Introduction
According to the cancer statistics for 2016, lung
cancer was the leading cause of cancer-associated mortality in
China (1). In addition, it is
difficult to diagnose patients with lung cancer at a very early
stage, and tumor biomarkers for early diagnosis and metastasis
identification are lacking (2).
Despite the improvement of treatments, the prognosis of non-small
cell lung cancer (NSCLC) is still poor and the 5-year survival rate
is only 11–15% (3,4).
Previous studies have confirmed the importance of
non-protein coding genes in carcinogenesis and metastasis (5,6). Among
those non-proteins coding RNAs, long noncoding RNAs (lncRNAs) have
participated in a great extent of cancer biological processes
(6–8).
lncRNA is a type of RNA without the ability of encoding protein and
>200 nucleotides in length (9,10).
According to the existing research results, the dysregulation of
lncRNA serves a critical role in human cancers (5,10,11). In lung cancer, a number of
cancer-associated lncRNAs have been demonstrated to be biomarkers
for metastasis or prognosis, including HOX transcript antisense RNA
(HOTAIR) (12,13), metastasis associated long antisense
transcript 1 (MALAT1) (14,15) and colon cancer-associated transcript 2
(16).
To identify functional lncRNAs, microarrays have
been frequently utilized to investigate lncRNA expression in tumor
tissues (17). In a previous study,
it was indicated that a lncRNA termed as actin filament associated
protein 1 antisense RNA 1 (AFAP1-AS1) was significantly
overexpressed in lung cancer, as demonstrated by microarrays
(17). Evidence has indicated that
AFAP1-AS1 is associated with tumor cell migration and invasion
(18,19). However, the prognostic role of
AFAP1-AS1 has not been fully explored in NSCLC (20). Therefore, the AFAP1-AS1 expression
level was analyzed by reverse transcription-quantitative polymerase
chain reaction (RT-qPCR) and in situ hybridization (ISH),
and the prognostic value of AFAPA-AS1 was investigated.
Materials and methods
Patients and tissue samples
The study was approved by the Ethics Boards of the
Cancer Institute of Jiangsu (Nanjing, China). The characteristics
of analyzed patients are presented in Tables I and II. Lung cancer tissues and adjacent normal
tissues were obtained from patients that received surgical
resection of lung cancer from January 2012 to December 2015 at the
Department of Thoracic Surgery, Cancer Institute of Jiangsu. A
tissue microarray (TMA) cohort of 74 patients (55 males and 19
females), with a median age of 61.1 years, and a PCR cohort of 52
patients with NSCLC (30 males and 22 females), with a median age of
59.0 years, were included in the present study. The TMA cohort data
was generated in 2015 and the PCR cohort data in 2016. The patient
samples were obtained from the Cancer Institute of Jiangsu. All
patients diagnosed with NSCLC who had never received any therapy
prior to surgery were collected. Clinical data including age, sex,
smoking history, stage, lymph node metastasis of these patients
were collected from all patients. In addition, informed written
consents were obtained from all patients included in the present
study.
 | Table I.Association between lncRNA AFAP1-AS1
expression levels to clinical, biological and histo-morphological
factors in the tissue microarray cohort. |
Table I.
Association between lncRNA AFAP1-AS1
expression levels to clinical, biological and histo-morphological
factors in the tissue microarray cohort.
|
|
| AFAP1-AS1 expression
level |
|
|---|
|
|
|
|
|
|---|
| Factor | Number of
patients | Low | High | P-value |
|---|
| Sex |
|
|
| 0.140 |
| Male | 55 | 11 | 44 |
|
|
Female | 19 | 7 | 12 |
|
| Age, years |
|
|
| 0.988 |
| ≤60 | 33 | 8 | 25 |
|
|
>60 | 41 | 10 | 31 |
|
| Smoking status |
|
|
| 0.136 |
| Yes | 44 | 8 | 36 |
|
| No | 30 | 10 | 20 |
|
| Histology |
|
|
| 0.981 |
| SCC | 17 | 4 | 13 |
|
| AC | 48 | 12 | 36 |
|
|
Others | 9 | 2 | 7 |
|
| Tissue
differentiation |
|
|
| 0.420 |
| Middle
and high | 27 | 8 | 19 |
|
|
Low | 47 | 10 | 37 |
|
| Stage |
|
|
| 0.423 |
| IA, IB,
IIA and IIB | 52 | 14 | 38 |
|
| IIIA,
IIIB and IV | 22 | 4 | 18 |
|
| Lymph node
metastasis |
|
|
| P.007a |
|
Present |
|
|
|
|
|
Absent |
|
|
|
|
 | Table II.Correlation of lncRNA AFAP1-AS1
expression levels to clinicopathological characteristic in the
polymerase chain reaction cohort. |
Table II.
Correlation of lncRNA AFAP1-AS1
expression levels to clinicopathological characteristic in the
polymerase chain reaction cohort.
| Characteristics of
all patients in this cohort | Number of
patients | AFAP1-AS1 level
(fold change) | P-value |
|---|
| Age, years |
|
|
|
|
≤60 | 27 | 11.28 |
|
|
>60 | 25 | 10.58 | 0.062 |
| Sex |
|
|
|
|
Male | 30 | 9.56 |
|
|
Female | 22 | 8.33 | 0.928 |
| Smoking |
|
|
|
| No | 38 | 9.93 |
|
|
Yes | 14 | 6.63 | 0.074 |
| Tumor size, cm |
|
|
|
|
1.0×1.0–3.0×4.0 | 35 | 10.44 |
|
|
3.0×4.0–8.0×5.0 | 17 | 6.16 | 0.071 |
| Histology |
|
|
|
|
SCC | 7 | 8.61 |
|
| AC | 45 | 9.11 | 0.771 |
| Tissue
differentiation |
|
|
|
| Middle
and high | 28 | 8.81 |
|
|
Low | 24 | 7.64 | 0.041a |
| Lymph node
metastasis |
|
|
|
|
Absent | 33 | 5.89 |
|
|
Present | 19 | 14.52 | 0.014a |
| Stage |
|
|
|
| IA, IB,
IIA and IIB | 36 | 11.1 |
|
| IIIA,
IIIB and IV | 16 | 4.4 | 0.064 |
RNA extraction and RT-qPCR
Total RNA was extracted from tissue samples with
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), following the manufacturer's protocol.
PrimerScript RT Master Mix (Takara Biotechnology Co., Ltd., Dalian,
China) was used to reverse transcribe RNA into a final volume of 20
µl. Then, RT-qPCR was executed using the SYBR® Select
Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.;
cat. no. 4472908) with 0.5 µl cDNA on the QuantStudio™ 6 flex
system (Applied Biosystems; Thermo Fisher Scientific, Inc.),
according to the manufacturer's instructions. GAPDH were used as
internal controls. The primers sequence are as follows: Forward,
5′-TCGCTCAATGGAGTGACGGCA-3′ and reverse,
5′-CGGCTGAGACCGCTGAGAACTT-3′ for AFAP1-AS1; forward,
5′-CCACATCGCTCAGACACCAT-3′ and reverse, 5′-ACCAGGCGCCCAATACG-3′ for
GAPDH. The RT-qPCR reaction was implemented with the following
conditions: 95°C for 10 min, 40 cycles of 95°C for 15 sec and 60°C
for 1 min. The expression fold changes were calculated by
2−ΔΔCq methods (21,22). Every
sample was performed in triplicate.
TMA and ISH analysis
TMAs included samples fixed with 10% formalin at
room temperature for 24–48 h and embedded in paraffin from 82 pairs
of NSCLC tissue and adjacent normal lung tissues, and were
constructed by Shanghai Biochip Co., Ltd (Shanghai, China). After
processing, unspotted slides from the TMA block were used for the
ISH with probes for AFAP1-AS1 (Exiqon A/S, Vedbaek, Denmark). The
TMA was placed in an oven at 60°C for 1 h then stored overnight at
4°C. Following that, slides were washed with xylene at 18°C and
rehydrated with 100% ethanol solutions at room temperature and then
incubated with Proteinase-K (Nanjing KeyGen Co., Ltd., Nanjing
China) for 7.5 min at 37°C. Additionally, 1,000 nmol/l AFAP1-AS1
probe (Shanghai Bogoo Biological Technology Co., Ltd. Shanghai
China) was used to hybridize slides in a SSC buffer (150 mM sodium
chloride and 15 mM trisodium citrate) for 20 min at 50°C.
Afterwards, the slides were washed with SSC buffers (150 mM sodium
chloride and 15 mM trisodium citrate). Following this, the slides
were stained with 3,3′-diaminobenzidine horseradish peroxidase
chromogenic liquid (Shanghai Bogoo Biological Technology Co., Ltd.)
at room temperature for 15 min. Following washing, slides were
ready for imaging. Visible colonies were counted using light
microscopy and a fluorescence microscope at magnification, ×40. The
software used for analysis was Aperio ImageScope v11.1.2.752.
(Leica Microsystems GmbH, Wetzlar, Germany). Following this, the
slides were scored comprehensively according to size and intensity
of the staining, as reported previously (23,24). Size:
<10%, 1 point; ≥10%-<30%, 2 points; ≥30%-<50%, 3 points;
≥50%-<75%, 4 points; ≥75%, 5 points. Intensity: Soft red, 1
point; sandy beige, 2 points; claybank, 3 points; brown, 4 points;
sepia, 5 points. The intensities were multiplied by the sizes and
the scores were calculated by taking the normal lung tissue's score
away from the NSCLC tissues' score. According to the final scores,
the ASAP1-AS1 expression level was divided into high (score ≥1) and
low (score <1). The evaluation was completed by two pathologists
blinded to the patient's outcome and clinical characteristics.
Statistical analysis
Data analysis was performed with SPSS 20.0 software
(IBM Corp., Armonk, NY, USA). Paired Student's t-test, one-way
analysis of variance (Bonferroni post-hoc test) and Spearman's rank
test were applied to analyze the association between AFAFP1-AS1
expression and clinical characteristics. The variables associated
with the prognostic values were tested with overall survival time
as the endpoint in the univariate and multivariate analysis, which
was conducted by Cox regression analysis. The hazard ratio (HR) and
its 95% confidence interval (CI) were derived from these results.
GraphPad (GraphPad Software, Inc., La Jolla, CA, USA) was used to
produce the Kaplan-Meier survival curve. Data are presented as the
mean ± standard error of the mean. P<0.05 was considered to
indicate a statistically significant difference.
Results
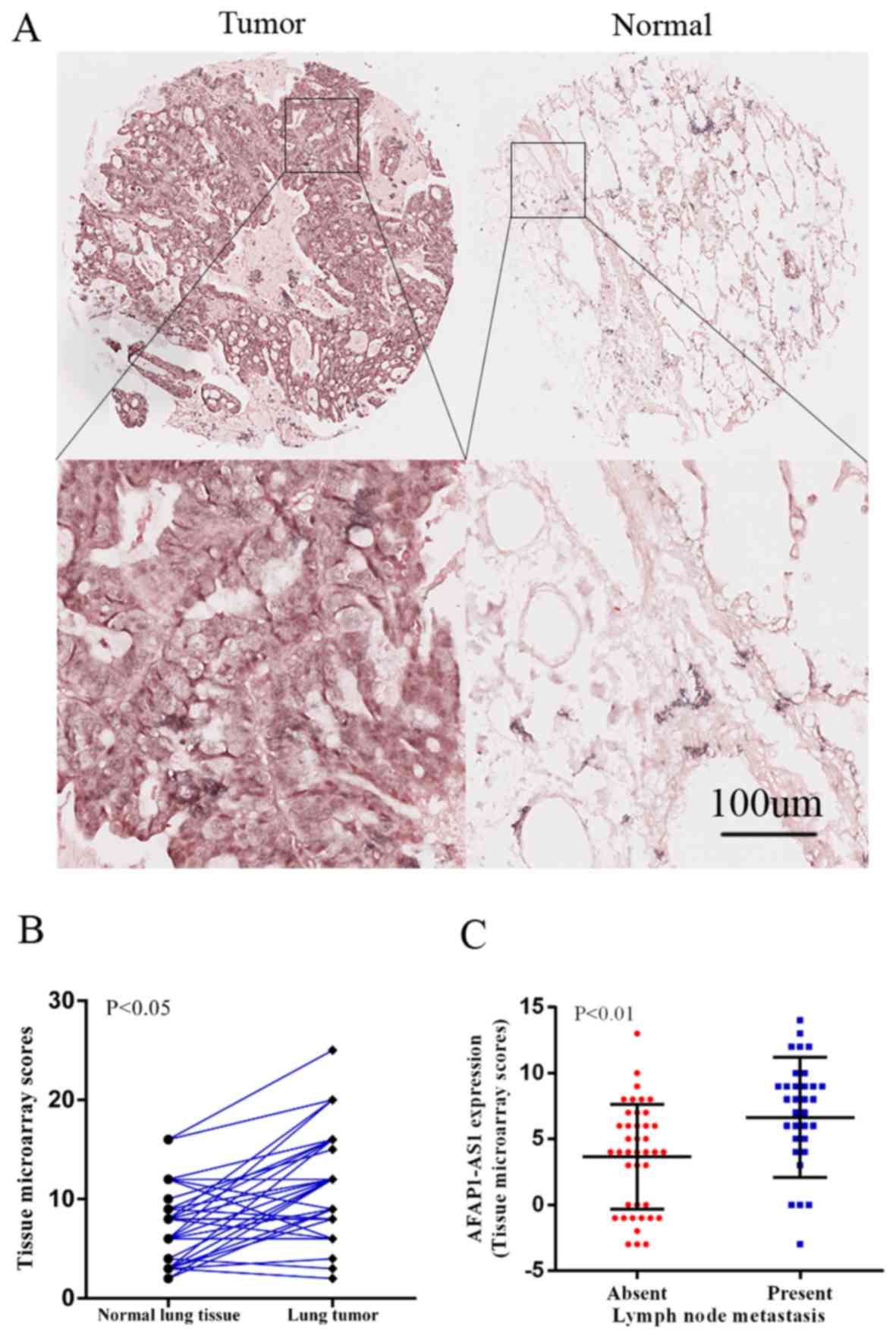
AFAP1-AS1 is overexpressed in NSCLC
tumor tissues and correlates with clinical characteristics
Firstly, the AFAP1-AS1 expression level was analyzed
via ISH in NSCLC tumor tissues. Subsequent to excluding 8 pairs for
missing data (4 tumor tissues and 4 normal tissues), the expression
of AFAP1-AS1 was compared between tumor tissues and normal tissues.
As indicated, AFAP1-AS1 was significantly overexpressed in 56 lung
tumor tissues, compared with paired adjacent normal lung tissues
(P<0.001; Fig. 1A and B). There
was a positive correlation between AFAP1-AS1 expression and lymph
node metastasis (P=0.007; Table I;
Fig. 1C). However, there were no
associations between AFAP1-AS1 expression and age, sex, smoking,
histology or stage.
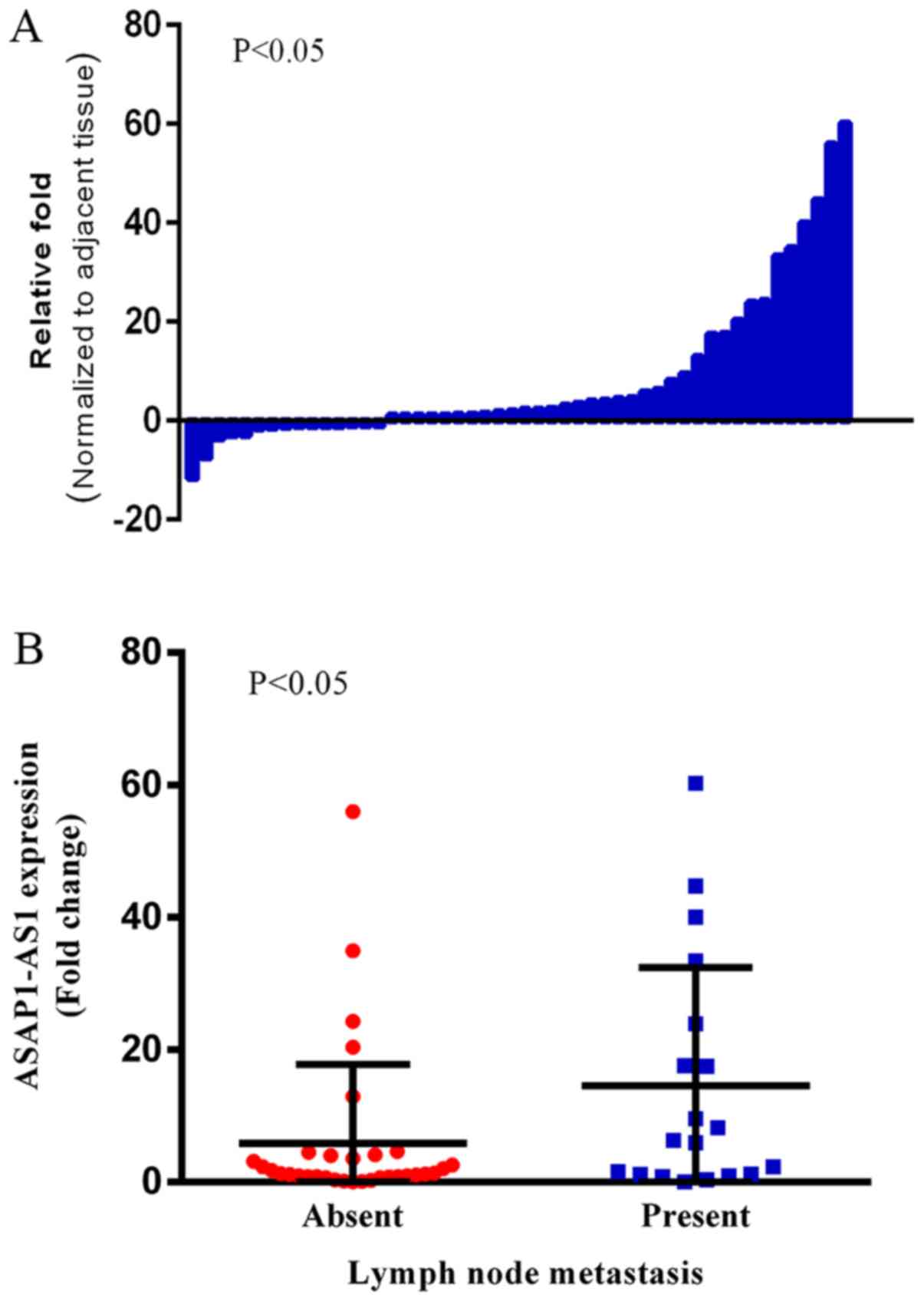
Following this, RT-qPCR was also performed in an
independent cohort of 52 patients with NSCLC. AFAP1-AS1 was
overexpressed in 77.0% (40/52) of patients with NSCLC, with mean
upregulation of 8.65-fold (P=0.040; Fig.
2A). Additionally, overexpression of AFAP1-AS1 was positively
correlated with tissue differentiation (P=0.041; Table II) and lymph node metastasis
(P=0.014; Table II; Fig. 2B).
Prognostic value of AFAP1-AS1 lncRNA
expression in patients with lung cancer
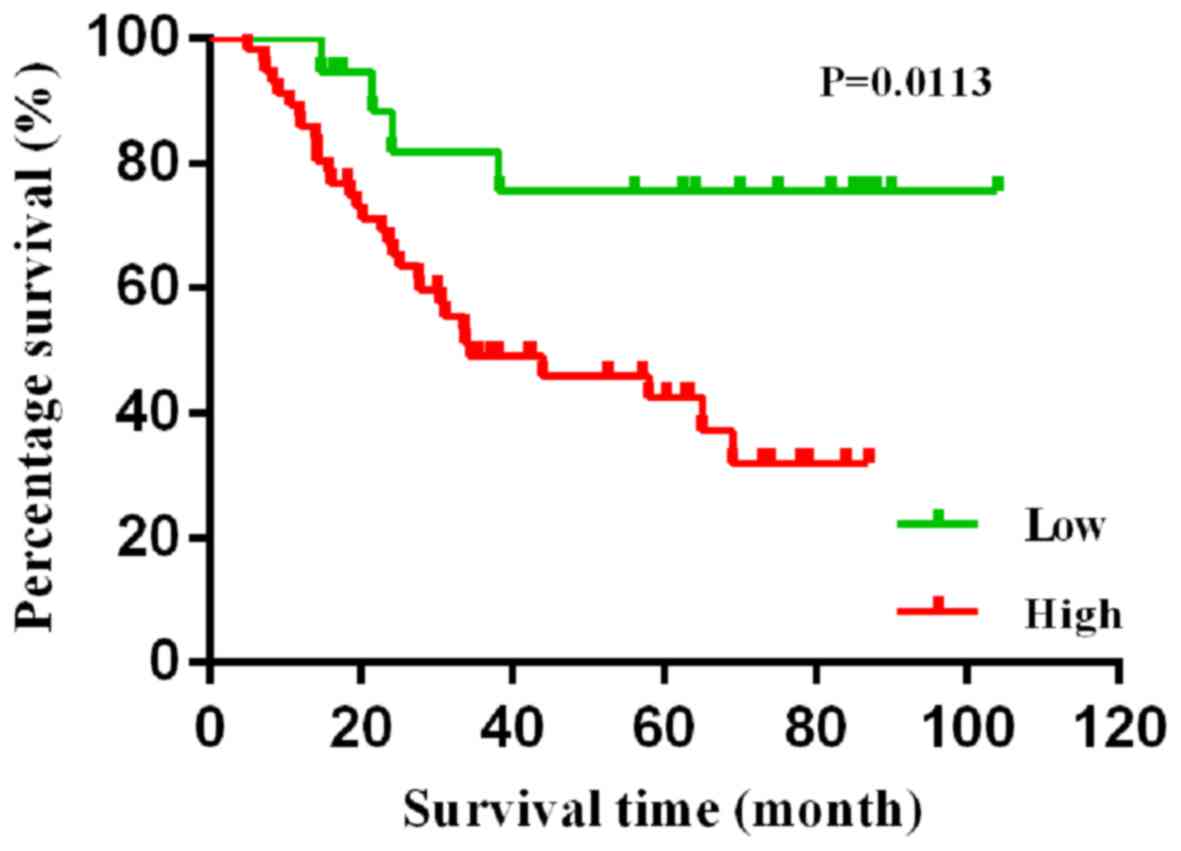
Kaplan-Meier survival analysis demonstrated that
patients with low expression level of AFAP1-AS1 had an improved
survival time (P=0.011; Fig. 3). To
additionally explore the association between lncRNA AFAP1-AS1 and
prognosis, Cox regression analysis was conducted. Kaplan-Meier
analysis showed that the high expression level of AFAP1-AS1 was
significantly associated with poor overall survival time (HR, 3.58;
95% CI, 1.25–10.24; P=0.0113; Table
III). Subsequently, AFAP1-AS1 was separately introduced to the
base multivariate model including age, sex, smoking, tissue
differentiation, stage and lymph node metastasis. High expression
level of ASAP1-S1 was an independent prognostic factor of poor
survival time for lung cancer (HR, 3.12; 95% CI, 1.05–9.25;
P=0.040; Table III).
 | Table III.AFAP1-AS1 expression levels in Cox
univariate and multivariate analysis for overall survival. |
Table III.
AFAP1-AS1 expression levels in Cox
univariate and multivariate analysis for overall survival.
|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|
|---|
| Factor | Number of
patients | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Sex |
|
|
|
|
|
|
|
|
Male | 55 | 1 |
|
| 1 |
|
|
|
Female | 19 | 0.61 | 0.26–1.40 | 0.238 | 0.82 | 0.26–2.62 | 0.740 |
| Age, years |
|
|
|
|
|
|
|
|
≤60 | 33 | 1 |
|
| 1 |
|
|
|
>60 | 41 | 1.18 | 0.60–2.31 | 0.629 | 1.29 | 0.62–2.69 | 0.499 |
| Smoking |
|
|
|
|
|
|
|
| No | 30 | 1 |
|
| 1 |
|
|
|
Yes | 44 | 1.11 | 0.56–2.20 | 0.758 | 1.10 | 0.42–2.87 | 0.839 |
| Tissue
differentiation |
|
|
|
|
|
|
|
| Middle
and high | 27 | 1 |
|
| 1 |
|
|
|
Low | 47 | 0.82 | 0.42–1.61 | 0.565 | 0.49 | 0.23–1.05 | 0.065 |
| Stage |
|
|
|
|
|
|
|
| IA, IB,
IIA and IIB | 52 | 1 |
|
| 1 |
|
|
| IIIA,
IIIB and IV | 22 | 1.41 | 0.70–2.84 | 0.333 | 0.68 | 0.26–1.73 | 0.416 |
| Lymph node
metastasis |
|
|
|
|
|
|
|
|
Absent | 33 | 1 |
|
| 1 |
|
|
|
Present | 41 | 2.33 | 1.19–4.56 | 0.014a | 3.54 | 1.36–9.22 | 0.009a |
| AFAP1-AS1
expression |
|
|
|
|
|
|
|
|
Low | 18 | 1 |
|
| 1 |
|
|
|
High | 56 | 3.58 | 1.25–10.24 | 0.017a | 3.12 | 1.05–9.25 | 0.040a |
Discussion
Recently, a number of studies have demonstrated that
lncRNAs serve an important role in cancer pathogenesis (20,25).
Additionally, the association between lncRNA and tumor development
and progression have been demonstrated (26,27). In
addition, lncRNA has been indicated to be a novel biomarker for
cancer diagnosis, prognosis and metastasis, and therefore have a
therapeutic effect (6).
Notably, associations between lncRNAs, including
HOTAIR and MALAT1, and human cancers have been previously reported
(28). Upregulation of HOTAIR in lung
tumor tissues is associated with metastasis, drug resistance and
poor survival time in patients with lung cancer (29). Furthermore, HOTAIR has been indicated
as a biomarker in lung cancer (12,13,30).
Additionally, high-expression of MALAT1 in primary tumors is a
biomarker of metastasis and poor survival time (14,15,31).
As indicated by the microarray data, AFAP1-AS1 was
upregulated in lung cancer tissues, compared with relative normal
tissues. In the present study, ISH and RT-qPCR was conducted to
analyze the expression of AFAP1-AS1 in lung cancer tissues.
Following this, the association between AFAP1-AS1 expression level
and clinical characteristics was investigated. Statistical analysis
revealed that the overexpression of AFAP1-AS1 was associated with
lymph node metastasis. Furthermore, the high expression level of
ASAP1-S1 also indicated poor survival time in patients with
NSCLC.
The present study indicated that the overexpression
of AFAP1-AS1 was notably associated with the poor survival time in
patients with NSCLC. However, the precise mechanism underlying this
effect remains unknown. Further experimental evidence is required
to explore the mechanism underlying AFAP1-AS1 leading to poor
outcomes.
To conclude, it was demonstrated that the expression
level of AFAP1-AS1 is upregulated in NSCLC and correlates with
lymph node metastasis. High AFAP1-AS1 expression level indicated a
poor prognosis for the patient with lung cancer. Therefore,
AFAP1-AS1 could be a potential biomarker for predicting NSCLC
progression and prognosis.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Technology
Projects of Jiangsu Provincial Commission of Health and Family
Planning (grant no. H201457), the Natural Science Foundation of
China (grant nos. 81201830 and 81472200 to Rong Yin), Natural
Science Foundation for High Education of Jiangsu (grant no.
13KJB320010 to Rong Yin) and Jiangsu Ordinary University Graduate
Student Research Innovation Project for 2013 (grant no. CXLX13_571
to Mantang Qiu).
Availability of data and materials
The data generated during the present study is
available upon reasonable request from the corresponding
author.
Authors' contributions
XL, XD and KX conceived the study. XL and XD
designed the study. TF, WS and RY coordinated the study. XL, XD and
SW performed the majority of the experiments and statistical
analyses. WS and TF obtained the clinical data. XL and RY drafted
the manuscript. KX and RY provided funds. WS, TF and WX revised the
manuscript and obtained the clinical data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Boards of the
Cancer Institute of Jiangsu Province. Informed written consent was
obtained from all patients included in this research.
Consent for publication
All patients provided written informed consent for
the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–32. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Aggarwal C: Targeted therapy for lung
cancer: Present and future. Ann Palliat Med. 3:229–235.
2014.PubMed/NCBI
|
|
3
|
Raungrut P, Wongkotsila A,
Lirdprapamongkol K, Svasti J, Geater SL, Phukaoloun M, Suwiwat S
and Thongsuksai P: Prognostic significance of 14–3-3γ
overexpression in advanced non-small cell lung cancer. Asian Pac J
Cancer Prev. 15:3513–3518. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Youlden DR, Cramb SM and Baade PD: The
International Epidemiology of lung cancer: Geographical
distribution and secular trends. J Thorac Oncol. 3:819–831. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Qiu MT, HU JW, Yin R and Xu L: Long
noncoding RNA: An emerging paradigm of cancer research. Tumour
Biol. 34:613–620. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zaratiegui M, Irvine DV and Martienssen
RA: Noncoding RNAs and gene silencing. Cell. 128:763–776. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ono H, Motoi N, Nagano H, Miyauchi E,
Ushijima M, Matsuura M, Okumura S, Nishio M, Hirose T, Inase N and
Ishikawa Y: Long noncoding RNA HOTAIR is relevant to cellular
proliferation, invasiveness, and clinical relapse in small-cell
lung cancer. Cancer Med. 3:632–642. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao W, An Y, Liang Y and Xie XW: Role of
HOTAIR long noncoding RNA in metastatic progression of lung cancer.
Eur Rev Med Pharmacol Sci. 18:1930–1936. 2014.PubMed/NCBI
|
|
14
|
Jeffers LK, Duan K, Ellies LG, Seaman WT,
Burger-Calderon RA, Diatchenko LB and Webster-Cyriaque J:
Correlation of transcription of MALAT-1, a novel noncoding RNA,
with deregulated expression of tumor suppressor p53 in small DNA
tumor virus models. J Cancer Ther. 4:2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Qiu M, Xu Y, Yang X, Wang J, Hu J, Xu L
and Yin R: CCAT2 is a lung adenocarcinoma-specific long non-coding
RNA and promotes invasion of non-small cell lung cancer. Tumour
Biol. 35:5375–5380. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Qiu M, Xu Y, Wang J, Zhang E, Sun M, Zheng
Y, Li M, Xia W, Feng D, Yin R and Xu L: A novel lncRNA, LUADT1,
promotes lung adenocarcinoma proliferation via the epigenetic
suppression of p27. Cell Death Dis. 6:e18582015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Deng J, Liang Y, Liu C, He S and Wang S:
The up-regulation of long non-coding RNA AFAP1-AS1 is associated
with the poor prognosis of NSCLC patients. Biomed Pharmacother.
75:8–11. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zeng Z, Bo H, Gong Z, et al: AFAP1-AS1, a
long noncoding RNA upregulated in lung cancer and promotes invasion
and metastasis. Tumour Biol. 37:729–737. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang F, Li J, Xiao H, Zou Y, Liu Y and
Huang W: AFAP1-AS1: A novel oncogenic long non-coding RNA in human
cancers. Cell Prolif. 51:123972018. View Article : Google Scholar
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
VanGuilder HD, Vrana KE and Freeman WM:
Twenty-five years of quantitative PCR for gene expression analysis.
Biotechniques. 44:619–626. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Krajewska M, Krajewski S, Epstein JI,
Shabaik A, Sauvageot J, Song K, Kitada S and Reed JC:
Immunohistochemical analysis of bcl-2, bax, bcl-X, and mcl-1
expression in prostate cancers. Am J Pathol. 148:1567–1576.
1996.PubMed/NCBI
|
|
24
|
Ding Y, Shimada Y, Maeda M, Kawabe A,
Kaganoi J, Komoto I, Hashimoto Y, Miyake M, Hashida H and Imamura
M: Association of CC chemokine receptor 7 with lymph node
metastasis of esophageal squamous cell carcinoma. Clin Cancer Res.
9:3406–3412. 2003.PubMed/NCBI
|
|
25
|
Wang ZY, Hu M, Dai MH, Xiong J, Zhang S,
Wu HJ, Zhang SS and Gong ZJ: Upregulation of the long non-coding
RNA AFAP1-AS1 affects the proliferation, invasion and survival of
tongue squamous cell carcinoma via the Wnt/β-catenin signaling
pathway. Mol Cancer. 17:32018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Negishi M, Wongpalee SP, Sarkar S, et al:
A new lncRNA, APTR, associates with and represses the CDKN1A/p21
promoter by recruiting polycomb proteins. PLoS One. 9:e952162014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang X, Gao L, Guo X, Shi X, Wu H, Song F
and Wang B: A network-based method for analysis of lncRNA-disease
associations and prediction of lncRNAs implicated in diseases. PLoS
One. 9:e877972014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sun W, Yang Y, Xu C and Guo J: Regulatory
mechanisms of long noncoding RNAs on gene expression in cancers.
Cancer Genet. 216-217:105–110. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yu X and Li Z: Long non-coding RNA HOTAIR:
A novel oncogene (Review). Mol Med Rep. 12:5611–5618. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Loewen G, Jayawickramarajah J, Zhuo Y and
Shan B: Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol.
7:902014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang HM, Yang FQ, Chen SJ, Che J and
Zheng JH: Upregulation of long non-coding RNA MALAT1 correlates
with tumor progression and poor prognosis in clear cell renal cell
carcinoma. Tumour Biol. 36:2947–2955. 2015. View Article : Google Scholar : PubMed/NCBI
|