Introduction
Prostate cancer (PCa) is the most common malignancy
in males worldwide, and the second-leading cause of
cancer-associated mortality (1,2). While the
prevalence of PCa in Arab countries is lower than that in Western
countries, the incidence is steadily increasing (3). Although the majority of PCa cases are
indolent and localized at diagnosis, localized tumors can develop
into aggressive tumors in the long term (4), the early detection of PCa, when
treatment is most effective, can significantly contribute to a
reduction in the mortality rate. Prostate-specific antigen (PSA)
has long been used for the detection and screening of PCa. However,
elevated levels of PSA have also been reported in non-malignant
conditions of the prostate (5),
including benign prostatic hyperplasia (BPH), which is commonly
misdiagnosed as PCa, leading to unnecessary biopsies (6). PSA also has a positive predictive value
of only ~35%, potentially leading to false-positive or
false-negative results (7). It has
been demonstrated that a significant proportion of males who
underwent a prostate biopsy due to an elevated PSA level did not
actually have PCa (8). PSA testing
and screening have been demonstrated to be associated with a high
rate of over-diagnosis and overtreatment in a number of clinical
trials (9–11). In addition, PSA testing and other
clinical parameters currently used for the stratification of
patients with PCa, including clinical stage and Gleason score (GS)
tumor grade have limitations in detecting and predicting the
disease outcome (12). Therefore, PSA
is neither an effective predictor of PCa, nor does it contribute to
risk stratification (13,14). Novel, non-invasive biomarkers that can
distinguish malignant from benign prostatic tumors, and detect PCa
at an early stage to improve disease diagnosis and management, are
urgently required (15).
Genetic and epigenetic modifications contribute
significantly to the pathogenicity and development of PCa (16). Among the alterations that affect the
regulation of gene expression, the disturbance of microRNA
(miRNA/miR) expression and function can affect several pathways
involved in the initiation and development of cancer, including
those of the cell cycle, proliferation, angiogenesis and apoptosis
(17). The differential expression
profiles of miRNAs in different types of cancer, compared with
those in normal tissue, support the hypothesis that miRNAs serve a
critical role in cancer (18). The
upregulation and downregulation of miRNAs in different types of
cancer suggest a dual role for miRNAs, acting as either tumor
suppressors or oncogenes (18).
Among several miRNAs implicated in cancer, the
expression levels of miR-15a, miR-126, miR-192 and miR-377 have
been identified as downregulated in a wide range of cancer types,
including myeloma, non-small cell lung carcinoma, pancreatic
cancer, renal cell carcinoma and osteosarcoma (19–24). These
four miRNAs have been described as tumor suppressors in PCa. Bonci
et al (25) reported that
miR-15a could function as a tumor suppressor in PCa via repression
of cyclin D1 and Wnt family member 3A (WNT3A) oncogene expression.
Song et al (26) observed that
miR-126 could inhibit cancer proliferation and metastasis by
targeting phosphoinositide-3-kinase regulatory subunit 2 (PIK3R2)
in PCa. Sun et al (27)
demonstrated that miR-192 could serve a tumor suppressive role and
impair cell tumorigenicity in PCa by targeting and repressing the
oncogene, nin one binding protein 1 (NOB1). Formosa et al
(28) also reported that miR-377
could affect the behavior of malignant cells in PCa cell lines by
targeting FZD4, a gene important in epithelial-to-mesenchymal
transition.
Previous studies have proposed miRNAs as diagnostic
and prognostic biomarkers for cancer (29,30) and
numerous other types of disease (31,32), due
to their high stability in bio-fluids, including blood (33,34).
Circulating miRNAs are considered to be either contained in
exosomes, microparticles and apoptotic bodies, or associated with
argonaute 2 and other RNA-binding-proteins and high-density
lipoproteins, enabling their transfer from one cell to another in
diverse biological processes (35).
Circulating miRNA profiling studies have demonstrated
differentially expressed miRNAs with diagnostic, prognostic and
predictive abilities in PCa (36,37). The
majority of these studies have evaluated the biomarker potential of
these miRNAs to distinguish early stage PCa from metastatic PCa
(36,37). However, the present study addresses
the feasibility of using circulating miRNAs as biomarkers to
distinguish localized PCa from benign prostatic hyperplasia (BPH),
and to discriminate between low- and high-risk patients at an early
stage.
Therefore, in the present study, the expression
levels of peripheral blood miR-15a, miR-126, miR-192 and miR-377,
selected based on their critical functions in PCa, were quantified
and their diagnostic value was investigated. These miRNAs may be
suitable biomarkers for the early detection of localized PCa, and
for PCa risk stratification.
Materials and methods
Participants
Ethical approval to conduct the present study was
obtained from the Medical Research and Ethics Committee of the
College of Medicine and Medical Sciences, Arabian Gulf University
(Manama, Kingdom of Bahrain). A total of 100 Bahraini participants
were recruited from the Urological Clinic of King Abdullah Medical
City Hospital between June 2013 and January 2014, and divided into
three groups: 35 patients with localized PCa, 35 patients with BPH
and 30 healthy control subjects. The mean age of the patients with
PCa was 73.8±4.2, and the mean age of BPH patients was 71.8±5.9,
whereas the mean age of healthy subjects was 71.6±4.8. All
participants provided informed consent for the extraction and use
of their blood samples and data. The diagnosis of localized PCa was
made based on elevated PSA levels and malignant histological
findings in the prostate biopsy specimen without evidence of
metastatic disease. Patients with PCa were categorized into two
groups, according to the D'Amico risk classification criteria for
localized PCa (38), tumor (T) stage,
PSA level and GS: Patients with low-risk localized PCa (n=20) had a
tumor stage of T1c or T2a, a PSA of ≤10 ng/ml or a GS of ≤7;
patients with high-risk localized PCa (n=15) had a tumor stage of
T2c, a PSA of >20 ng/ml or a GS of ≥8. The diagnosis of BPH was
made based on digital rectal examination, and a benign histological
finding of the prostate biopsy specimen if PSA was elevated. The
healthy control subjects were those who underwent routine physical
examinations with no underlying prostate disease.
Blood sampling
Whole blood samples (5 ml) were obtained from
patients with PCa one day prior to radical prostatectomy surgery,
as well as from patients with BPH and healthy control subjects. All
blood samples were collected in EDTA tubes. RNAlater (1.3 ml;
Ambion; Thermo Fisher Scientific, Inc., Waltham, MA, USA), an RNA
stabilization reagent, was added to each tube, and the blood
samples were stored at −80°C until RNA extraction.
Selection of miRNAs as candidate blood
biomarkers
According to previous reports, a number of
PCa-associated miRNAs were selected, including miR-15a, miR-126,
miR-192 and miR-377, to evaluate their potential as diagnostic
biomarkers for PCa. The 4 selected miRNAs were previously reported
to be dysregulated in prostate tumor tissue and PCa cell lines
(25–28).
miRNA extraction and reverse
transcription (RT)
The extraction of RNA, including small RNA, was
performed using the miRNeasy kit (Qiagen GmbH, Hilden, Germany)
according to the manufacturer's protocols, as previously described
(30,31,39). The
quality and concentration of RNA extracted from the clinical
samples was evaluated using a Nanodrop ND-100 spectrophotometer
(Thermo Fisher Scientific, Inc.). The concentrations of RNA ranged
between 50 and 75 ng/µl; aliquots from all RNA samples were diluted
to identical final concentrations of 20 ng/µl. cDNA was prepared
from 20 ng RNA using a TaqMan miRNA Reverse Transcription kit
(Applied Biosystems; Thermo Fisher Scientific, Inc.) as previously
described (30,31,39), with
specific stem-loop RT primers. All cDNA samples were stored at
−20°C until further analysis.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
The expression of miRNAs was quantified by RT-qPCR
using TaqMan Universal PCR Master mix II (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The kit contained 1.33 µl cDNA, 10
µl TaqMan 2X Universal PCR Master mix II (Applied Biosystems;
Thermo Fisher Scientific, Inc.), 1 µl gene-specific primers and
7.67 µl nuclease-free water to a final volume of 20 µl. RT-qPCR was
performed on a 7900HT Real Time PCR system (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The PCR primer sequences for the
targets were as follows: miR-15a, 5′-UAGCAGCACAUAAUGGUUUGUG-3′;
miR-126, 5′-UCGUACCGUGAGUAAUAAUGC-3′; miR-192,
5′-CUGCCAAUUCCAUAGGUCACAG-3′; and miR-377,
5′-AUCACACAAAGGCAACUUUUGU-3′. The PCR primer sequence for reference
was as follows: U6B,
5′-CGCAAGGATGACACGCAAATTCGTGAAGCGTTCCATATTTTT-3′.
The qPCR was performed using the following cycling
conditions: 95°C for 10 min, followed by 95°C for 15 sec and 60°C
for 60 sec for a total of 40 cycles. The 2−ΔΔCq method
was used to determine the expression of miRNAs relative to U6B
(40) using SDS software version 1.4
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The mean fold
change of duplicate qPCR amplifications was used in statistical
analysis.
Statistical analysis
Statistical analysis was performed using SPSS
version 23 (IBM Corp., Armonk, NY, USA). Differences in the
clinical variables and the relative expression of blood miRNAs
between patients with PCa and those with BPH, as well as between
patients with low-risk and high-risk localized PCa were analyzed
with Student's t test. Comparisons of the clinical variables among
more than 2 groups were evaluated using one-way analysis of
variance. Data for miRNA expression, age and PSA are presented as
the mean ± standard deviation. Data for GS, tumor stage, lymph node
stage and metastasis stage are presented as numbers and
percentages. The association between individual miRNAs and PCa was
determined with multivariate logistic regression analysis by
obtaining the odds ratios (OR) and 95% confidence intervals (CIs)
for either a crude model (1a and 2a), an age-adjusted model (1b),
or an age- and PSA-adjusted model (2b). Receiver operating
characteristic (ROC) curves and the area under the ROC curve (AUC)
were used to assess the diagnostic accuracy of miRNAs as
biomarkers. P<0.05 was considered to represent a statistically
significant difference.
Results
Clinical characteristics of
participants
Table I lists the
clinical characteristics of the participants, including 35 patients
with localized PCa, 35 patients with BPH and 30 healthy control
subjects. Patients with localized PCa were divided into two groups:
Low-risk (n=20) and high-risk (n=15), based on T stage, serum PSA
and GS (35).
 | Table I.Clinical characteristics of
participants. |
Table I.
Clinical characteristics of
participants.
| Characteristic | All PCa | Low-risk PCa | High-risk PCa | BPH | Healthy |
|---|
| Total number | 35 | 20 | 15 | 35 | 30 |
| Age, years | 73.8±4.2 | 73.0±4.6 | 74.9±3.5 | 71.8±5.9 | 71.6±4.8 |
| PSA, ng/ml |
15.6±7.6a | 9.15±0.51 |
21.7±5.98b | 8.7±1.5 | 3.7±0.7 |
| Gleason score |
|
|
|
|
|
| ≤7 | 20 (57.1) | 20 (100) |
| – | – |
| ≥8 | 15 (42.9) |
| 15 (100) | – | – |
| Tumor stage |
|
|
|
|
|
|
T1/2 | 20 (57.1) | 20 (100) |
| – | – |
|
T3/4 | 15 (42.9) |
| 15 (100) | – | – |
| Lymph node
stage |
|
|
|
|
|
| N0 | 32 (91.4) | 20 (100) |
| – | – |
| N1 | 3 (8.6) |
| 3 (20) | – | – |
| Metastasis
stage |
|
|
|
|
|
| M0 | 35 (100) | 20 (100) | 15 (100) | – | – |
| M1 | – | – | – | – | – |
As shown in Table I,
there was no significant difference in the mean age among the three
subjects groups (P>0.05). Patients with PCa or BPH had
significantly higher levels of serum PSA, with mean values of 15.6
and 8.7 ng/ml, respectively, compared with the healthy subjects
(3.7 ng/ml; P<0.05). Furthermore, the mean serum PSA level was
significantly higher in patients with PCa than in patients with BPH
(P<0.05).
In the localized PCa group, 20 patients presented
with low-risk PCa, with a tumor stage of T1c or T2a, a PSA of ≤10
ng/ml and a GS of ≤7 and 15 patients presented with high-risk PCa,
with a tumor stage of T2c or greater, a PSA of >20 ng/ml and a
GS of ≥8. There was no significant difference in mean age between
the groups (P>0.05). The mean serum PSA levels differed
significantly (P<0.05) and were higher in the high-risk group
than in the low-risk group. Three patients in the high-risk group
presented with regional lymph node involvement (stage N1), and none
of the patients presented with distant metastasis.
Expression of blood miRNAs in patients
with localized PCa, patients with BPH and healthy subjects
Using RT-qPCR, the expression of miR-15a, miR-126,
miR-192 and miR-377 relative to the internal control U6B were
quantitated in peripheral whole blood and compared between patients
with localized PCa or BPH, and healthy control subjects.
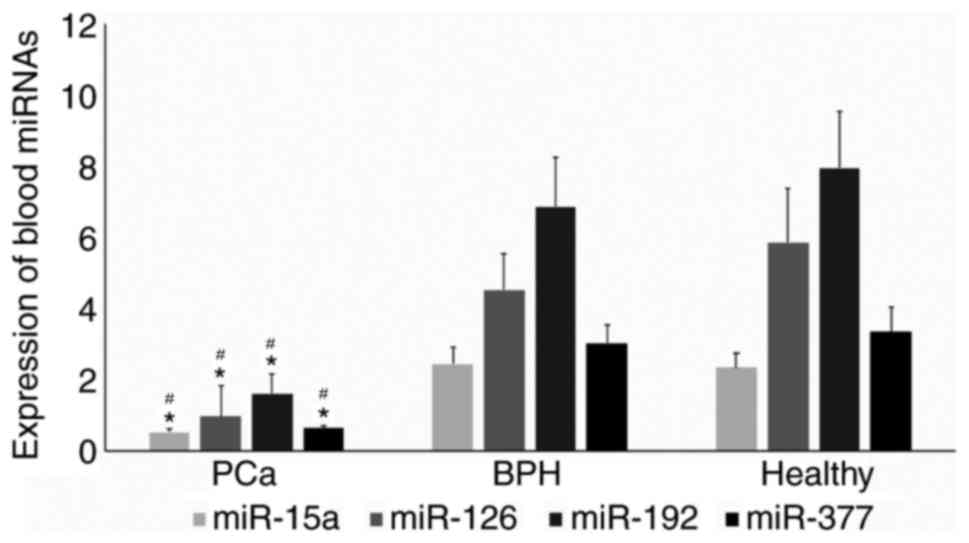
Fig. 1 demonstrates
the overall expression of the 4 miRNAs, which were all
significantly lower in patients with PCa than in patients with BPH
or healthy subjects (P<0.05). miR-15a expression was lower in
patients with PCa (0.54±0.1) compared with either patients with BPH
(2.44±0.47) or healthy subjects (2.35±0.4). miR-126 expression was
also lower in patients with PCa (0.97±0.88) with respect to
patients with BPH (4.53±1.0) and healthy subjects (5.85±1.5).
Similarly, patients with PCa demonstrated a decreased expression of
miR-192 (1.61±0.45) compared with patients with BPH (6.85±1.4) or
healthy subjects (7.93±1.6). Finally, patients with PCa exhibited a
lower expression of miR-377 (0.64±0.07) compared with patients with
BPH (3.04±0.5) or healthy subjects (3.34±0.7).
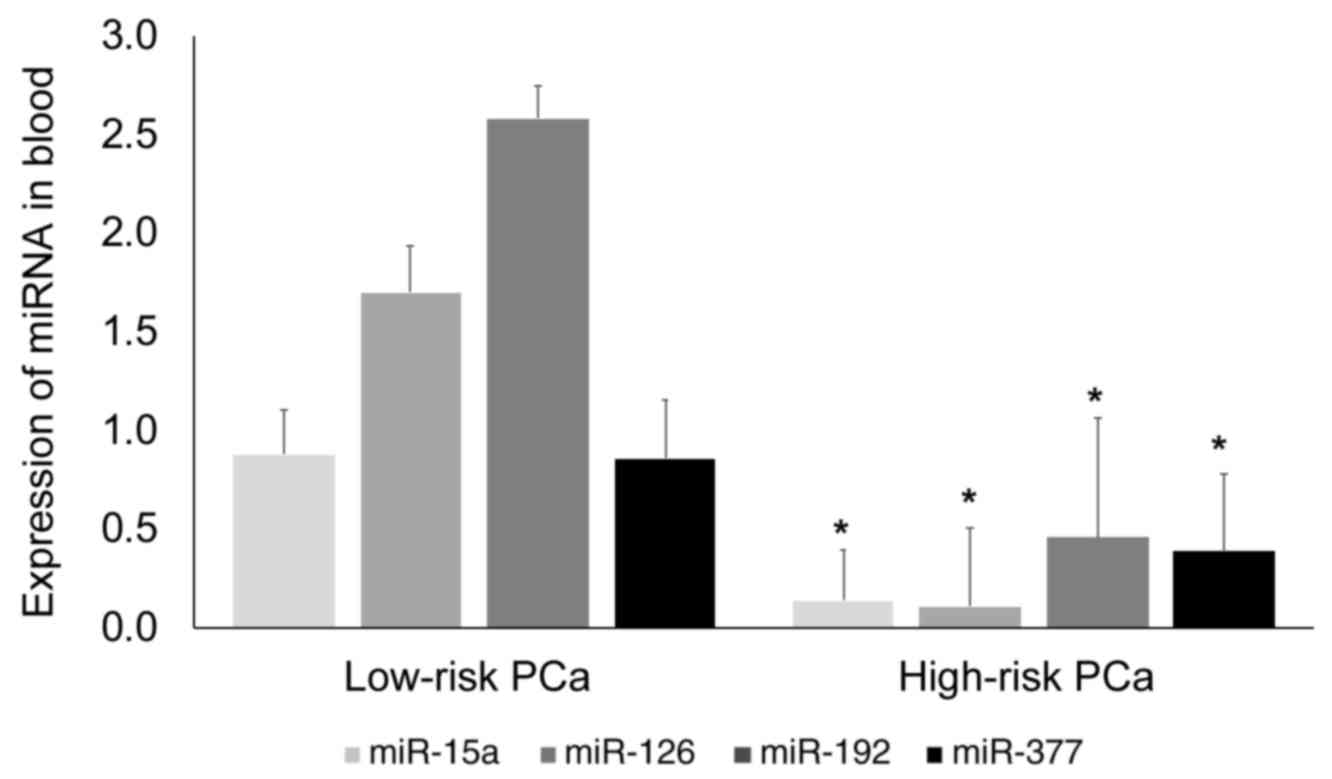
Expression of blood miRNAs in low-risk
and high-risk patients with localized PCa
A subgroup analysis was performed to determine
whether the expression of miR-15a, miR-126, miR-192 and miR-377
differed between patients with localized PCa categorized as
low-risk (T1c or T2a, PSA ≤10 ng/ml and GS ≤7) or high-risk (T2c or
greater, PSA >20 ng/ml and GS ≥8) using the D'Amico
classification. As illustrated in Fig.
2, expression levels of the 4 miRNAs were significantly
(P<0.05) lower in patients with high-risk localized PCa than in
patients with low-risk localized PCa.
miR-15a expression was 6.3-fold lower in high-risk
PCa (0.11±0.24) than in low-risk PCa (0.88±0.34). miR-126
expression was 15.5-fold lower in high-risk PCa (0.11±0.17) than in
low-risk PCa (1.7±0.45). miR-192 expression was 5.6-fold lower in
high-risk PCa (0.46±0.17) than in low-risk PCa (2.58±0.5; P=0.01).
miR-377 expression was lower by 2.2-fold in high-risk PCa
(0.39±0.3) than in low-risk PCa (0.86±0.4). Amongst the 4 miRNAs
analyzed, miR-126 was the most downregulated.
Multivariate regression analysis of
association
Associations between the expression of blood
miR-15a, miR-126, miR-192 and miR-377 and the presence of localized
PCa were further analyzed. Analysis using multivariate logistic
regression models based on single miRNAs confirmed a significant,
independent association of each miRNA with PCa. When the group of
healthy control subjects was used as the reference category,
(Table II), crude and age-adjusted
regression models demonstrated that the 4 miRNAs were significantly
and independently associated with PCa (P<0.05), whereas none of
the miRNAs were significantly associated with BPH (P>0.05). In
addition, when the group of patients with BPH was used as the
reference category, the association of the four miRNAs with the
presence of PCa was statistically significant (P<0.05) prior to
and following adjustment for age and PSA (Table III).
 | Table II.Multivariate logistic regression
analysis of miRNAs for discriminating PCa in the healthy control
group reference category (Model 1). |
Table II.
Multivariate logistic regression
analysis of miRNAs for discriminating PCa in the healthy control
group reference category (Model 1).
|
| PCa | BPH |
|---|
|
|
|
|
|---|
| miRNA | OR | 95% CI | P-value | OR | 95% CI | P-value |
|---|
| miR-15a |
|
|
|
|
|
|
| 1a | 0.39 | 0.21–0.72 | 0.003 | 1.01 | 0.84–1.23 | 0.89 |
| 1b | 0.34 | 0.21–0.74 | 0.004 | 1.02 | 0.84–1.24 | 0.86 |
| miR-126 |
|
|
|
|
|
|
| 1a | 0.67 | 0.53–0.85 | 0.001 | 0.97 | 0.91–1.04 | 0.44 |
| 1b | 0.69 | 0.55–0.87 | 0.002 | 0.97 | 0.90–1.03 | 0.45 |
| miR-192 |
|
|
|
|
|
|
| 1a | 0.77 | 0.64–0.93 | 0.005 | 0.71 | 0.95–1.01 | 0.71 |
| 1b | 0.76 | 0.64–0.92 | 0.004 | 0.99 | 0.95–1.11 | 0.69 |
| miR-377 |
|
|
|
|
|
|
| 1a | 0.51 | 0.31–0.85 | 0.009 | 0.98 | 0.87–1.11 | 0.76 |
| 1b | 0.50 | 0.30–0.84 | 0.009 | 0.97 | 0.83–1.20 | 0.77 |
 | Table III.Multivariate logistic regression
analysis of miRNAs for the presence of prostate cancer in the
benign prostatic hyperplasia group reference category (Model
2). |
Table III.
Multivariate logistic regression
analysis of miRNAs for the presence of prostate cancer in the
benign prostatic hyperplasia group reference category (Model
2).
| miRNA | Odds ratio | 95% confidence
interval | P-value |
|---|
| miR-15a |
|
|
|
| 2a | 0.45 | 0.25–0.80 | 0.013 |
| 2b | 0.35 | 0.13–0.95 | 0.024 |
| miR-126 |
|
|
|
| 2a | 0.60 | 0.46–0.78 | 0.001 |
| 2b | 0.54 | 0.35–0.83 | 0.046 |
| miR-192 |
|
|
|
| 2a | 0.63 | 0.49–0.82 | 0.001 |
| 2b | 0.66 | 0.49–0.89 | 0.030 |
| miR-377 |
|
|
|
| 2a | 0.46 | 0.27–0.81 | 0.008 |
| 2b | 0.36 | 0.13–0.97 | 0.040 |
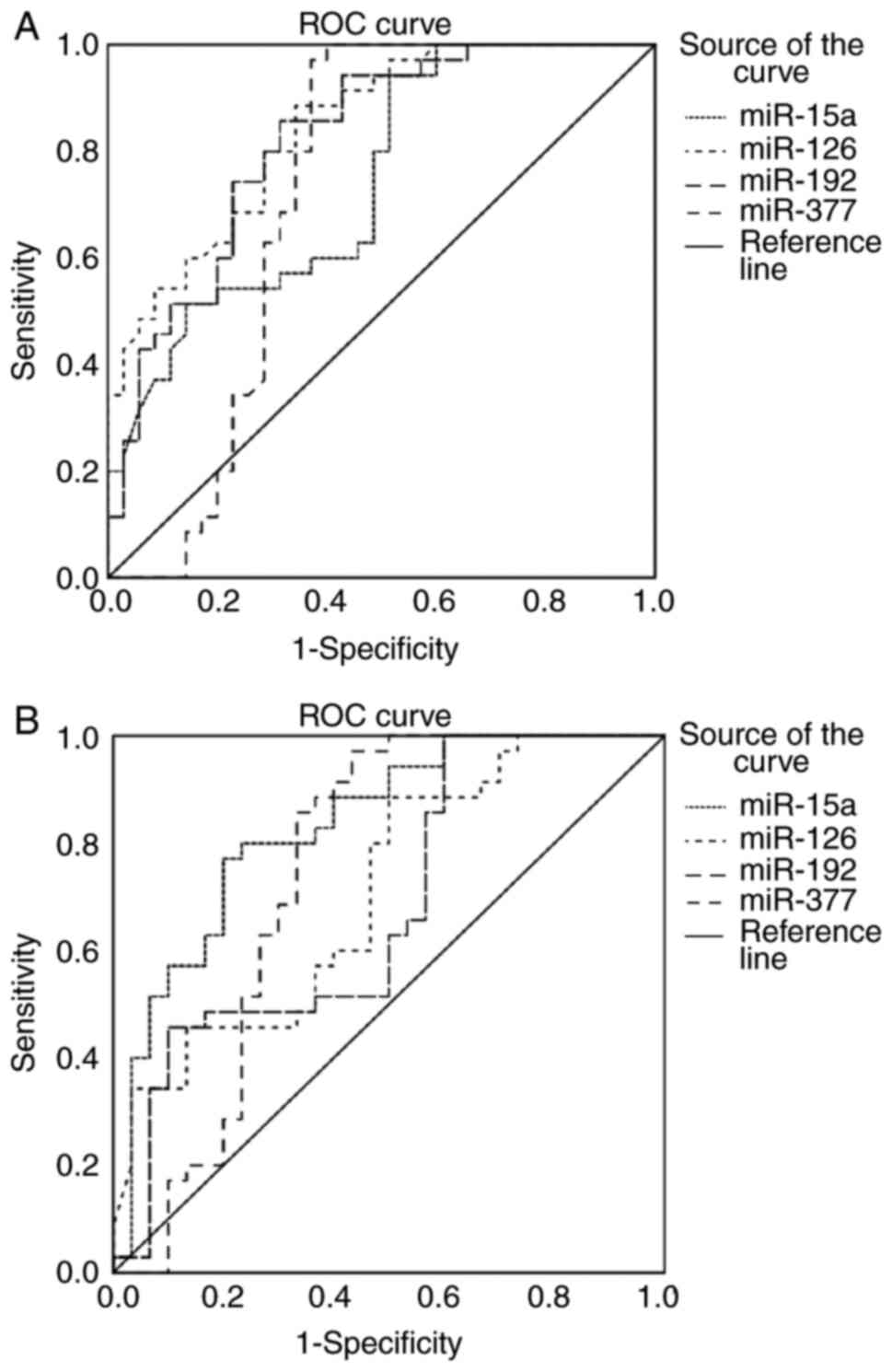
Discriminative ability of miRNAs for
localized PCa
To determine the discriminative ability of blood
miR-15a, miR-126, miR-192 and miR-377 for localized PCa, ROC curve
analysis was performed and the AUC was reported for each miRNA.
The ROC analysis was first applied for the four
miRNAs in patients with PCa vs. patients with BPH. The results
demonstrated good diagnostic capabilities in discriminating between
the groups, with an AUC of 0.744 (P<0.001), 0.844 (P<0.001),
0.82 (P<0.001) and 0.73 (P=0.001) for miR-15a, miR-126, miR-192
and miR-377, respectively (Fig. 3A
and Table IV).
 | Table IV.Discriminative ability of miRNAs for
localized PCa. |
Table IV.
Discriminative ability of miRNAs for
localized PCa.
|
| PCa vs. benign
prostatic hyperplasia | PCa vs.
healthy |
|---|
|
|
|
|
|---|
| miRNA | AUC | 95% CI | P-value | AUC | 95% CI | P-value |
|---|
| miR-15a | 0.744 | 0.63–0.86 | <0.001 | 0.79 | 0.67–0.90 | <0.001 |
| miR-126 | 0.844 | 0.76–0.93 | <0.001 | 0.70 | 0.56–0.82 | 0.009 |
| miR-192 | 0.82 | 0.72–0.92 | <0.001 | 0.66 | 0.52–0.78 | 0.024 |
| miR-377 | 0.73 | 0.61–0.86 | 0.001 | 0.75 | 0.63–0.87 | <0.001 |
Likewise, when the ROC analysis was applied for the
4 miRNAs in patients with PCa vs. healthy subjects (Fig. 3B and Table
IV), the results revealed significant diagnostic abilities in
differentiating between the groups, with an AUC of 0.79
(P<0.001), 0.70 (P=0.009), and 0.75 (P<0.001) for miR-15a,
miR-126 and miR-377, respectively, and a more moderate but
significant ability for miR-192 (AUC, 0.66; P=0.024).
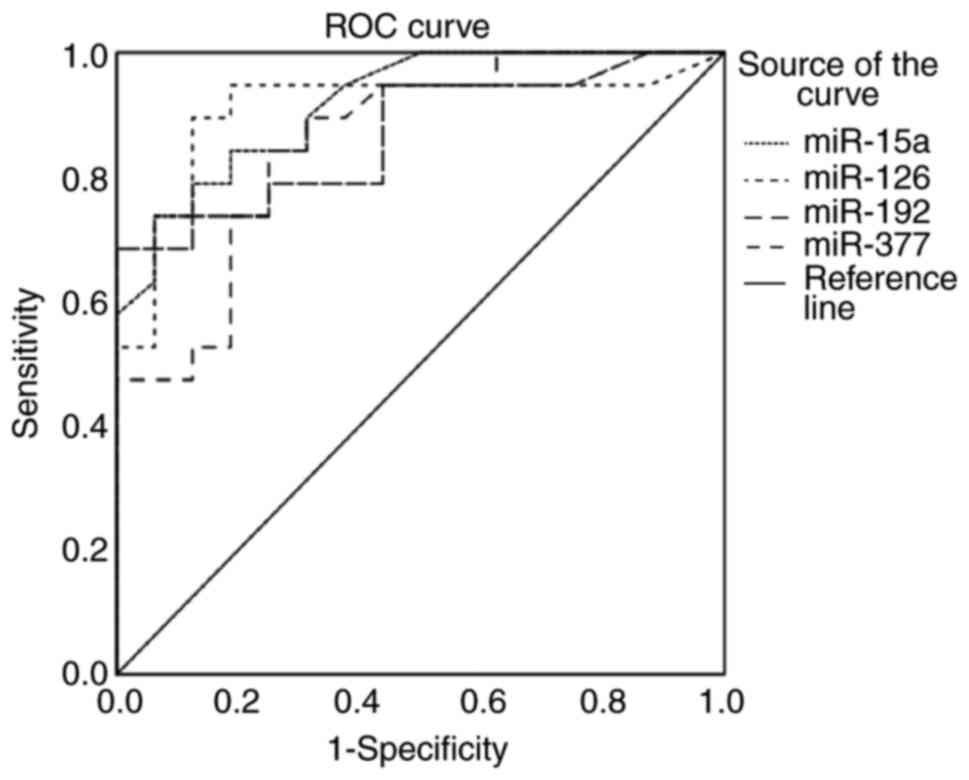
Discriminative ability of miRNAs for
PCa risk stratification
Next, the abilities of blood miR-15a, miR-126,
miR-192 and miR-377 to accurately risk-stratify patients with
localized PCa were evaluated. The ROC analysis was applied to
patients with localized PCa categorized as low-risk or high-risk
using the D'Amico classification.
As demonstrated in Fig.
4 and Table V, the 4 miRNAs
displayed high diagnostic capabilities for differentiating between
low- and high-risk PCa. The AUCs were 0.92 (P<0.001), 0.91
(P<0.001), 0.87, P<0.001) and 0.86 (P<0.001) for miR-15a,
miR-126, miR-192 and miR-377, respectively. Notably, the highest
predictive values were obtained for miR-15a and miR-126.
 | Table V.Discriminative ability of miRNAs for
localized PCa-risk stratification. |
Table V.
Discriminative ability of miRNAs for
localized PCa-risk stratification.
|
| Low-risk vs.
high-risk PCa |
|---|
|
|
|
|---|
| miRNA | Area under the
curve | 95% confidence
interval | P-value |
|---|
| miR-15a | 0.92 | 0.83–1.00 | <0.001 |
| miR-126 | 0.91 | 0.8–1.00 | <0.001 |
| miR-192 | 0.87 | 0.75–0.99 | <0.001 |
| miR-377 | 0.86 | 0.73–0.98 | <0.001 |
Discussion
The introduction of PSA testing into clinical
practice has aided the early detection and screening of PCa
(2). However, despite the high
sensitivity of PSA, the test is not specific for PCa (5,7,13,14), as
PSA levels may also increase in non-malignant conditions of the
prostate, including BPH; this has led to over-diagnosis and
over-treatment (5,6). Furthermore, PSA levels are poorly
correlated with tumor aggressiveness and poorer disease outcomes
(15). The limitations in the
diagnostic accuracy of PSA raise an urgent requirement for
alternative rapid and accurate markers for the detection of PCa,
and the identification of high-risk patients at an early stage. The
aberrant expression of miRNAs in various physiopathological
conditions has led to the intense investigation of their potential
role as a novel class of markers (17). As they are highly stable in human
biofluids (33,34), miRNAs exhibit promise as non-invasive
biomarkers for a variety of diseases (31,32,39).
Circulating miRNAs secreted by tumor cells into the blood may
reflect the tumor of origin (41) and
the extent of tumor progression, including in PCa (42,43), thus
representing promising diagnostic or prognostic disease biomarkers
(29–31,36).
Among the growing number of miRNAs that have been
identified as serving a key role in cancer, the expression levels
of miR-15a, miR-126, miR-192 and miR-377 have been frequently
identified as downregulated in a wide range of cancer types
(19–24). Particularly in PCa, these miRNAs have
been described as tumor suppressors that target multiple oncogenes
(25–28). Since the majority of studies have
focused on the potential value of miRNAs as biomarkers for advanced
and metastatic PCa (36,37), the present study focused on the
utility of circulating miRNAs as biomarkers to distinguish
localized PCa from BPH, and also to discriminate between patients
with low- and high-risk localized PCa. Therefore, the expression
levels of peripheral blood miR-15a, miR-126, miR-192 and miR-377,
selected based on their critical functions in PCa (25–28), were
quantified, and their diagnostic value investigated; they may be
novel biomarkers for the early detection of localized PCa, and for
PCa risk stratification.
Significantly lower expression of miR-15a, miR-126,
miR-192 and miR-377 was identified in patients with PCa, compared
with in patients with BPH or healthy subjects (Fig. 1). Subgroup analysis of patients with
localized PCa categorized as high- or low-risk using the D'Amico
classification (38) revealed that
the four miRNAs investigated were significantly lower in patients
with high-risk PCa than in patients with low-risk PCa (Fig. 2). Among the miRNAs, miR-126 was the
most downregulated, with a 15.5-fold decrease in patients with
high-risk PCa vs. in patients with low-risk PCa (Fig. 2). These results may suggest that
miR-15a, miR-126, miR-192 and miR-377 participate in the
pathogenicity and development of PCa. Furthermore, analysis using
multivariate logistic regression models based on single miRNAs
confirmed a significant, independent association between each of
the 4 miRNAs and PCa, with or without adjustments for age, whereas
none of the miRNAs were significantly associated with BPH (Table II). When these models were applied in
multivariate logistic regression analysis using patients with BPH
as the reference category, the 4 miRNAs were significantly
associated with PCa even after adjustment for age and PSA (Table III). These data indicated that the
reduced expression of circulating miR-15a, miR-126, miR-192 and
miR-377 in the blood may represent predictive biomarkers for the
presence of PCa. To the best of our knowledge, this is the first
study to identify the downregulation of blood miR-15a, miR-126,
miR-192 and miR-377 in patients with localized PCa, compared with
in patients with BPH and healthy individuals, and to demonstrate
that these four miRNAs are predictive for PCa.
miR-15a is located near miR-16-1 at chromosome
13q14, a region that is frequently altered in cancer (19). miR-15a serves an important role in
cell differentiation, proliferation, apoptosis and angiogenesis
(19). In PCa, miR-15a was reported
to target and suppress the expression of the cyclin D1 and WNT3A
oncogenes (25).
miR-126 (also referred to as miR-126-3p) and its
complement miR-126* (miR-126-5p) are derived from the epidermal
growth factor-like domain 7 gene on chromosome 9 located within
intron 7 (44,45). It is highly expressed in vascular
endothelial cells, and represses negative regulators of vascular
endothelial growth factor signaling pathways, thus promoting
pro-angiogenic processes and enhancing blood vessel formation
(44,45). The function of miR-126 as a tumor
suppressor was proposed based on its ability to inhibit cancer cell
growth, adhesion, migration and invasion (20,46–49). Song
et al (26) identified that
miR-126 could inhibit cancer proliferation and metastasis by
targeting PIK3R2 in PCa. However, miR-126 may also act as an
oncogene by promoting blood vessel formation and supporting cancer
progression (50). These observations
suggested that the behavior of miR-126 in cancer biology is
complex, indicating that the role of miR-126 in different tissue
types may vary, as miR-126 mediates different signaling pathways in
different tissues (51).
miR-192, another cancer regulator, was demonstrated
to impair cell tumorigenicity in PCa by targeting and repressing
the oncogene NOB1 (27).
miR-377 could also affect the malignant cell
behavior of PCa cell lines by targeting FZD4, an important gene in
the epithelial-to-mesenchymal transition (28). The downregulation of miR-377 was also
reported to be associated with a higher PSA and Gleason score, and
lymph node invasion, in specimens of primary and metastatic
prostate tumors (28).
In the present study, the downregulation of blood
miR-15a, miR-126, miR-192 and miR-377 in patients with localized
PCa compared with in patients with BPH and healthy controls
supports the hypothesis that these miRNAs may function as tumor
suppressers and have an important role in disease pathogenicity.
Additionally, the low blood expression of these four miRNAs in
patients with high-risk localized PCa compared patients with
low-risk localized PCa may suggest that they serve a role in tumor
aggressiveness.
The results of the present study indicated that
blood miR-15a, miR-126, miR-192 and miR-377 levels could be used to
discriminate PCa from BPH with high diagnostic accuracy, as
determined by ROC analysis (Fig. 3A
and Table IV). In the
differentiation between patients with PCa and healthy subjects,
miR-15a, miR-126 and miR-377 displayed relatively high diagnostic
values, while miR-192 exhibited a more moderate, though
significant, ability (Fig. 3B and
Table IV). Of note, ROC analysis
demonstrated the significant diagnostic power of miR-15a, miR-126,
miR-192 and miR-377 to discriminate patients with low-risk
localized PCa from patients with high-risk localized PCa, and
miR-15a and miR-126 were the best predictors of high-risk disease
(Fig. 4 and Table V). A previous study by Watahiki et
al (37) demonstrated the
decreased expression of miR-126, and other miRNAs, including
miR151-3p, miR423-3p, miR152 and miR-21, in the plasma of patients
with localized PCa. However, in their study, only the combination
of these miRNAs differentiated the two groups of patients rather
than any individual miRNA (37).
Peripheral blood is easily accessible and its
extraction is non-invasive. The development of miRNA biomarkers
that allow the differentiation between patients with malignant and
benign prostate tumors could aid in avoiding invasive biopsies in
Patients with BPH (15).
Additionally, miRNA biomarkers for indolent localized PCa, which
may remain unrecognized for a number of years, could allow early
diagnosis when the cancer is treatable and had not progressed to a
more clinically significant disease (15).
The present study clearly demonstrated that
expression of miR-15a, miR-126, miR-192 and miR-377 could be
developed as blood-based non-invasive biomarkers to distinguish PCa
from BPH, and to identify patients with low-risk and high-risk
localized PCa. The small sample size in the present study may limit
the statistical power of the results, and further validation
studies for the clinical implementation of the results are
warranted with a larger sample size. Furthermore, the present study
focused solely on the potential of miR-15a, miR-126, miR-192 and
miR-377 as biomarkers for the early detection of localized PCa or
PCa risk stratification. Additional studies are underway in our
laboratory to investigate the clinical significance of other
PCa-associated miRNA biomarkers.
Taken together, the results of the present study
revealed that the expression of circulating miR-15a, miR-126,
miR-192 and miR-377 in blood was significantly lower in patients
with localized PCa than in patients with BPH and healthy subjects,
lower in patients with high-risk localized PCa than in patients
with low-risk localized PCa, and independently associated with the
occurrence of localized PCa. These results demonstrated that the
expression of miR-15a, miR-126, miR-192 and miR-377 holds promise
as early blood-based biomarkers for determining the presence of
localized PCa, and for PCa risk assessment.
Acknowledgements
Not applicable.
Funding
The present study was supported by a research grant
from the College of Medicine and Medical Sciences, Arabian Gulf
University (grant no. 93).
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author, on reasonable
request.
Authors' contributions
GAK was responsible for project development, data
management, data analysis, manuscript writing and manuscript
editing. HMS was responsible for project development, data
management and manuscript writing. MAA was responsible for data
collection and data analysis. ZTAN was responsible for data
collection and manuscript writing. All authors have read and
approved the manuscript.
Ethical approval and consent to
participate
Ethical approval to conduct the present study was
obtained from the Medical Research and Ethics Committee of the
College of Medicine and Medical Sciences, Arabian Gulf University.
All participants provided written informed consent for the use of
their blood samples and data.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
ROC
|
receiver operating characteristic
|
|
AUC
|
area under the curve
|
References
|
1
|
Greenlee RT, Hill-Harmon MB, Murray T and
Thun M: Cancer statistics, 2001. CA Cancer J Clin. 51:15–36. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hilal L, Shahit M, Mukherji D,
Charafeddine M, Farhat Z, Temaraz S, Khauli R and Shamseddine A:
Prostate cancer in the Arab world: A view from the inside. Clin
Genitourin Cancer. 13:505–511. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Popiolek M, Rider JR, Andrén O, Andersson
SO, Holmberg L, Adami HO and Johansson JE: Natural history of
early, localized prostate cancer: A final report from three decades
of follow-up. Eur Urol. 63:428–435. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hoffman RM, Gilliland FD, Adams-Cameron M,
Hunt WC and Key CR: Prostate-specific antigen testing accuracy in
community practice. BMC Fam Pract. 3:192002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nadler RB, Humphrey PA, Smith DS, Catalona
WJ and Ratliff TL: Effect of inflammation and benign prostatic
hyperplasia on elevated serum prostate specific antigen levels. J
Urol. 154:407–413. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Adhyam M and Gupta AK: A review on the
clinical utility of PSA in cancer prostate. Indian J Surg Oncol.
3:120–129. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Boddy JL, Pike DJ and Malone PR: A
seven-year follow up of men following a benign prostate biopsy. Eur
Urol. 44:17–20. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hugosson J, Carlsson S, Aus G, Bergdahl S,
Khatami A, Lodding P, Pihl CG, Stranne J, Holmberg E and Liljia H:
Mortality results from the Göteborg randomised population-based
prostate-cancer screening trial. Lancet Oncol. 11:725–732. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schröder FH, Hugosson J, Roobol MJ,
Tammela TL, Ciatto S, Nelen V, Kwiatkowski M, Lujan M, Lilja H,
Zappa M, et al: Screening and prostate-cancer mortality in a
randomized european study. N Engl J Med. 360:1320–1328. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Andriole GL, Crawford ED, Grubb RL III,
Buys SS, Chia D, Church TR, Fouad MN, Gelmann EP, Kuale PA, Reding
DJ, et al: Mortality results from a randomized prostate-cancer
screening trial. N Engl J Med. 360:1310–1319. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Stamey TA, McNeal JE, Yemoto CM, Sigal BM
and Johnstone IM: Biological determinants of cancer progression in
men with prostate cancer. JAMA. 281:1395–1400. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Oesterling JE: Prostate specific antigen:
A critical assessment of the most useful tumor marker for
adenocarcinoma of the prostate. J Urol. 145:907–923. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Thompson IM, Pauler DK, Goodman PJ, Tangen
CM, Lucia MS, Parnes HL, Minasian LM, Ford LG, Lippman SM, Crawford
ED, et al: Prevalence of prostate cancer among men with a
prostate-specific antigen level <or =4.0 ng per milliliter. N
Engl J Med. 350:2239–2246. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Velonas VM, Woo HH, Remedios CG and
Assinder SJ: Current status of biomarkers for prostate cancer. Int
J Mol Sci. 14:11034–11060. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kgatle MM, Kalla AA, Islam MM, Sathekge M
and Moorad R: Prostate cancer: Epigenetic alterations, risk
factors, and therapy. Prostate Cancer. 2016:56538622016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li F, Xu Y, Deng S, Li Z, Zou D, Yi S, Sui
W, Hao M and Qiu L: MicroRNA-15a/16-1 cluster located at chromosome
13q14 is down-regulated but displays different expression pattern
and prognostic significance in multiple myeloma. Oncotarget.
6:38270–38282. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ebrahimi F, Gopalan V, Smith RA and Lam
AK: miR-126 in human cancers: Clinical roles and current
perspectives. Exp Mol Pathol. 96:98–107. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yunxia Z and Hongying D: Low expression of
miR-192 in NSCLC and its tumor suppressor functions in metastasis
via targeting ZEB2. Open Life Sci. 11:293–297. 2016.
|
|
22
|
Botla SK, Savant S, Jandaghi P, Bauer AS,
Mücke O, Moskalev EA, Neoptolemos JP, Costello E, Greenhalf W,
Scarpa A, et al: Early epigenetic downregulation of microRNA-192
expression promotes pancreatic cancer progression. Cancer Res.
76:4149–4159. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang R, Ma Y, Yu D, Zhao J and Ma P:
miR-377 functions as a tumor suppressor in human clear cell renal
cell carcinoma by targeting ETS1. Biomed Pharmacother. 70:64–71.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang L, Shao J, Zhang X, Xu M and Zhao J:
microRNA-377 suppresses the proliferation of human osteosarcoma
MG-63 cells by targeting CDK6. Tumour Biol. 36:3911–3917. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bonci D, Coppola V, Musumeci M, Addario A,
Giuffrida R, Memeo L, D'Urso L, Pagliuca A, Biffoni M, Labbaye C,
et al: The miR-15a-miR-16-1 cluster controls prostate cancer by
targeting multiple oncogenic activities. Nat Med. 14:1271–1277.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Song L, Xie X, Yu S, Peng F and Peng L:
MicroRNA-126 inhibits proliferation and metastasis by targeting
pik3r2 in prostate cancer. Mol Med Rep. 13:1204–1210. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sun J, Fan Z, Lu S, Yang J, Hao T and Huo
Q: miR-192 suppresses the tumorigenicity of prostate cancer cells
by targeting and inhibiting nin one binding protein. Int J Mol Med.
37:485–492. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Formosa A, Markert EK, Lena AM, Italiano
D, Finazzi-Agro' E, Levine AJ, Bernardini S, Garabadgiu AV, Melino
G and Candi E: MicroRNAs, miR-154, miR-299-5p, miR-376a, miR-376c,
miR-377, miR-381, miR-487b, miR-485-3p, miR-495 and miR-654-3p,
mapped to the 14q32.31 locus, regulate proliferation, apoptosis,
migration and invasion in metastatic prostate cancer cells.
Oncogene. 33:5173–5182. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ferracin M, Veronese A and Negrini M:
Micromarkers: miRNAs in cancer diagnosis and prognosis. Expert Rev
Mol Diagn. 10:297–308. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Al-Kafaji G, Al Naieb ZT and Bakhiet M:
Increased oncogenic microRNA-18a expression in the peripheral blood
of patients with prostate cancer: A potential novel non-invasive
biomarker. Oncol Lett. 11:1201–1206. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Al-Kafaji G, Al-Mahroos G, Abdulla
Al-Muhtaresh H, Sabry MA, Abdul Razzak R and Salem AH: Circulating
endothelium-enriched microRNA-126 as a potential biomarker for
coronary artery disease in type 2 diabetes mellitus patients.
Biomarkers. 22:268–278. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Keller A, Leidinger P, Bauer A, Elsharawy
A, Hass J, Backes C, Wendschlag A, Giese N, Tjaden C, Ott K, et al:
Toward the blood-borne miRNome of human diseases. Nat Methods.
8:841–843. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mitchell PS, Parkin RK, Kroh EM, Frit BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Weber JA, Baxter DH, Zhang S, Huang KH,
Lee MJ, Galas DJ and Wang K: The microRNA spectrum in 12 body
fluids. Clin Chem. 56:1733–1741. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Turchinovich A, Samato TR, Tonevitsky AG
and Burwinkel B: Circulating miRNAs: Cell-cell communication
function? Front Genet. 4:1192013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sita-Lumsden A, Dart DA, Waxman J and
Bevan CL: Circulating microRNAs as potential new biomarkers for
prostate cancer. Br J Cancer. 108:1925–1930. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Watahiki A, Macfarlane RJ, Gleave ME, Crea
F, Wang Y, Helgason CD and Chi KN: Plasma miRNAs as biomarkers to
identify patients with castration-resistant metastatic prostate
cancer. Int J Mol Sci. 14:7757–7770. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
D'Amico AV, Whittington R, Malkowicz SB,
Schultz D, Blank K, Broderick GA, Tomaszewski JE, Renshaw AA,
Kaplan I, Beard CJ and Wein A: Biochemical outcome after radical
prostatectomy, external beam radiation therapy, or interstitial
radiation therapy for clinically localized prostate cancer. JAMA.
280:969–974. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Al-Kafaji G, Al-Mahroos G, Alsayed NA,
Hasan ZA, Nawaz S and Bakhiet M: Peripheral blood microRNA-15a as a
potential biomarker for type 2 diabetes mellitus and pre-diabetes.
Mol Med Rep. 12:7485–7490. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Coppola V, de Maria R and Bonci D:
MicroRNAs and prostate cancer. Endocr Relat Cancer. 17:F1–F17.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fang YX and Gao WQ: Roles of microRNAs
during prostatic tumorigenesis and tumor progression. Oncogene.
33:135–147. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang S, Aurora AB, Johnson BA, Qi X,
McAnally J, Hill JA, Richardson JA, Bassel-Duby R and Olson EN: The
endothelial-specific microRNA miR-126 governs vascular integrity
and angiogenesis. Dev Cell. 15:261–271. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Fish JE, Santoro MM, Morton SU, Yu S, Yeh
RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY and Srivastava D:
miR-126 regulates angiogenic signaling and vascular integrity. Dev
Cell. 15:272–284. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Miko E, Margitai Z, Czimmerer Z, Varkonyi
I, Dezso B, Lányi A, Bacsó Z and Scholtz B: miR-126 inhibits
proliferation of small cell lung cancer cells by targeting SLC7A5.
FEBS Lett. 585:1191–1196. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang J, Du YY, Lin YF, Chen YT, Yang L,
Wang HJ and Ma D: The cell growth suppressor, mir-126, targets
IRS-1. Biochem Biophys Res Commun. 377:136–140. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yang X, Wu H and Ling T: Suppressive
effect of microRNA-126 on oral squamous cell carcinoma in vitro.
Mol Med Rep. 10:125–130. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Li XM, Wang AM, Zhang J and Yi H:
Down-regulation of miR-126 expression in colorectal cancer and its
clinical significance. Med Oncol. 28:1054–1057. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Sasahira T, Kurihara M, Bhawal UK, Ueda N,
Shimomoto T, Yamamoto K, Kirita T and Kuniyasu H: Downregulation of
miR-126 induces angiogenesis and lymphangiogenesis by activation of
VEGF-A in oral cancer. Br J Cancer. 107:700–706. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Meister J and Schmidt MH: miR-126 and
miR-126*New players in cancer. ScientificWorldJournal.
10:2090–2100. 2010. View Article : Google Scholar : PubMed/NCBI
|