Introduction
MicroRNAs (miRNAs/miRs) are a class of endogenous,
small and single-stranded RNA molecules, ~22 nucleotides in length
and highly conserved, encoded by the genome (1,2). MiRNAs
promote the degradation and translational inhibition of specific
target mRNAs in order to regulate various processes, including cell
proliferation and differentiation, and organism growth and
development (3–5). MiRNAs have been identified as a mode of
gene expression regulation in tumor development via the modulation
of oncogenes or tumor suppressor gene expression levels (6–9). Certain
studies have identified that miRNAs are expressed at abnormal
levels in a variety of cancer tissues, and similar to the role of
proto-oncogenes or tumor suppressor genes, aberrantly expressed
miRNAs may affect tumor proliferation and apoptosis (10,11).
Gastric cancer is the fourth leading type of cancer
worldwide, and is endemic in certain Asian countries, including
China, Japan, Iran and India (12).
In particular, gastric cancer has a high incidence in China, where
the morbidity and mortality rates are approximately twice the world
average (13). The abnormal
expression of a number of miRNAs, including miR-23a and miR-27a,
has already been identified in gastric cancer; these miRNAs may
function as oncogenes, based on their upregulated expression in
gastric cancer (14,15). Consequently, the mechanism of action
of miRNAs in gastric cancer has been a focus for research in recent
years. In our previous study, it was identified that miR-27a
functions as an oncogene in gastric adenocarcinoma by targeting
prohibitin (15).
In the present study, in order to further clarify
the role of miR-23a in gastric cancer, gene recombination
technology was used to construct a eukaryotic expression vector,
pcDNA3/pri-23a, for miR-23a. This vector was transfected into the
human gastric epithelial GES-1 cell line followed by continuous
clonal selection with G418 (Geneticin). The expression of miR-23a
was assessed by reverse transcription-semi-quantitative polymerase
chain reaction (RT-sqPCR) until a human gastric epithelial
monoclonal cell line was obtained, and this cell line stably
expressed miR-23a. Furthermore the expression of miR-23a in gastric
adenocarcinoma and normal gastric tissues was considered, and
whether miR-23a promotes the growth and invasion of the human
gastric epithelium GES-1 cell line was assessed.
Materials and methods
Human gastric cancer tissue
samples
In September 2004, four pairs of human gastric
cancer tissue samples (males; 26, 45, 46 and 62 years old) and
adjacent non-cancerous tissue samples were obtained from the Tumor
Bank Facility of Tianjin Medical University Cancer Institute and
Hospital and the National Foundation of Cancer Research (Tianjin,
China). All samples were confirmed by pathological analysis. The
study was performed with the written informed consent of all
participants, was approved by the Tianjin Ethics Committee, and was
in accordance with the ethical standards of the Declaration of
Helsinki and its later amendments.
The miRNA profiles of the tissue samples and matched
normal gastric tissue samples were determined using an
oligonucleotide microarray. Carcinoma tissue RNAs were labeled with
cyanine 5 (Cy5) and normal tissue RNAs were labeled with cyanine 3
(Cy3). The labeled samples were hybridized in a microarray (Version
3.0.0.0016; PerkinElmer, Inc., Waltham, MA, USA) containing 243
Homo sapiens miRNA probes. The ScanArray™ Express Microarray
scanner (PerkinElmer, Inc.) to scan the hybridized microarray, and
the figures were processed, normalized and analyzed using the
ScanArray® Express Microarray Analysis system. The data
were analyzed using the ScanArray® Express Microarray
Analysis System, and the Cy5/Cy3 value was calculated (16), and the value of miR-23a in every pair
of samples was calculated.
Materials and reagents
The normal human gastric epithelial GES-1 cell line
and 293 cells were purchased from the Beijing Institute of Cancer
Research (Beijing, China). GES-1 cells were maintained in RPMI-1640
with 10% fetal bovine serum at 37°C in a humidified chamber
supplemented with 5% CO2. The pcDNA3 plasmid was
provided by Professor Kenzo Takada (Japan Tumor Virology
Department, Institute of Genetic Medicine, Hokkaido University
School of Medicine, Sapporo). Lipofectamine™ 2000, Dulbecco's
modified Eagle's medium (DMEM) and RPMI-1640 medium were purchased
from Thermo Fisher Scientific, Inc. (Gibco; Waltham, MA, USA).
Fetal bovine serum was purchased from Tianjin Saierbio (Tianjin,
China). The restriction enzymes, T4 DNA ligase, PCR kit, plasmid
mini preparation kit and gel extraction purification kit were
purchased from Takara Biotechnology Co., Ltd. (Dalian, China). G418
(Geneticin) reagents and primers were synthesized by Shanghai
Biology Engineering Technology Service Ltd. (Shanghai, China).
Construction of plasmid
pcDNA3/pri-23a
The miR-23a precursor (pri-23a) sequence, which
refers to its coding site in the human genome, was identified in
the Rfam database (17).
Simultaneously, the gene sequence for pri-23a was retrieved from
the GenBank database provided by National Center for Biotechnology
Information (18). Primer Premier 5.0
software (version 5.0; Premier Biosoft International, Palo Alto,
CA, USA) was used to analyze these pri-23a sequences from the Rfam
and GenBank databases, and to design specific PCR primers to
amplify the miR-23a sequence. Specific PCR primers to amplify the
miR-23a sequence and to introduce restriction sites for cloning
were designed using Primer Premier5 and were as follows: Upstream
of pri-23a-Bgl II, the forward sequence was,
5′-CTCATATGCAGGAGCCAGATCTCGC-3′ and the reverse sequence was,
5′-GCGAGATCTGGCTCCTGCATATGAG-3′; downstream of
pri-23a-EcoRI, the forward sequence was,
5′-CCGAAGCCTGTGCCTGAATTCATC-3′ and the reverse sequence was,
5′-GATGAATTCAGGCACAGGCTTCGG-3′. The BglII and EcoRI restriction
enzyme sites were introduced into the upstream and downstream
primers, respectively. HEK293 cell genomic DNA was used as a
template for PCR reactions and purification. The PCR products and
pcDNA3 were digested with restriction endonuclease, the fragments
were recovered and they were ligated with T4 ligase. These
connecting fragments were transformed into competent XL1-blue
bacteria and white, medium-sized colonies were selected on
ampicillin plates. The selected colonies were cultured overnight
with agitation in lysogeny broth medium (Takara Biotechnology Co.,
Ltd.) us ampicillin, and plasmids were extracted. The recombinant
plasmid, pcDNA3/pri-23a, was verified by restriction enzyme
digestion with 1% gel electrophoresis and ethidium bromide
staining, followed by gene sequencing.
Cell transfection and screening of
stable GES-1 cell derivatives
GES-1 cells in the logarithmic phase were harvested
1 day prior to transfection and were seeded at a density of
1×106 cells/ml onto 24-well plates with DMEM, prior to
being incubated in a humidified incubator supplemented with 5%
CO2 at 37°C. When the cells reached 70% confluence, they
were transfected with Lipofectamine 2000 according to the
manufacturer's protocol. The cells were seeded onto 96-well plates
following transfection for 48 h. G418, also known as Geneticin, is
a 2-deoxystreptamine antibiotic from Micromonospora echinospora,
which inhibits protein synthesis by preventing the elongation step
in prokaryotic and eukaryotic cells. Complete medium (Takara
Biotechnology Co., Ltd.) with 300 g/ml G418 was used to select
clones for between 20 to 25 days. The monoclonal culture was
selected, the cultivation was further expanded, and the stable
GES-1/miR-23a (transfection group) and GES-1/EV (empty vector
group) cell lines were established.
Detection of miR-23a expression by
RT-sqPCR
Total RNA was extracted using TRIzol (Invitrogen;
Thermo Fisher Scientific, Inc.), reverse transcribed to cDNA using
a QuantiTect Reverse Transcription kit (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol, and
amplified with the PTC 200 machine (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). The cDNA was amplified using the following PCR
primers: Pri-23a forward, 5′-GCGAGATCTGGCTCCTGCATATGAG-3′ and
reverse, 5′-GATGAATTCCAGGCACAGGCTTCGG-3′; U6 forward,
5′-TGCGGGTGCTCGCTTCGGCAGC-3′ and reverse,
5′-CCAGTGCAGGGTCCGAGGT-3′. The length of the amplification product
was 324 bp. The reaction conditions of PCR were as follows: 94°C
for 4 min, followed by 33 cycles of 94°C for 30 sec, 42°C for 1 min
and 72°C for 30 sec. The PCR products were detected by 3% agarose
gel electrophoresis and the images were assessed using Quantity One
software version 4.6.2 (Bio-Rad Laboratories, Inc.). The ratio of
intensity of the target band versus the internal reference, U6
rRNA, was used to measure the levels of miR-23a expression.
Cell counting assay
GES-1/miR-23a, GES-1/EV and GES-1 cells in the
logarithmic growth phase were seeded onto 48-well plates at a
density of 1×103 cells/well. Three wells were selected
each day for 7 days to count the total number of cells, and the
means were calculated for each group in order to generate growth
curves.
MTT assay
GES-1/miR-23a, GES-1/EV and GES-1 cells in the
logarithmic growth phase were seeded onto 96-well plates at a
density of 1×103 cells/well and cultured in a humidified
incubator with 5% CO2 at 37°C. Following incubation for
24, 48 or 72 h, the culture plates were incubated with 20 µl
MTT/well, with a final concentration of 5 mg/ml for a further 4 h.
The culture plates were centrifuged at 2,000 × g for 5 min at room
temperature, the supernatant was carefully removed, and 100 µl DMSO
was added to each well to end the reaction. Blue-violet formazan
particles were dissolved for ~10 min, in the dark at 25°C. The
absorbance at 570 nm was detected using a µQuant Universal
microplate spectrophotometer (BioTek Instruments, Inc., Winooski,
VT, USA). The experiment was repeated three times in the same
conditions.
Transwell invasion assay
At 48 h after transfection, GES-1/miR-23a, GES-1/EV
and GES-1 cells were seeded at a density of 1×105
cells/ml onto 24-well plates. RPMI-1640 medium supplemented with 5%
fetal bovine serum and without 100 IU/ml penicillin and 100 mg/ml
streptomycin was placed into the upper chamber of an insert coated
with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA). RPMI-1640
medium supplemented with 10% fetal bovine serum, 100 IU/ml
penicillin and 100 mg/ml streptomycin was placed into the lower
chamber. The cells were incubated in a humidified incubator
supplemented with 5% CO2 at 37°C for 5 h. Cells that
invaded through the membrane were fixed with 75% methanol and 25%
glacial acetic acid at 37°C for 30 min, and were then stained with
0.1% crystal violet at 37°C for 5 min. The cells were imaged and
counted in four random fields/well using an inverted microscope at
×100 magnification (Olympus Corporation, Tokyo, Japan). The
experiment was repeated independently three times in the same
conditions.
Colony formation assay
GES-1/miR-23a, GES-1/EV and GES-1 cells were
evaluated using colony formation analysis. Cells were harvested at
3,000 × g at 37°C for 10 min and seeded onto 12-well plates at a
density of 1×102 cells/well. The plates were incubated
in a humidified incubator supplemented with 5% CO2 at
37°C for 2 weeks. During colony growth, the culture medium was
replaced every 3 days. The culture medium was removed and the cells
were stained with 0.1% crystal violet at 37°C for 5 min at the end
of the 2 weeks. The colonies were counted using an inverted
microscope at ×100 magnification. Each assay was performed in
triplicate.
Statistical analysis
SPSS 17.0 statistical software (SPSS, Inc., Chicago,
IL, USA) and GraphPad Prism 5.0 (GraphPad Software, Inc., La Jolla,
CA, USA) were used for statistical analysis. All data are expressed
as the mean ± standard deviation. One-way analysis of variance
followed by a Student Newman-Keuls post hoc test was used to
compare the data measurement groups. A two-tailed P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-23a is overexpressed in gastric
cancer
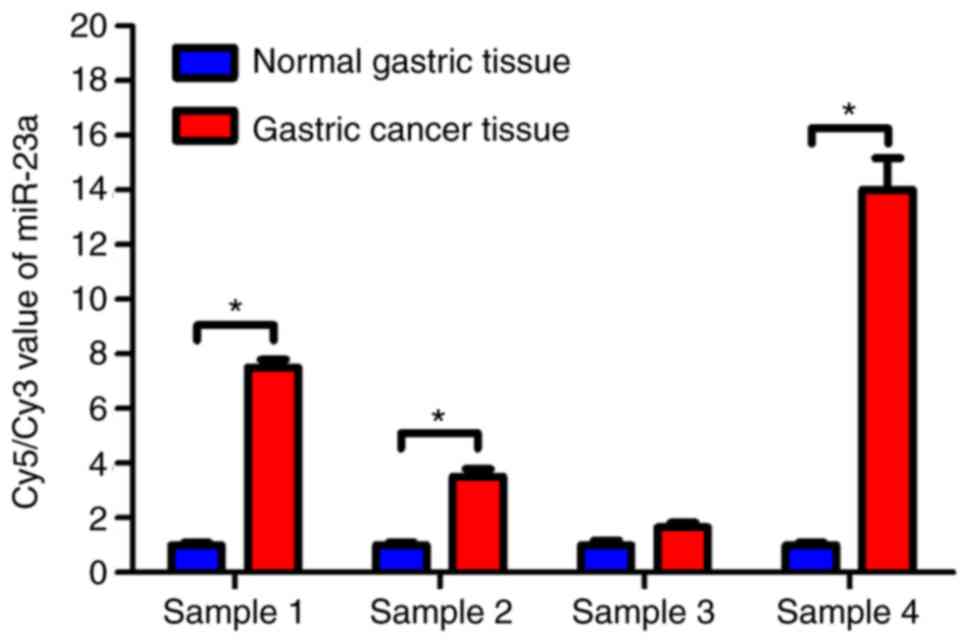
Based on the oligonucleotide microarray analysis of
four pairs of cancer and normal gastric tissues, miR-23a was
overexpressed in the gastric cancer tissues (Fig. 1). Compared with the normal gastric
tissue samples, miR-23a was significantly upregulated in gastric
cancer tissues (P<0.05). These results indicated that miR-23a
may promote gastric cancer development and influence the gene
regulation of gastric cancer cells.
Preparation of the miR-23a expression
plasmid
A plasmid expressing miR-23a was prepared by PCR
amplification of the miR-23a sequence and cloning into pcDNA3 to
yield recombinant plasmid pcDNA3/pri-23a. The recombinant plasmid
pcDNA3/pri-23a and the empty vector were identified by restriction
endonuclease BglII and EcoRI. For the pcDNA3/pri-23a plasmid, a
pcDNA vector fragment of 5,337 bp and a miR-23a precursor fragment
of 324 bp was obtained. For the empty vector, only a vector
fragment of 5,337 bp was obtained. The presence of the 324 bp
miR-23a precursor gene fragment demonstrated that the recombinant
pcDNA3/pri-23a plasmid was successfully constructed.
Selection of stable GES-1 cells
expressing miR-23a
To prepare stable control and miR-23a expressing
cell lines, the pcDNA3 and pcDNA3/pri-23a plasmids were transfected
into GES-1 cells. Following selection with G418, stable clones with
the empty pcDNA3 vector (GES-1/EV cells) and pcDNA3/pri-23a
(GES-1/miR-23a cells) were selected (Fig.
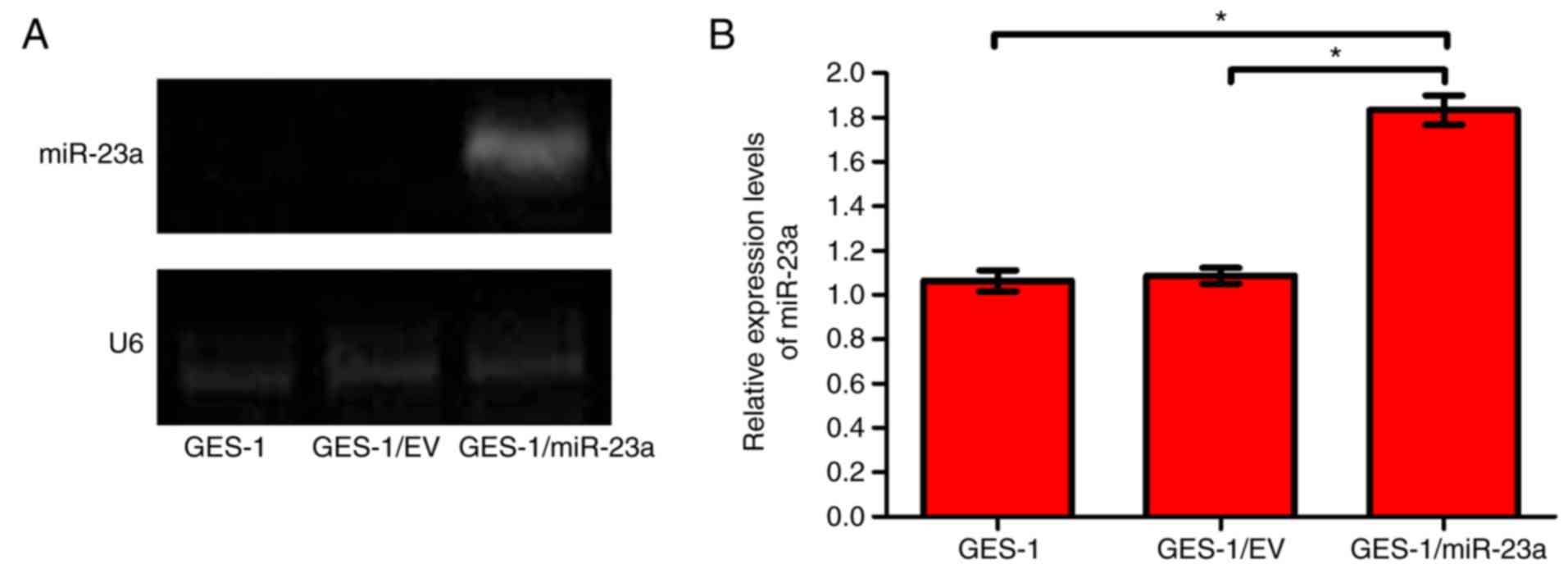
2). The expression of miR-23a in GES-1, GES-1/EV and
GES-1/miR-23a cells was measured by RT-sqPCR. The gel
electrophoresis image and quantification demonstrated that miR-23a
was expressed in GES-1/miR-23a cells, but not at observable levels
in parental GES-1 cells or the GES-1/EV control (Fig. 3A). The relative expression level of
miR-23a in GES-1/miR-23a cell was significantly higher compared
with that in the GES-1and GES-1/EV cells (P<0.05; Fig. 3B). This indicated that the
GES-1/miR-23a cell line effectively expressed miR-23a.
miR-23a expression promotes cell
proliferation and viability
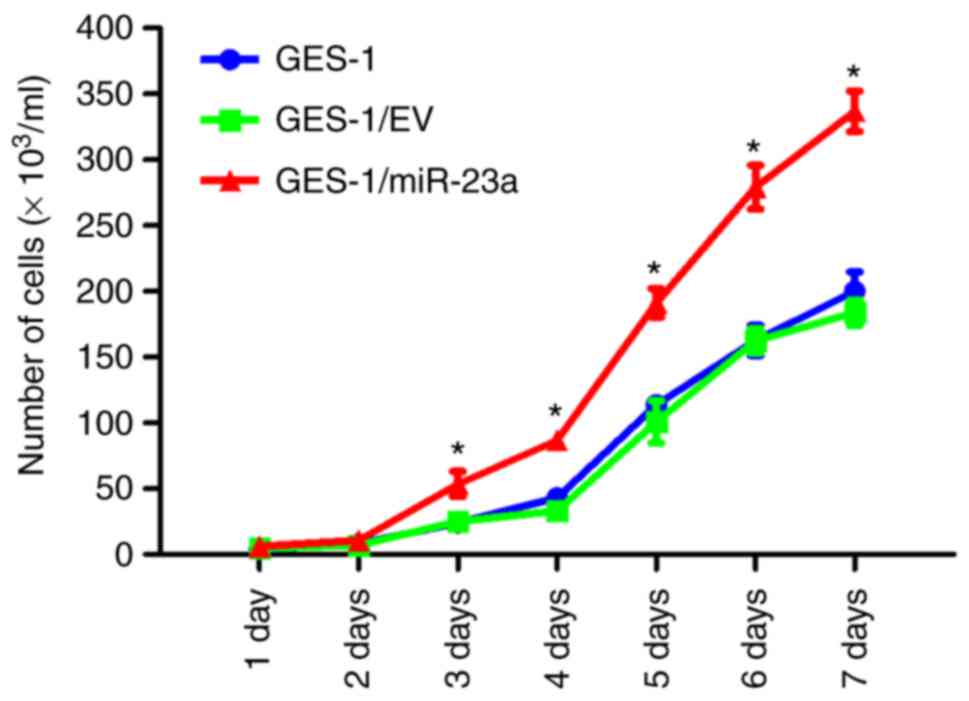
To determine whether miR-23a expression affects the
proliferation of GES-1 cells, cell counting and MTT assays were
performed. Cell counting demonstrated that GES-1/miR-23a cells
proliferated significantly faster compared with cells in the
control groups (P<0.05) (Fig. 4).
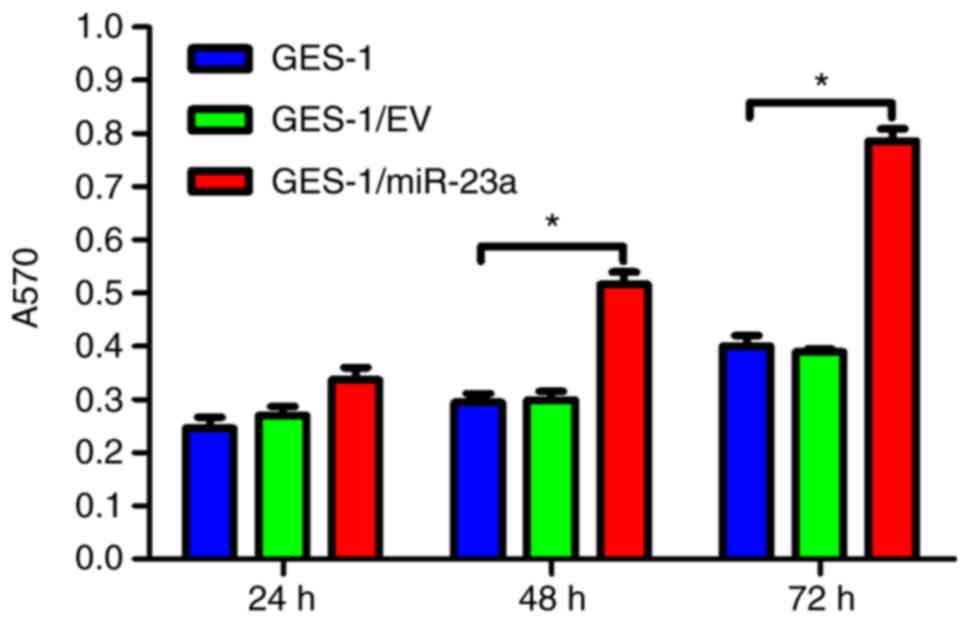
Furthermore, compared with the control groups, the viability of
GES-1/miR-23a cells was significantly increased at 48 and 72 h
(P<0.05), as determined with an MTT assay (Fig. 5).
miR-23a expression promotes cell
invasion
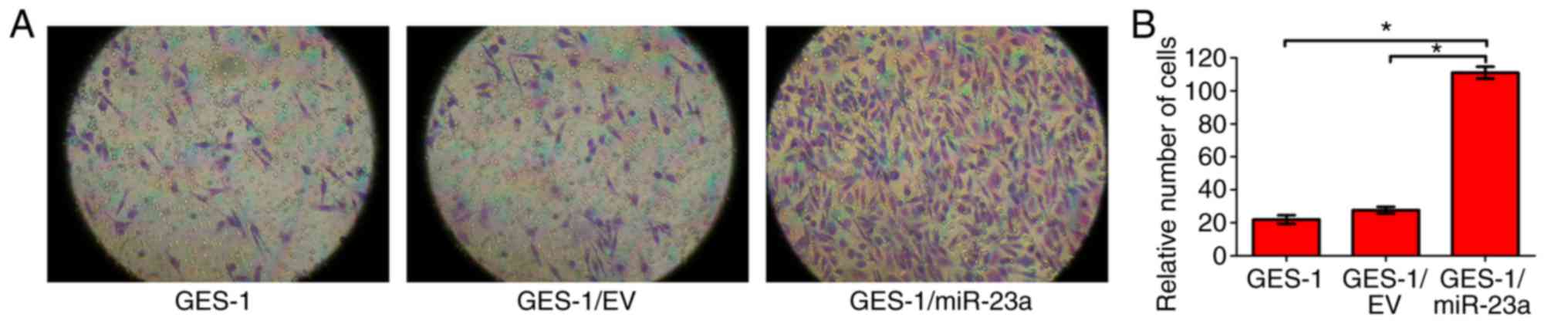
To identify whether miR-23a expression affects the
invasive ability of GES-1 cells, a Transwell invasion assay was
performed. The results indicated that the invasive ability of
GES-1/miR-23a cells was significantly enhanced compared with that
of the control groups (P<0.05; Fig.
6). It was concluded that miR-23a expression enhanced the
invasive ability of GES-1 cells.
miR-23a expression promotes
colony-forming ability
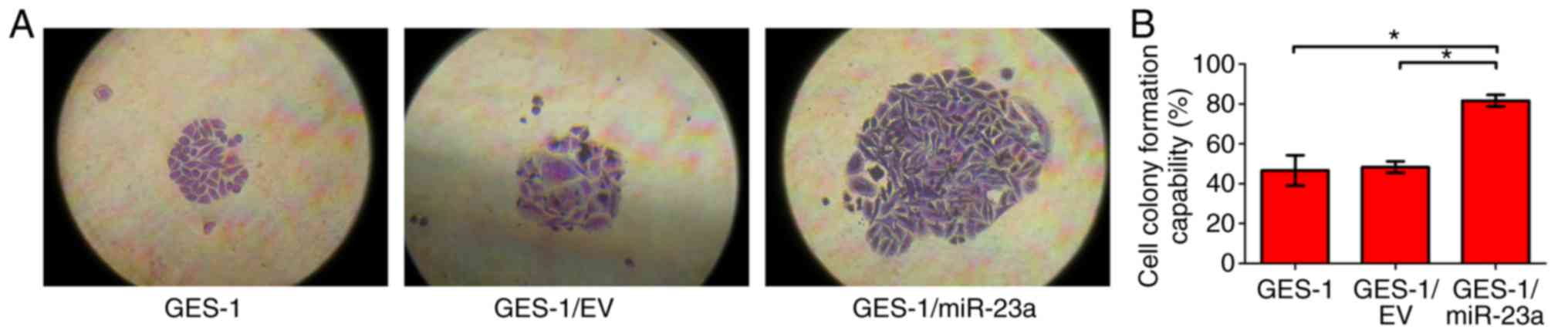
A colony formation assay was used to detect the
effect of miR-23a on the colony-forming ability of GES-1 cells.
After growth for 2 weeks, the cell clones were counted. The results
revealed that the colony forming capability of GES-1/miR-23a was
significantly enhanced compared with that of the control groups
(P<0.05; Fig. 7). It was concluded
that miR-23a enhanced the colony forming ability of GES-1
cells.
Discussion
miRNAs are a novel class of endogenous and
non-coding small RNAs, which function predominantly as
sequence-targeted modifiers of gene expression through
translational repression (19–21), and
were first described as a regulator of gene expression in
Caenorhabditis elegans (22,23). It
has since been identified that miRNAs are prevalent among plants
and animals, including in humans. Increasing evidence has
demonstrated that the alteration of miRNA expression profiles is
associated with several human diseases, including diabetes
(24), liver disease (25), inflammation (26), and cardiac development and pathologies
(27). miRNAs serve an essential role
in various biological and pathological processes, including cell
proliferation, stem cell differentiation, tumorigenesis, neuronal
development, apoptosis and carcinogenesis (28,29). These
studies suggest that miRNAs serve an important role during the
growth and development of an organism.
Emerging studies have shown that miRNA regulation is
associated with the development of a number of types of malignant
tumor. Therefore, an increasing focus of research is the screening
of tumor-associated miRNAs, the identification of miRNA target
genes and the mechanism of their regulation. The understanding of
the role of miRNAs in gastric cancer is an emerging area of
research. miR-34 may be involved in the negative regulation of p53
in gastric cancer (30). miR-125a-5p
is an independent positive prognostic factor in gastric cancer and
inhibits proliferation in vitro (31). miRNA-27a may regulate the growth of
gastric cancer cells by functioning as an oncogene through its
effects on prohibitin (15). miR-650
promotes the proliferation and growth of gastric cancer cells
(32). miR-663 contributes to
hyperplasia, leading to the development of gastric cancer (33). Furthermore, miR-9, miR-16 and miR-21
may regulate the growth of gastric cancer cells in human gastric
cancer pathogenesis (34,35). miR-29 inhibits the proliferation,
migration and invasion of gastric cancer cells by targeting cell
division cycle 42 (36). In addition,
miR-544 is an essential regulator of cell cycle control in gastric
cancer (37). miR-223 appears to
regulate apoptosis, proliferation and invasion in gastric cancer
(38). In addition, miR-200b
regulates zinc finger E-box binding homeobox 2 expression and
metastasis in gastric cancer (39).
The trio of miR-23a/27a/24 has growth-promoting and anti-apoptotic
roles (40).
The miRNA expression differences were analyzed in
four pairs of gastric cancer tissue samples and matched adjacent
normal tissue samples to identify candidate miRNAs that may be
associated with gastric cancer. Previous studies have demonstrated
that miR-23a was significantly upregulated in gastric cancer
tissues (14,40). In the present study, of the
differentially expressed miRNAs identified, miR-23a exhibited the
highest upregulation fold in gastric cancer tissues compared with
normal tissues, and thus, we hypothesized that it may have a
significant role. In our previous study, it was identified that the
genes coding miR-23a and miR-27a were located in the same cluster,
and that miR-27a acted as an oncogenic miRNA in gastric cancer
cells through targeting prohibitin (14). Hence GES-1, a normal human gastric
epithelial cell line, was selected for determining the role of
miR-23a, using the pcDNA3/pri-23a plasmid to introduce exogenous
expression.
The stable expression of miR-23a in GES-1 cells was
confirmed following cloning and selection. The primary transcript
(pri-23a), which was overexpressed in GES-1/miR-23a cells, was
processed in the cell to obtain mature miR-23a, thereby
facilitating its ability to serve a negative regulatory role in the
expression of target genes. Using cell counting and MTT methods, it
was identified that the overexpression of miR-23a significantly
increased the proliferation rate and viability of gastric
epithelial cells. In addition, the results of the Transwell assay
demonstrated that the invasive ability of GES-1/miR-23a cells was
significantly enhanced compared with the controls. Furthermore, a
colony formation assay was used to detect the effect of miR-23a on
the colony-forming ability of GES-1 cells. The colony-forming
ability of GES-1/miR-23a was significantly increased compared with
the controls. Based on these observations, the present study
demonstrated that miR-23a may serve a role as an oncogene in
gastric carcinogenesis.
With further research, additional tumor-associated
miRNAs and target genes, as well as downstream regulatory pathways,
will be identified. These studies will facilitate the further
understanding of the specific mechanisms associating miRNAs and
tumor formation. In addition, the establishment of miRNA-expressing
cell lines will assist in the discovery of novel methods for the
prevention and treatment of malignant tumors.
Acknowledgements
Not applicable.
Funding
The present study was supported by the project of
Science and Technology of Hebei Province of China (grant no.
16277782D).
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
LC designed the experiments and edited the
manuscript; YG conducted the statistical analyses; LHZ and HS
conceived and designed the experiments; LLZ and AL performed the
experiments; GZ and GS collected the data and provided the
technical assistance. All authors have read and approved the
manuscript.
Ethics approval and consent to
participate
The study was performed following written informed
consent from all participants; the study was approved by the
Tianjin Ethics Committee in accordance with the ethical standards
of the Declaration of Helsinki and its later amendments.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Garzon R, Marcucci G and Croce CM:
Targeting microRNAs in cancer: Rationale, strategies and
challenges. Nat Rev Drug Discov. 9:775–789. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Carthew RW and Sontheimer EJ: Origins and
mechanisms of miRNAs and siRNAs. Cell. 136:642–655. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Iorio MV and Croce CM: microRNA
involvement in human cancer. Carcinogenesis. 33:1126–1133. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Santovito D, Mezzetti A and Cipollone F:
MicroRNAs and atherosclerosis: New actors for an old movie. Nutr
Metab Cardiovasc Dis. 22:937–943. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhou H, Wang K, Hu Z and Wen J: The effect
of miR-193b on the metastatic and invasive capacity of gastric
cancer cells. Chin J Gen Surg. 20:377–382. 2011.
|
|
7
|
Tang H, Liu X, Wang Z, She X, Zeng X, Deng
M, Liao Q, Guo X, Wang R, Li X, et al: Interaction of hsa-miR-381
and glioma suppressor LRRC4 is involved in glioma growth. Brain
Res. 1390:21–32. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xiong X and Zheng CL: The effect of
miRNA-214 on cell cycle and apoptosis of gastric cancer cell lines
BGC823 and MKN45. Chin J Gen Surg. 19:1311–1315. 2010.
|
|
9
|
Zhang L, Deng T, Li X, Liu H, Zhou H, Ma
J, Wu M, Zhou M, Shen S, Li X, et al: microRNA-141 is involved in a
nasopharyngeal carcinoma-related genes network. Carcinogenesis.
31:559–566. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang J, Han C and Wu T: MicroRNA-26a
promotes cholangiocarcinoma growth by activating β-catenin.
Gastroenterology. 143:246–56.e8. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Frampton AE, Castellano L, Colombo T,
Giovannetti E, Krell J, Jacob J, Pellegrino L, Roca-Alonso L, Funel
N, Gall TM, et al: MicroRNAs cooperatively inhibit a network of
tumor suppressor genes to promote pancreatic tumor growth and
progression. Gastroenterology. 146:268–277. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mir MR, Shabir N, Wani KA, Shaff S,
Hussain I, Banday MA, Chikan NA, Bilal S and Aejaz S: Association
between p16, hMLH1 and E-cadherin promoter hypermethylation and
intake of local hot salted tea and sun-dried foods in Kashmiris
with gastric tumors. Asian Pac J Cancer Prev. 13:181–186. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen L, Yan Y, Zhu L, Cong X, Li S, Song
S, Song H and Xue Y: Systemic immune-inflammation index as a useful
prognostic indicator predicts survival in patients with advanced
gastric cancer treated with neoadjuvant chemotherapy. Cancer Manag
Res. 9:849–867. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhu LH, Liu T, Tang H, Tian RQ, Su C, Liu
M and Li X: MicroRNA-23a promotes the growth of gastric
adenocarcinoma cell line MGC803 and downregulates interleukin-6
receptor. FEBS J. 277:3726–3734. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu T, Tang H, Lang Y, Liu M and Li X:
MicroRNA-27a functions as an oncogene in gastric adenocarcinoma by
targeting prohibitin. Cancer Lett. 273:233–242. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Evans CA, Tonge R, Blinco D, Pierce A,
Shaw J, Lu Y, Hamzah HG, Gray A, Downes CP, Gaskell SJ, et al:
Comparative proteomics of primitive hematopoietic cell populations
reveals differences in expression of proteins regulating motility.
Blood. 103:3751–3759. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gardner PP, Daub J, Tate JG, Nawrocki EP,
Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S,
Eddy SR, et al: Rfam: Updates to the RNA families database. Nucleic
Acids Res. 37:D136–D140. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nikolic I, Elsworth B, Dodson E, Wu SZ,
Gould CM, Mestdagh P, Marshall GM, Horvath LG, Simpson KJ and
Swarbrick A: Discovering cancer vulnerabilities using
high-throughput micro-RNA screening. Nucleic Acids Res.
45:12657–12670. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Creemers EE, Tijsen AJ and Pinto YM:
Circulating microRNAs: Novel biomarkers and extracellular
communicators in cardiovascular disease? Circ Res. 110:483–495.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Topkara VK and Mann DL: Role of microRNAs
in cardiac remodeling and heart failure. Cardiovasc Drugs Ther.
25:171–182. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Divakaran V and Mann DL: The emerging role
of microRNAs in cardiac remodeling and heart failure. Circ Res.
103:1072–1083. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993.
|
|
23
|
Ambros V: microRNAs: Tiny regulators with
great potential. Cell. 107:823–826. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Guay C, Roggli E, Nesca V, Jacovetti C and
Regazzi R: Diabetes mellitus, a microRNA-related disease? Transl
Res. 157:253–264. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kerr TA, Korenblat KM and Davidson NO:
MicroRNAs and liver disease. Transl Res. 157:241–252. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dai R and Ahmed SA: MicroRNA, a new
paradigm for understanding immunoregulation, inflammation, and
autoimmune diseases. Transl Res. 157:163–179. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Small EM and Olson EN: Pervasive roles of
microRNAs in cardiovascular biology. Nature. 469:336–342. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Olde Loohuis NF, Kos A, Martens GJ, van
Bokhoven H, Nadif Kasri N and Aschrafi A: MicroRNA networks direct
neuronal development and plasticity. Cell Mol Life Sci. 69:89–102.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang S, He X, Ding J, Liang L, Zhao Y,
Zhang Z, Yao X, Pan Z, Zhang P, Li J, et al: Upregulation of
miR-23a approximately 27a approximately 24 decreases transforming
growth factor-beta-induced tumor-suppressive activities in human
hepatocellular carcinoma cells. Int J Cancer. 123:972–978. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ji Q, Hao X, Meng Y, Zhang M, Desano J,
Fan D and Xu L: Restoration of tumor suppressor miR-34 inhibits
human p53-mutant gastric cancer tumorspheres. BMC Cancer.
8:2662008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nishida N, Mimori K, Fabbri M, Yokobori T,
Sudo T, Tanaka F, Shibata K, Ishii H, Doki Y and Mori M:
MicroRNA-125a-5p is an independent prognostic factor in gastric
cancer and inhibits the proliferation of human gastric cancer cells
in combination with trastuzumab. Clin Cancer Res. 17:2725–2733.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang X, Zhu W, Zhang J, Huo S, Zhou L, Gu
Z and Zhang M: MicroRNA-650 targets ING4 to promote gastric cancer
tumorigenicity. Biochem Biophys Res Commun. 395:275–280. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pan J, Hu H, Zhou Z, Sun L, Peng L, Yu L,
Sun L, Liu J, Yang Z and Ran Y: Tumor-suppressive mir-663 gene
induces mitotic catastrophe growth arrest in human gastric cancer
cells. Oncol Rep. 24:105–112. 2010.PubMed/NCBI
|
|
34
|
Shin VY, Jin H, Ng EK, Cheng AS, Chong WW,
Wong CY, Leung WK, Sung JJ and Chu KM: NF-κB targets miR-16 and
miR-21 in gastric cancer: involvement of prostaglandin E receptors.
Carcinogenesis. 32:240–245. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wan HY, Guo LM, Liu T, Liu M, Li X and
Tang H: Regulation of the transcription factor NF-kappaB1 by
microRNA-9 in human gastric adenocarcinoma. Mol Cancer. 9:162010.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lang N, Liu M, Tang QL, Chen X, Liu Z and
Bi F: Effects of microRNA-29 family members on proliferation and
invasion of gastric cancer cell lines. Chin J Cancer. 29:603–610.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhi QM, Guo X, Guo L, Zhang R, Jiang J, Ji
J, Zhang J, Zhang J, Chen X, Cai Q, et al: Oncogenic miR-544 is an
important molecular target in gastric cancer. Anticancer Agents Med
Chem. 13:270–275. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li J, Guo Y, Liang X, Sun M, Wang G, De W
and Wu W: MicroRNA-223 functions as an oncogene in human gastric
cancer by targeting FBXW7/hCdc4. J Cancer Res Clin Oncol.
138:763–774. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kurashige J, Kurashige J, Kamohara H,
Watanabe M, Hiyoshi Y, Iwatsuki M, Tanaka Y, Kinoshita K, Saito S,
Baba Y and Baba H: MicroRNA-200b regulates cell proliferation,
invasion, and migration by directly targeting ZEB2 in gastric
carcinoma. Ann Surg Oncol. 19(Suppl 3): S656–S664. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhou R, O'Hara SP and Chen XM: MicroRNA
regulation of innate immune responses in epithelial cells. Cell Mol
Immunol. 8:371–379. 2011. View Article : Google Scholar : PubMed/NCBI
|