differentiated liver
cancer and same field. (C1) The tumor tissue exhibited a solid
pattern with marked atypical cells (HE at magnification, ×200).
(C2) Tumor tissue indicated strong staining with GPC3 (GPC3
immunostaining at magnification, ×200). (C3) Positive cells were
>50% (Ki-67 immunostaining at magnification, ×200). HE,
hematoxylin and eosin; GPC3, glypican 3; Ki-67, antigen Ki-67. |
 | Table II.Association between GPC3 expression
and the differentiation in liver cancer samples. |
Table II.
Association between GPC3 expression
and the differentiation in liver cancer samples.
|
| GPC3 |
|
|---|
|
|
|
|
|---|
| Degree of
differentiation | − | + | ++ | +++ | Total |
|---|
|
Well-differentiated | 6 | 2 | 2 | 2 | 12 |
|
Moderately-differentiated | 18 | 32 | 13 | 9 | 72 |
|
Poorly-differentiated | 4 | 6 | 9 | 11 | 30 |
GPC3 expression is significantly
correlated with the expression of Ki-67
Ki-67 IHC staining usually represents the cell
proliferation index (12). Among the
39 patients with liver cancer that were Ki-67-negative, 21 cases
(53.85%) expressed GPC3, but only 1 (2.56%) case was strong
positive (+++). There was 94.12% (32/34) positive expression for
GPC3 in liver cancer samples with low-grade cell proliferation
(Ki-67+), 76% (19/25) of samples with intermediate-grade cell
proliferation (Ki-67++) were positive for GPC3 and 87.5% (14/16) of
samples with high-grade cell proliferation (Ki-67+++) exhibited
GPC3 expression. The numbers of liver cancer samples with marked
GPC3 expression with low, intermediate and high grades of cell
proliferation were 23.53 (8/34), 28 (7/25) and 37.5% (6/16),
respectively. There was a marked positive correlation between GPC3
and cell proliferation (r=0.316, P=0.01). The results indicated
that cancer cell proliferation was positively correlated with the
expression of GPC3 (r=0.316, P=0.001; Table III; Figs.
1 and 2B).
 | Table III.GPC3 expression is significantly
associated with the expression of Ki-67. |
Table III.
GPC3 expression is significantly
associated with the expression of Ki-67.
|
| GPC3 |
|
|---|
|
|
|
|
|---|
| Ki-67
expression | − | + | ++ | +++ | Total |
|---|
| − | 18 | 13 | 7 | 1 | 39 |
| + | 2 | 13 | 11 | 8 | 34 |
| ++ | 3 | 6 | 9 | 7 | 25 |
| +++ | 2 | 5 | 3 | 6 | 16 |
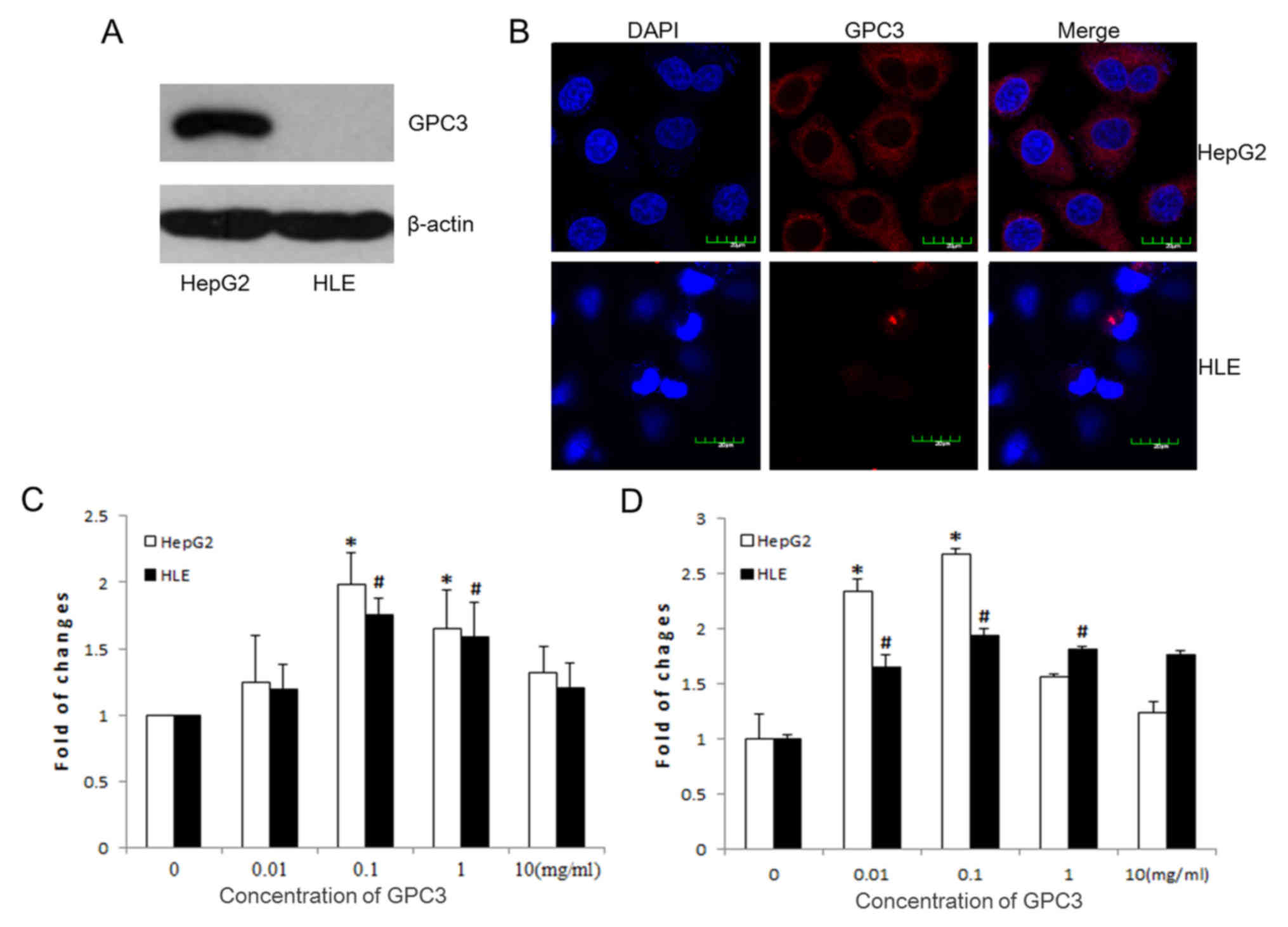
Verification of the effect of GPC3 on
cell proliferation in HepG2 and HLE cells
To additionally analyze the potential effect of GPC3
on cell growth, cell culture-based assays were performed with the
HepG2 and HLE hepatoma cell lines. As expected, the western
blotting results indicated that GPC3 was expressed in the HepG2
cells, but not in the HLE cells (Fig.
3A). Morphological images captured by the fluorescence
microscope additionally confirmed that GPC3 was present in the
HepG2 cells and present throughout the cytoplasm, but not in the
HLE cells (Fig. 3B). CCK-8 and MTT
assays were performed to verify the effect of GPC3 on tumor growth.
Following treatment of the HepG2 and HLE cells with various
concentrations of GPC3 (0, 0.01, 0.1, 1 and 10 mg/ml) for 24 h, the
CCK-8 (Fig. 3C) and MTT (Fig. 3D) results were similar, demonstrating
that that the most effective GPC3 dosage in promoting growth in the
HepG2 and HLE cells was 0.1 mg/l.
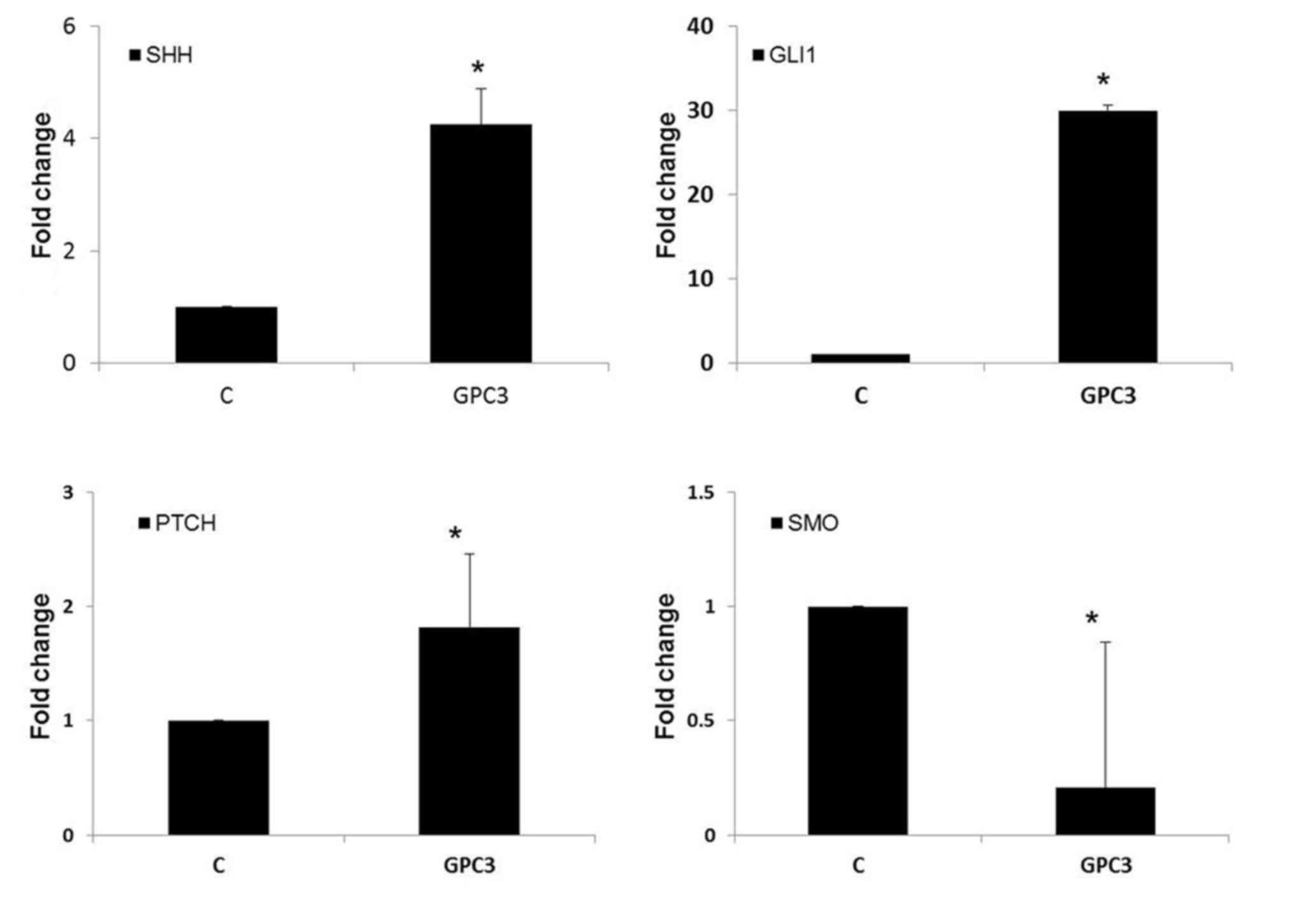
GPC3 promote HepG2 cells proliferation
through the Hedgehog (Hh) pathway
As aforementioned, GPC3 was able to stimulate HepG2
and HLE cell proliferation. The precise mechanism of how GPC3
promotes cell proliferation remains unclear. Based on the
aforementioned experimental results, HepG2 cells were treated with
0.1 mg/ml GPC3 for 24 h. RT-qPCR results indicated that
GPC3-treated HepG2 cells exhibited an increase in the mRNA
expression of the Hh signal pathway members Sonic hedgehog protein,
Zinc finger protein GLI1 and Protein patched homolog 1 (PTCH), and
the negative regulator of Hh signaling, Smoothened homolog, was
downregulated (Fig. 4). These results
suggested that GPC3 promoted HepG2 cell proliferation by
stimulating the Hh signaling pathway.
Discussion
In view of its high mortality rate and its
prevalence in several countries including China, liver cancer is a
malignancy of global importance. The prognosis of patients with
liver cancer is generally poor, with a 5-year survival rate of
<10-15% (21). It has been
demonstrated that GPC3 mRNA and protein were overexpressed in
patients with liver cancer compared with healthy people and
patients with benign liver lesions (22). The role that GPC3 serves in liver
cancer is controversial, but the present study focused on data from
clinical specimens, supporting the hypothesis that GPC3 may promote
liver cancer cell proliferation through the Hh signaling
pathway.
GPC3 is expressed ubiquitously in the embryo, but
the expression level is reduced in adults (23). The overexpression of GPC3 has been
detected in a number of human malignancies, including liver cancer,
melanoma and ovarian clear-cell carcinoma (24). GPC3 has an important role in cell
proliferation, differentiation, adhesion and migration, and its
function in tumorigenesis is tissue-dependent (25). Previous studies indicated that GPC3
protein expression was increased with lower degrees of tumor
differentiation in lung squamous cell carcinoma (26,27).
Similar results were also identified in liver cancer: Suzuki et
al (25) identified that GPC3 was
preferentially stained in poorly differentiated liver cancer when
compared with the well-differentiated liver cancer samples. The
present study identified that poorly differentiated liver cancer
was more likely to exhibit high expression of GPC3 (r=0.295,
P=0.001), that 30 liver cancer cases were poorly differentiated in
the total 114 specimens and that 36.67% (11/30) liver cancer
samples exhibited strong positive GPC3 (3+) staining; an additional
previous study suggested that GPC3 exhibited a significant
correlation with levels of differentiation in liver cancer only,
but did not correlate with tumor size (28).
Loss-of-function mutations in GPC3 cause SGBS, an
overgrowth syndrome also involving multiple embryonal neoplasia.
The study of Valsechi et al (29) suggested that GPC3 reduces the rate of
cell proliferation through cell cycle arrest during the G1 phase in
renal cell carcinoma. GPC3 may also inhibit breast cancer cells
growth in vitro (30). In the
present study, it was identified that 39/114 cases of liver cancer
were negative for Ki-67 staining, of which 18/39 (50%) cases were
also negative for GPC3. Only 1/39 cases were strong positive for
GPC3 (3+). Statistical analysis suggested that the expression of
Ki-67 was positively correlated with the expression of GPC3
(r=0.316, P=0.001).
GPC3 is frequently upregulated in liver cancer, but
its mechanism is largely unclear and is currently debated (25). One of the more well-studied pathways
associated with the biological functions of GPC3 is the Wnt
signaling pathway. GPC3 stimulates liver cancer cell growth in
vitro and in vivo by increasing autocrine or paracrine
canonical Wnt signaling (31). A
previous study suggested that the overexpression of GPC3 in Huh7
and SK-HEP-1 liver cancer cell lines effectively inhibited cell
proliferation through induction of apoptosis (32). An additional previous study indicated
that GPC3 suppressed cell growth in ovarian clear cell carcinoma
(CCC) cells via the insulin-like growth factor II signaling
pathway. Previous data also suggested that GPC3 has the potential
to become a novel therapeutic target for patients with ovarian CCC
(33). However, an additional study
indicated that the knockdown of GPC3 inhibits Huh7 cells
proliferation through the downregulation of Yes-associated protein,
which is a key effector molecule in the Hippo pathway (34). It has also been indicated that GPC3
binds Hh at the cell membrane and competes with PTCH, suggesting
that GPC3 regulates embryonic growth, perhaps by inhibiting the Hh
signaling pathway (4). The
significance of GPC3 in cell proliferation is of particular
interest. In the present study, the results demonstrated that
exogenous GPC3 may promote HepG2 and HLE hepatoma cell
proliferation, and that it is possible that HepG2 cells expressing
GPC3 are more sensitive to GPC3 compared with HLE cells, thereby
exhibiting increased rates of proliferation. Additional
experimental results suggested that exogenous GPC3 may promote
HepG2 cell proliferation by stimulating Hh signaling.
Taken together, the results of the present study
emphasize the significance and importance of GPC3 level in liver
cancer differentiation and proliferation. This function of GPC3 is
consistent with the observations of the present study, in that
patients with liver cancer with higher GPC3 levels appeared to
exhibit poorer levels of differentiation when compared with
patients with lower GPC3 levels. It appears that GPC3 is not only a
marker for diagnosis; it is also a growth factor in tumor
progression. Nonetheless, additional clinical studies are required
to confirm the function and molecular mechanism of GPC3 in liver
cancer.
Acknowledgements
The authors would like to thank Professor Gang Li
(Department of Biochemistry and Molecular Biology, Peking
University Health Science Center, Beijing 100191, P.R. China) for
gifting the HepG2 and HLE cells and assistance with the
experiments.
Funding
The present study was supported by National Natural
Science Foundation of China (No. 81401970) and Beijing Municipal
Institute of Public Medical Research Development and Reform Pilot
Project (grant no. JING YI YAN 2016-2).
Availability of data and materials
All data generated and analyzed during this study
are included in this published article.
Authors' contributions
WS contributed significantly in the collection and
analysis all the data; CN performed the in vitro experiment;
CY and LL undertook collection of patient information; SL and LH
were responsible for the pathological diagnosis; LH was responsible
for this project and wrote the manuscript.
Ethics approval and consent to
participate
This study was approved by the Beijing You'An
Hospital, Capital Medical University, Human Research Protection
Program (Beijing, China). Patients provided written informed
consent.
Consent for publication
Written informed consent was obtained from patients
for their inclusion in the present study and the publication of any
accompanying data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Boily G, Ouellet S, Langlois S, Larivière
M, Drouin R and Sinnett D: In vivo footprinting analysis of the
Glypican 3 (GPC3) promoter region in neuroblastoma cells. Biochim
Biophys Acta. 1769:182–193. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Filmus J and Selleck SB: Glypicans:
Proteoglycans with a surprise. J Clin Invest. 108:497–501. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Filmus J, Capurro M and Rast J: Glypicans.
GENOME BIOL 2008 2008-01-20. 9:224
|
|
4
|
Capurro MI, Xu P, Shi W, Li F, Jia A and
Filmus J: Glypican-3 inhibits Hedgehog signaling during development
by competing with patched for Hedgehog binding. Dev Cell.
14:700–711. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Filmus J and Capurro M: The role of
glypican-3 in the regulation of body size and cancer. Cell Cycle.
7:2787–2790. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Song HH and Filmus J: The role of
glypicans in mammalian development. Biochim Biophys Acta.
1573:241–246. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pilia G, Hughes-Benzie RM, MacKenzie A,
Baybayan P, Chen EY, Huber R, Neri G, Cao A, Forabosco A and
Schlessinger D: Mutations in GPC3, a glypican gene, cause the
Simpson-Golabi-Behmel overgrowth syndrome. Nat Genet. 12:241–247.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li M, Shuman C, Fei YL, Cutiongco E,
Bender HA, Stevens C, Wilkins-Haug L, Day-Salvatore D, Yong SL,
Geraghty MT, et al: GPC3 mutation analysis in a spectrum of
patients with overgrowth expands the phenotype of
Simpson-Golabi-Behmel syndrome. Am J Med Genet. 102:161–168. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nakatsura T, Kageshita T, Ito S, Wakamatsu
K, Monji M, Ikuta Y, Senju S, Ono T and Nishimura Y: Identification
of glypican-3 as a novel tumor marker for melanoma. Clin Cancer
Res. 10:6612–6621. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Maeda D, Ota S, Takazawa Y, Aburatani H,
Nakagawa S, Yano T, Taketani Y, Kodama T and Fukayama M: Glypican-3
expression in clear cell adenocarcinoma of the ovary. Mod Pathol.
22:824–832. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zynger DL, Dimov ND, Luan C, Teh BT and
Yang XJ: Glypican 3: A novel marker in testicular germ cell tumors.
Am J Surg Pathol. 30:1570–1575. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Schmilovitz-Weiss H, Tobar A, Halpern M,
Levy I, Shabtai E and Ben-Ari Z: Tissue expression of squamous
cellular carcinoma antigen and Ki67 in hepatocellular
carcinoma-correlation with prognosis: a historical prospective
study. Diagn Pathol. 6:1212001. View Article : Google Scholar
|
|
13
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gao JD, Shao YF, Xu Y, Ming LH, Wu ZY, Liu
GT, Wang XH, Gao WH, Sun YT, Feng XL, et al: Tight association of
hepatocellular carcinoma with HBV infection in North China.
Hepatobiliary Pancreat Dis Int. 4:46–49. 2005.PubMed/NCBI
|
|
15
|
Wang HL, Anatelli F, Zhai QJ, Adley B,
Chuang ST and Yang XJ: Glypican-3 as a useful diagnostic marker
that distinguishes hepatocellular carcinoma from benign
hepatocellular mass lesions. Arch Pathol Lab Med. 132:1723–1728.
2008.PubMed/NCBI
|
|
16
|
Bosman FT, Carneiro F, Hruban RH and
Theise ND: WHO classification of tumours of the digestive system.
4th edition. Int Agen Res Cancer Lyon. 1089:2010.
|
|
17
|
Shirakawa H, Kuronuma T, Nishimura Y,
Hasebe T, Nakano M, Gotohda N, Takahashi S, Nakagohri T, Konishi M,
Kobayashi N, et al: Glypican-3 is a useful diagnostic marker for a
component of hepatocellular carcinoma in human liver cancer. Int J
Oncol. 34:649–656. 2009.PubMed/NCBI
|
|
18
|
López-Terrada D, Cheung SW, Finegold MJ
and Knowles BB: Hep G2 is a hepatoblastoma-derived cell line. Hum
Pathol. 40:1512–1525. 2009. View Article : Google Scholar
|
|
19
|
Wang SS, Chen YH, Ning C, Wang LJ, Chen
DX, Weng HL, Dooley S and Ding HG: Hydrogen sulfide promotes
autophagy of hepatocellular carcinoma cells through the
PI3K/Akt/mTOR signaling pathway. Cell Death Dis. 8:e26882017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kawano Y, Sasaki A, Kai S, Endo Y, Iwaki
K, Uchida H, Shibata K, Ohta M and Kitano S: Short- and long-term
outcomes after hepatic resection for hepatocellular carcinoma with
concomitant esophageal varices in patients with cirrhosis. Ann Surg
Oncol. 15:1670–1676. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu H, Li P, Zhai Y, Qu CF, Zhang LJ, Tan
YF, Li N and Ding HG: Diagnostic value of glypican-3 in serum and
liver for primary hepatocellular carcinoma. World J Gastroenterol.
16:4410–4415. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Iglesias BV, Centeno G, Pascuccelli H,
Ward F, Peters MG, Filmus J, Puricelli L and de Kier Joffé EB:
Expression pattern of glypican-3 (GPC3) during human embryonic and
fetal development. Histol Histopathol. 23:1333–1340.
2008.PubMed/NCBI
|
|
24
|
Jian R, Zheng DY and Luo RC: The
expression of GPC3 in human malignant cancers and its potential
clinical application. Tumor. 31:863–866. 2011.
|
|
25
|
Suzuki M, Sugimoto K, Tanaka J, Tameda M,
Inagaki Y, Kusagawa S, Nojiri K, Beppu T, Yoneda K, Yamamoto N, et
al: Up-regulation of glypican-3 in human hepatocellular carcinoma.
Anticancer Res. 30:5055–5061. 2010.PubMed/NCBI
|
|
26
|
Lin Q, Xiong LW, Pan XF, Gen JF, Bao GL,
Sha HF, Feng JX, Ji CY and Chen M: Expression of GPC3 protein and
its significance in lung squamous cell carcinoma. Med Oncol.
29:663–669. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
An SJ, Huang YS, Chen ZH, Su J, Yang Y,
Chen JG, Yan HH, Lin QX, Yang JJ, Yang XN, et al: Posttreatment
plasma VEGF levels may be associated with the overall survival of
patients with advanced non-small cell lung cancer treated with
bevacizumab plus chemotherapy. Med Oncol. 29:627–632. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mounajjed T, Zhang L and Wu TT: Glypican-3
expression in gastrointestinal and pancreatic epithelial neoplasms.
Hum Pathol. 44:542–550. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Valsechi MC, Oliveira AB, Conceição AL,
Stuqui B, Candido NM, Provazzi PJ, de Araújo LF, Silva WA Jr,
Calmon Mde F and Rahal P: GPC3 reduces cell proliferation in renal
carcinoma cell lines. BMC Cancer. 14:6312014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Stigliano I, Puricelli L, Filmus J,
Sogayar MC, Bal de Kier Joffé E and Peters MG: Glypican-3 regulates
migration, adhesion and actin cytoskeleton organization in mammary
tumor cells through Wnt signaling modulation. Breast Cancer Res
Treat. 114:251–262. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gao W and Ho M: The role of glypican-3 in
regulating Wnt in hepatocellular carcinomas. Cancer Rep. 1:14–19.
2011.PubMed/NCBI
|
|
32
|
Pan Z, Chen C, Long H, Lei C, Tang G, Li
L, Feng J and Chen F: Overexpression of GPC3 inhibits
hepatocellular carcinoma cell proliferation and invasion through
induction of apoptosis. Mol Med Rep. 7:969–974. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sakurai M, Shibata K, Umezu T, Kajiyama H,
Yamamoto E, Ino K, Nawa A and Kikkawa F: Growth-suppressing
function of glypican-3 (GPC3) via insulin like growth factor II
(IGF-II) signaling pathway in ovarian clear cell carcinoma cells.
Gynecol Oncol. 119:332–336. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Miao HL, Pan ZJ, Lei CJ, Wen JY, Li MY,
Liu ZK, Qiu ZD, Lin MZ, Chen NP and Chen M: Knockdown of GPC3
inhibits the proliferation of Huh7 hepatocellular carcinoma cells
through down-regulation of YAP. J Cell Biochem. 114:625–631. 2013.
View Article : Google Scholar : PubMed/NCBI
|