Introduction
Hepatocellular carcinoma (HCC), a primary liver
cancer, is one of the most prevalent causes of cancer-associated
mortality globally and originates from hepatocyte cells in the
liver (1). Various risk factors,
including chronic viral infection (including with hepatitis B and C
viruses), aflatoxin exposure, alcohol ingestion and iron overload,
contribute to the development of HCC (2). The incidence rate of HCC is increasing
in numerous Asian countries, including Thailand (1). A majority of patients with HCC have a
poor prognosis due to the first detection of disease at an advanced
stage. Numerous different therapies have been devised in order to
improve the prognosis and long-term survival of patients; however,
limitations remain, including severe side effects such as fatigue,
pain, diarrhea, nausea, vomiting, hair loss and the resistance of
cancer cells to treatment (3). At
present, there is a great deal of interest in traditional plant
medicines, which may be used for cancer therapy or in combination
with other cancer treatments.
Derris scandens, locally known as
‘Thao-wan-Priang’ in Thailand, belongs to the family of Leguminosae
or Fabaceae. It is well-known as an Asian medicinal plant, which
grows as a woody vine throughout Southeast Asia, including in
Thailand. Dried stems of D. scandens have been used as an
expectorant, antitussive, diuretic and anti-dysentery agent
(4), and additionally for the
treatment of several diseases including osteoarthritis,
inflammation and muscle pains (4,5). A
previous study revealed that D. scandens ethanolic extract
has potential anti-metastatic activity in cholangiocarcinoma and
hepatoma cell lines equal to paclitaxel (Taxol; 10−9 M),
which was used as positive control (6). Furthermore, extracts from D.
scandens have been revealed to exert anti-proliferative effects
against colon cancer by upregulating B-cell lymphoma 2-associated X
protein (Bax), which is pro-apoptotic, and downregulating B-cell
lymphoma 2 (Bcl-2) anti-apoptotic proteins (7).
Proteomic analysis is used extensively in the field
of cancer research. The main principle of this technique is to
separate proteins in two dimensions according to their isoelectric
point and molecular mass. Mass spectrometry is used for protein
identification following two-dimensional (2D) electrophoresis
(8). These processes of proteomic
analysis have been used as crucial tools in order to
comprehensively monitor, identify and characterize the variations
of proteins for numerous different diseases (9,10). Thus,
proteomic analysis is a useful tool to examine and identify the
changes in protein expression in a HCC cell line in response to
traditional plant treatment.
In the present study, the cytotoxicity of D.
scandens ethanolic extract on a human HCC cell line, HCC-S102,
was examined. Induction of cytotoxicity via apoptosis was
additionally studied. As the mechanisms underlying the
anti-proliferative properties of D. scandens on this cell
line are yet to be reported, 2D electrophoresis was performed to
identify protein alterations which will improve understanding of
the mode of action.
Materials and methods
Plant preparation
D. scandens was purchased from a Thai medicinal herb
shop in Bangkok, Thailand. Stem parts were selected and prepared
using ethanol extraction. Briefly, the stem parts of dry plants
were chopped and ground into small pieces. Dried ground plant
materials (50 g) were percolated with absolute ethanol and then
shaken with an orbital shaker at 60 rev/min for 22 h at room
temperature. The plant materials in absolute ethanol solution were
then filtered using Whatman filter paper no. 4 (Hyclone; GE
Healthcare Life Sciences, Logan, UT, USA), followed by drying under
decreased pressure to yield 62.93 mg. Ethanolic plant extracts were
dissolved in dimethylsulfoxide (DMSO) at 200 mg/ml and stored as a
stock solution at −20°C.
Cell culture
The HCC-S102 cell line, established from a Thai
patient (11), was kindly provided by
Dr Sumalee Tungpradabkul (Department of Biochemistry, Faculty of
Science, Mahidol University, Bangkok, Thailand). Cells were
maintained in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum
(Hyclone; GE Healthcare Life Sciences), 1% penicillin/streptomycin
(Gibco; Thermo Fisher Scientific, Inc.) and 125 ng/ml amphotericin
B (Gibco; Thermo Fisher Scientific, Inc.). All cultures were
incubated in a CO2 incubator at 37°C in a humidified
atmosphere of 5% CO2. Culture medium was replenished
three times per week.
Cytotoxic activity
HCC-S102 cells were used to examine the cytotoxic
effect of D. scandens using an MTT assay (12). Cells in culture medium were plated in
96-well plates at a density of 5×103 cells/well and
incubated at 37°C overnight. The cells were then treated with
various concentrations of D. scandens ethanolic extract (0–50
µg/ml) for 24, 48 and 72 h in addition to DMSO (negative control)
and doxorubicin (positive control) at various concentrations (0–2.7
µg/ml) for 24, 48 and 72 h. Subsequently, medium was discarded and
replaced with 100 µl fresh medium containing tetrazolium dye
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) solution (0.5
mg/ml). All plates were incubated for 2 h at 37°C. Then, the medium
was removed and formazan product was dissolved by the addition of
100 µl DMSO to each well. Absorbance was measured at 550 nm with
subtracted background at 650 nm using a microplate reader
(Molecular Devices, LLC, Sunnyvale, CA, USA).
Protein extraction
Cells treated with 20 and 30 µg/ml D. scandens
ethanolic extract for 72 h and control cells (treated with 0.5%
DMSO) were separately washed with 0.25 M sucrose to remove salt.
Cells were then scraped into 450 µl 0.25 M sucrose containing
cOmplete™, Mini Protease Inhibitor Cocktail Tablets
(ratio of 1:10, Sigma-Aldrich; Merck KGaA) and centrifuged at 778 ×
g for 15 min at 4°C. The pellets were resuspended in lysis buffer
containing 9 M urea, 2%
3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate, 2%
dithiothreitol (DTT), 2% ampholine [pH 3–10, the same range as that
of the immobilized pH gradient (IPG) strip] and 1% protease
inhibitor cocktail, followed by sonication on ice and
centrifugation at 13,800 × g for 10 min at 4°C. Protein
concentrations were determined using a Bradford assay (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) as previously described
(13).
2D gel electrophoresis and image
analysis
Amounts of 200 µg of each sample were applied onto
Immobiline™ Drystrips (7 cm, non-linear, pH 3–10 gradient; Bio-Rad
Laboratories). IPG strips were incubated at room temperature
overnight. First-dimension isoelectric focusing was performed at
6,500 V/h, 55 µA per gel strip using an Ettan IPGphor 3 (GE
Healthcare, Little Chalfont, UK). For second-dimension SDS-PAGE,
the IPG strips were equilibrated in an equilibration buffer at room
temperature for 15 min for each step. Electrophoresis was performed
in a SE 600 Ruby apparatus (GE Healthcare Life Sciences) at 10
mA/gel, followed by Coomassie blue R-250 staining as previously
described (14). Gels were scanned
using Labscan software (version 5.0; GE Healthcare Life Sciences)
and analyzed using ImageMaster 2D Platinum (version 7.0; GE
Healthcare Life Sciences). All experiments were performed
independently in triplicate. Protein spots revealing a significant
difference in volume ratio (P<0.05) were selected for protein
identification by mass spectrometry analysis.
In-gel digestion
Protein spots were subjected to in-gel tryptic
digestion as described previously (14). Briefly, the gels were cut into small
pieces and destained using 0.1 M NH4HCO3 in
50% acetonitrile at 30°C for 20 min, followed by reduction with 10
mM DTT at 60°C for 45 min and alkylation with 100 mM iodoacetamide
at 25°C for 30 min, respectively. Following the removal of
reagents, gel pieces were completely dried and digested using 0.3
µg trypsin (Promega Corporation, Madison, WI, USA) in 30 µl
digestion buffer at 37°C overnight. The digestion buffer was
collected for protein identification.
Protein identification by liquid
chromatography-tandem mass spectrometry (LC-MS/MS)
Identification of protein spots were performed by
nanoflow liquid chromatography coupled with amaZon speed ion trap
mass spectrometry (Bruker Corporation, Billerica, MA, USA). All
trypsinized peptides were concentrated and desalted on a 75 µm
internal diameter ×100 mm C18 EASY-nLC™ column (Thermo
Fisher Scientific, Inc.). Subsequently, the peptides were eluted by
gradient separation using eluents A and B, which were 0.1% formic
acid in water and 0.1% formic acid in acetonitrile, respectively.
In total, 6 µl sample was injected into the nano-LC system and
separated at a flow rate of 0.5 µl/min for 30 min using the
following gradient: 0 min 95% A, 20 min 60% A, 20.5 min 5% A, 29
min 5% A and 29.5 min 95% A, followed by MS/MS equipped with the
CaptiveSpray™ source using 1.0 sec automatic scan rate with 0.1 sec
interscan delay. For MS/MS analysis, parent mass peaks with a range
between 50 and 3,000 m/z were selected. Collision energy was fixed
at 1,300 V. MS/MS data were processed and converted into mgf files
using Compass software (version 1.4; Bruker Corporation) and
proteins were identified using the MASCOT search engine (www.matrixscience.com). Search parameters were set as
follows: Database, Swiss-Prot and NCBInr; taxonomy, Homo sapiens;
enzyme, trypsin; two missed cleavages allowed. Peptide and MS/MS
fragment ion mass tolerance were set at 1.2 and 0.6 Da,
respectively. Proteins with a consistent molecular mass and
isoelectric point gel spot with a MASCOT score of >25 using a
P-value of ≤0.05 were considered to be positively identified.
Western blot analysis
Protein samples were extracted using
radioimmunoprecipitation assay buffer that contained cOmplete™,
Mini Protease Inhibitor Cocktail Tablets. A total of 10 µM NaF (New
England BioLabs, Inc., Ipswich, MA, USA), 1 mM of
Na3VO4 (Sigma-Aldrich; Merck KGaA) and 20 mM
of β-glycerophosphate (Merck KGaA) were used as phosphatase
inhibitors. A total of 15 µg protein was loaded into each lane and
was separated by SDS-PAGE (12.5% gel) and electrophoretically
transferred onto polyvinylidene difluoride membranes (EMD
Millipore, Billerica, MA, USA). The membranes were blocked with 3%
bovine serum albumin for 1 h at room temperature and subsequently
incubated at 4°C overnight with primary antibodies as follows:
Rabbit polyclonal anti-poly(ADP-ribose) polymerase (PARP; 1:1,000;
cat. no. 9542; Cell Signaling Technology, Inc., Danvers, MA, USA),
mouse monoclonal anti-α-tubulin (1:3,000; cat. no. 3873; Cell
Signaling Technology, Inc.), rabbit polyclonal anti-heterogeneous
ribonucleoprotein K (hnRNP K, 1:1,000; cat. no. 4675; Cell
Signaling Technology), rabbit monoclonal
anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 1:1,000; cat.
no. ab75834; Abcam, Cambridge, UK), mouse monoclonal anti-hnRNP
A2/B1 (1:1,500; cat. no. ab6102; Abcam), rabbit polyclonal
anti-peroxiredoxin-4 (Prx4; 1:10,000; cat. no. ab15574; Abcam),
rabbit monoclonal anti-oxygen-regulated protein 150 (ORP150, also
known as HYOU1 or hypoxia-upregulated protein 1; 1:1,000; cat. no.
ab134944; Abcam) and rabbit monoclonal anti-stomatin-like protein 2
(STOML2; 1:2,000; cat. no. ab191884; Abcam). The membranes were
then washed 3 times with Tris-buffered saline with 0.1% Tween 20
and was incubated with a horseradish peroxidase (HRP)-conjugated
polyclonal rabbit anti-mouse immunoglobulin (cat no. P0260; Dako;
Agilent Technologies, Inc., Santa Clara, CA, USA) or polyclonal
swine anti-rabbit immunoglobulin (cat no. P0217; Dako; Agilent
Technologies, Inc.) (dilution, 1:2,000) at room temperature for 1
h. Bands were detected using electrochemiluminescence reagent (GE
Healthcare Life Sciences) and images were captured using the
ImageQuant™ LAS 4000 digital imaging system (GE Healthcare Life
Sciences).
Annexin V and dead cell assay (flow
cytometry)
A total of 2×105 cells (2 ml/well) was
seeded in 6-well plates at 37°C overnight. The control condition
group contained 0.5% DMSO as a vehicle control. For treated cells,
cells were treated with D. scandens ethanolic extract at various
concentrations (20 and 30 µg/ml) and further incubated at 37°C in a
humidified atmosphere of 5% CO2 for 24, 48 and 72 h.
Following incubation, cells were trypsinized and resuspended at
1×106 cells/ml prior to staining with a 1:1 ratio of
Muse® Annexin V and Dead Cell Assay kit (EMD Millipore,
Billerica, MA, USA) at room temperature in the dark for 20 min.
Apoptotic analysis was performed using Muse™ Cell Analyzer (EMD
Millipore), with Muse™ Analysis Software version 1.3.0.0.
Statistical analysis
All data are presented as the mean ± standard
deviation. Statistical analysis was performed by two-way analysis
of variance with Tukey's Honest Significant Difference post-hoc
analysis), using GraphPad Prism (version 7; GraphPad Software,
Inc., La Jolla, CA, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
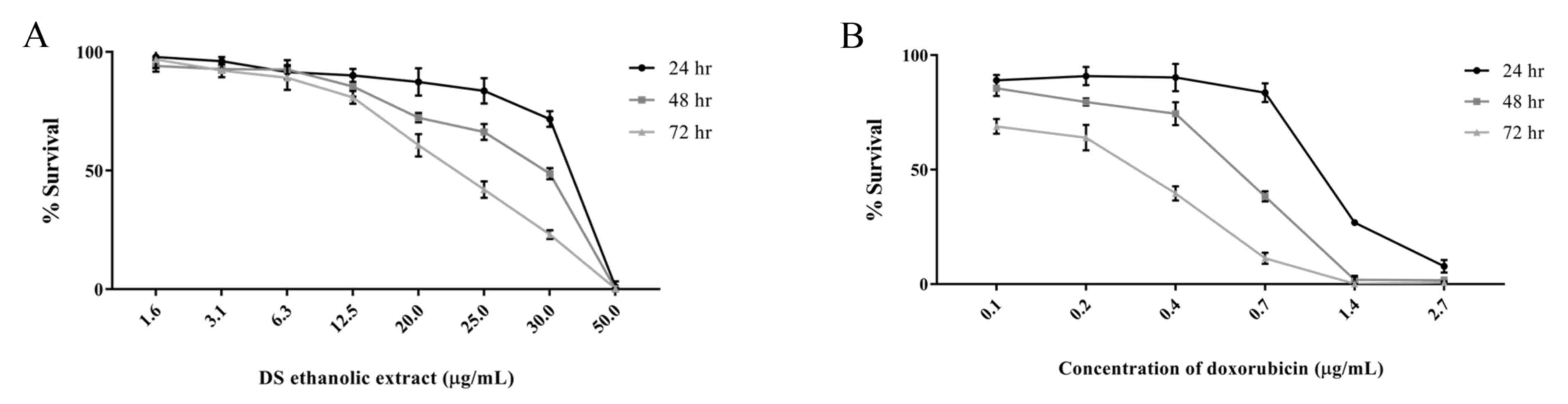
Cytotoxic activity of D. scandens
ethanolic extract on HCC-S102 cells
To estimate the cytotoxic effect of D. scandens
ethanolic extract on HCC-S102 cells, an MTT assay was performed.
Half-maximal inhibitory concentration (IC50) values were
calculated, and the IC50 values were 36.0±1.0, 29.6±0.6,
and 22.6±1.5 µg/ml at 24, 48 and 72 h, respectively (Fig. 1A). Additionally, doxorubicin was used
as a positive control, and the IC50 values for the
doxorubicin-treated cells were 1.1±0.03, 0.6±0.01 and 0.3±0.02
µg/ml at 24, 48 and 72 h, respectively (Fig. 1B).
These results reveal that D. scandens
ethanolic extract exerts a cytotoxic effect on HCC-S102 cells in
dose- and time-dependent manner compared with the control. On the
basis of the inhibitory concentration of D. scandens
ethanolic extract, the IC50 and IC80 were 20
and 30 µg/ml, respectively, and so these doses were selected for
further studies.
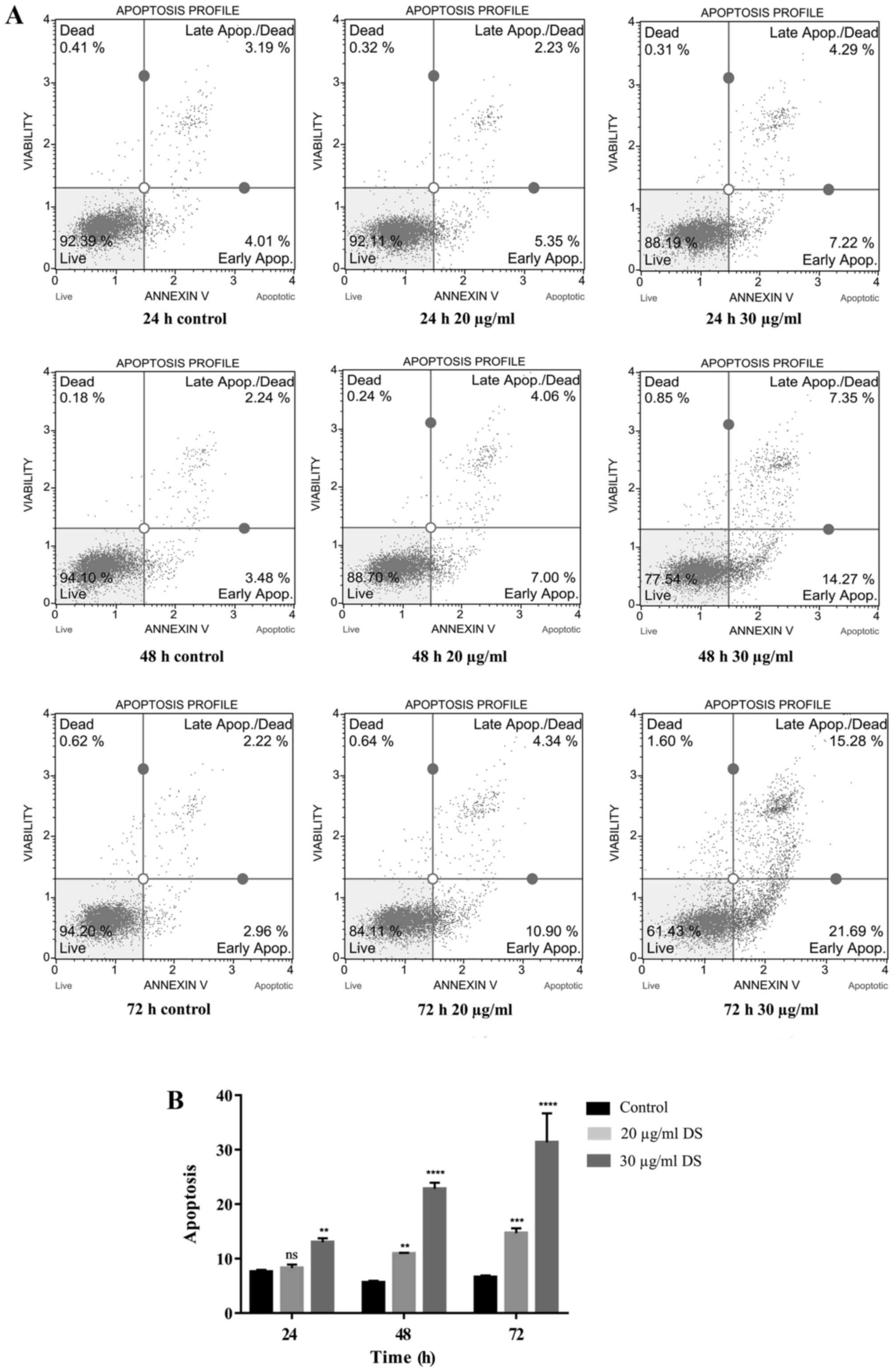
Detection of apoptosis of HCC-S102
cells induced by D. scandens ethanolic extract
The apoptosis-inducing effect of D. scandens
ethanolic extract on HCC-S102 cells was quantified using Annexin V
and dead cell assays. HCC-S102 cells were exposed to D. scandens
ethanolic extract at concentrations of 20 and 30 µg/ml for 24, 48
and 72 h. Following treatment, a gradual increase in Annexin
V-positive-staining was detected in cells treated with 20 and 30
µg/ml D. scandens ethanolic extract compared with the control
(Fig. 2A).
The proportions of total apoptotic cells
demonstrated that the treatment of D. scandens ethanolic
extract on HCC-S102 induced apoptosis in a dose- and time-dependent
manner. Thus, total apoptotic cells were significantly increased
(P<0.01) in cell groups treated with 20 and 30 µg/ml D.
scandens ethanolic extract for 48 and 72 h compared with the
control, and significantly increased (P<0.01) in the group
treated with 30 µg/ml D. scandens ethanolic extract for 24 h
compared with the control (Fig.
2B).
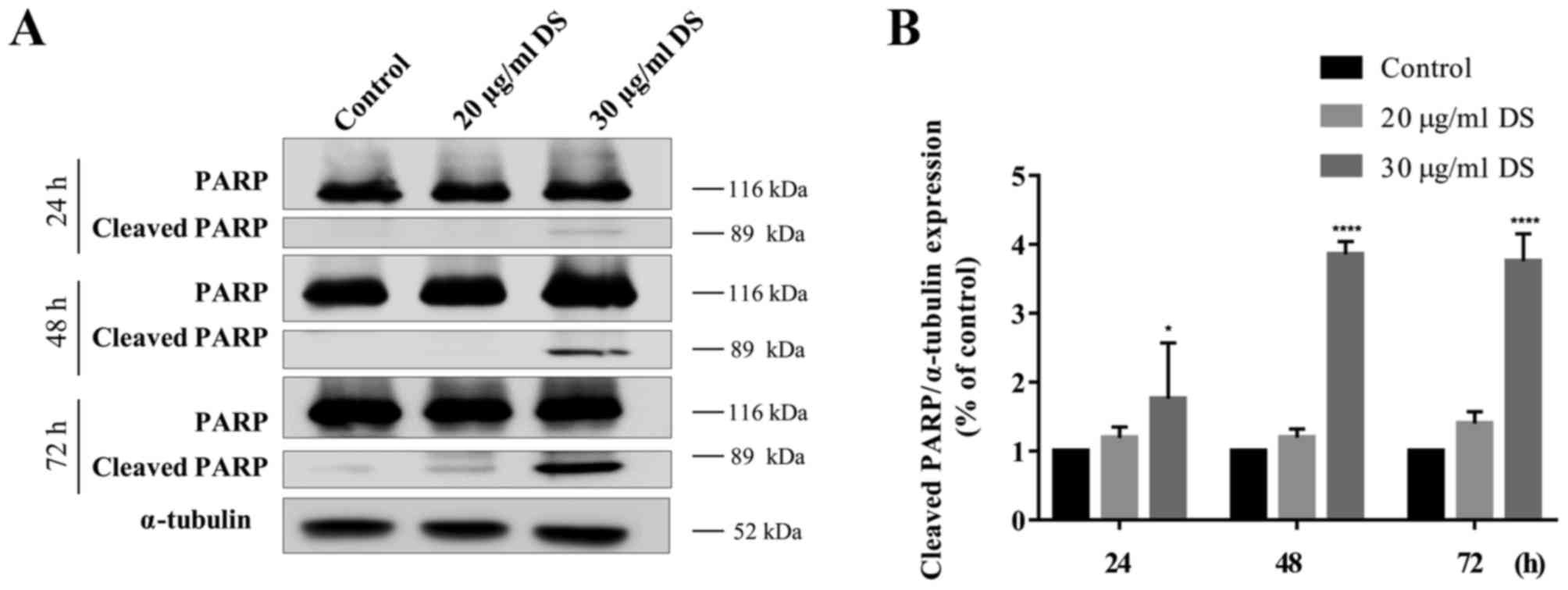
To confirm whether D. scandens ethanolic
extract was associated with an induction of apoptosis in HCC-S102
cells, the expression levels of cleaved PARP were determined using
western blot analysis. The results revealed that treatment of
HCC-S102 cells with D. scandens ethanolic extract increased
the band intensity of cleaved PARP, particularly at 30 µg/ml (at 89
kDa; Fig. 3A). Therefore, 30 µg/ml
D. scandens ethanolic extract resulted in significantly
(P<0.01) increased expression levels of cleaved PARP after 24,
48, and 72 h of treatment (Fig. 3B).
An increase in cleaved PARP expression levels confirmed the
hypothesis that D. scandens ethanolic extract induced the
death of HCC-S102 cells via the apoptotic pathway.
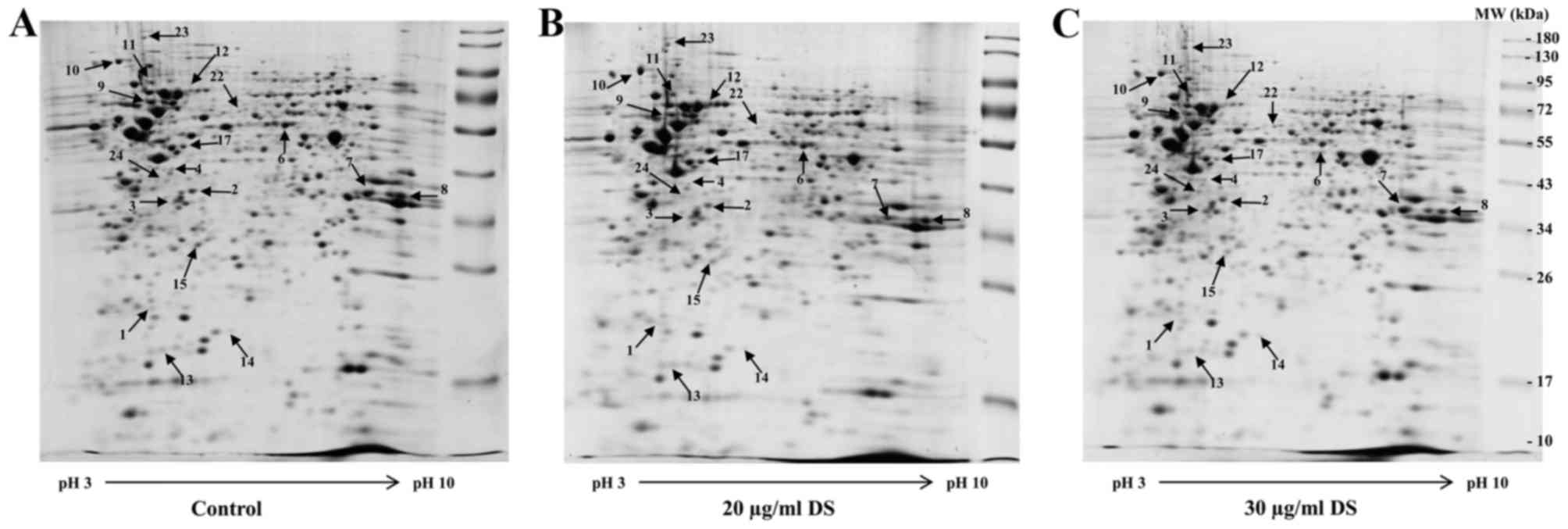
Effects of D. scandens ethanolic
extract treatment by proteomic analysis
In order to identify any differences in protein
expression, proteins were extracted from HCC-S102 cells treated
with 20 and 30 µg/ml D. scandens ethanolic extract for 72 h. The 2D
patterns of control and D. scandens ethanolic extract-treated cells
(Fig. 4A-C) were then analyzed using
ImageMaster 2D Platinum program. As a result, ~466 protein spots
were detected in D. scandens ethanolic extract-treated (either 20
or 30 µg/ml) cells and, the majority of proteins were revealed to
have statistically significant differences in their expression
levels >1.5-fold change (P<0.05; data not shown), compared
with the equivalent spots in control cells. Among these, 18 protein
spots that were identified to have significantly different
expression levels were selected for further analysis by LC-MS/MS
and subsequently identified using the MASCOT search engine and
SwissProt database.
In total, 18 proteins were identified, as presented
in Table I. In total, 6 upregulated
proteins and 12 downregulated proteins were identified in D.
scandens ethanolic extract-treated (20 and 30 µg/ml) HCC-S102
cells compared with the control. The upregulated proteins included
endoplasmin (Hsp90B1), heat-shock cognate 71 kDa protein (Hsc70),
cytoplasmic dynein 1 intermediate chain 2 (DC1I2),
UDP-N-acetylhexosamine pyrophosphorylase-like protein 1, ORP150 and
type I cytoskeletal keratin 9.
 | Table I.Differentially expressed proteins in
D. scandens ethanolic extract treated HCC-S102 cells identified by
liquid chromatography-tandem mass spectrometry. |
Table I.
Differentially expressed proteins in
D. scandens ethanolic extract treated HCC-S102 cells identified by
liquid chromatography-tandem mass spectrometry.
|
|
|
|
|
|
|
| Fold change of
protein volume |
|---|
|
|
|
|
|
|
|
|
|
|---|
| Spot no. | SwissProt entry
name | Protein name | Molecular mass, kDa
kDa/isoelectric point | Protein score | Peptides
matched | Sequence coverage,
% | Control and 20
µg/ml D. scandens ethanolic extract | Control and 30
µg/ml D. scandens ethanolic extract |
|---|
| Upregulated
proteins |
| 10 | ENPL_HUMAN | Endoplasmin | 92.4/4.8 | 609 | 13 | 15 | (+) 1.68 | (+) 1.17 |
| 11 | DC1I2_HUMAN | Cytoplasmic dynein
1 intermediate chain 2 | 71.4/5.1 | 226 | 3 | 7 | (+) 1.90 | (+) 1.37 |
| 12 | HSP7C_HUMAN | Heat-shock cognate
71 kDa protein | 73.6/5.9 | 60 | 1 | 2 | (+) 2.03 | (+) 1.76 |
| 14 | K1C9_HUMAN | Type I cytoskeletal
keratin 9 | 62.0/5.1 | 90 | 1 | 3 | (+) 0.77 | (+) 1.54 |
| 22 | UAP1L_HUMAN |
UDP-N-acetylhexosamine
pyrophosphorylase-like protein 1 | 56.9/5.9 | 61 | 1 | 3 | (+) 1.23 | (+) 1.63 |
| 23 | HYOU1_HUMAN | Hypoxia-upregulated
protein 1 | 111.3/5.2 | 435 | 7 | 9 | (+) 1.34 | (+) 1.81 |
| Downregulated
proteins |
| 1 | CBX3_HUMAN | Chromobox protein
homolog 3 | 20.8/5.2 | 292 | 4 | 22 | (−) 1.42 | (−) 1.46 |
| 2 | RLA0_HUMAN | 60S acidic
ribosomal protein P0 | 34.3/5.7 | 66 | 2 | 9 | (−) 1.53 | (−) 1.20 |
| 3 | CAZA1_HUMAN | Filamentous
actin-capping protein subunit α-1 | 32.9/5.5 | 186 | 3 | 15 | (−) 1.62 | (−) 1.05 |
| 4 | GLRX3_HUMAN | Glutaredoxin-3 | 37.4/5.3 | 119 | 2 | 8 | (−) 1.50 | (−) 1.07 |
| 6 | AL1A1_HUMAN | Retinal
dehydrogenase 1 | 54.8/6.3 | 654 | 11 | 29 | (−) 1.56 | (−) 1.55 |
| 7 | AK1C1_HUMAN | Aldo-keto reductase
family 1 member C1 | 36.8/8.0 | 133 | 3 | 9 | (−) 1.80 | (−) 1.15 |
| 7 | ROA2_HUMAN | Heterogeneous
nuclear ribonucleoprotein A2/B1 | 37.4/8.9 | 131 | 2 | 8 | (−) 1.80 | (−) 1.15 |
| 8 | G3P_HUMAN |
Glyceraldehyde-3-phosphate
dehydrogenase | 36.0/8.6 | 146 | 3 | 13 | (−) 1.09 | (−) 1.56 |
| 9 | HNRPK_HUMAN | Heterogeneous
nuclear ribonucleoprotein K | 50.9/5.4 | 311 | 6 | 19 | (−) 1.76 | (−) 1.57 |
| 13 | NCALD_HUMAN | Neurocalcin-δ | 22.2/5.2 | 58 | 1 | 6 | (−) 1.05 | (−) 1.98 |
| 15 | PRDX4_HUMAN |
Peroxiredoxin-4 | 30.5/5.8 | 104 | 2 | 12 | (−) 1.28 | (−) 1.93 |
| 17 | QCR1_HUMAN | Cytochrome bc1
complex subunit 1 | 52.6/5.9 | 160 | 4 | 10 | (−) 1.03 | (−) 1.57 |
| 24 | STML2_HUMAN | Stomatin-like
protein 2 | 38.5/6.8 | 143 | 3 | 13 | (−) 1.12 | (−) 1.83 |
Downregulated proteins included chromobox protein
homolog 3 (CBX3), 60S acidic ribosomal protein P0, filamentous
actin-capping protein subunit α-1, retinal dehydrogenase 1
(RALDH1), aldo-keto reductase family 1 member C1 (AKR1C1), GAPDH,
neurocalcin-δ, cytochrome bc1 complex subunit 1
(UQCRC1), glutaredoxin-3 (TXNL2), peroxiredoxin-4 (Prx4), hnRNP K,
hnRNP A2/B1 and STOML2.
Validation of differential 2D
electrophoresis protein expression levels by western blot
analysis
To validate the difference of the expression levels
of the proteins of interest, which have been reported to be
involved in the survival of cancer cells (15–20),
western blot analysis was used to confirm the differences of
protein expression levels between D. scandens ethanolic
extract-treated cells and untreated cells. ORP150 was revealed to
be significantly increased (P<0.0001) in D. scandens ethanolic
extract-treated cells compared with the control, which is
consistent with the 2D electrophoresis result. Similarly, STOML2
was revealed to be significantly decreased in 20 µg/ml D. scandens
ethanolic extract-treated cells (P<0.05), and Prx4, STOML2,
hnRNP K and hnRNP A2/B1 were all significantly decreased in 30
µg/ml treated cells (P<0.05) compared with the control (Fig. 5A and B).
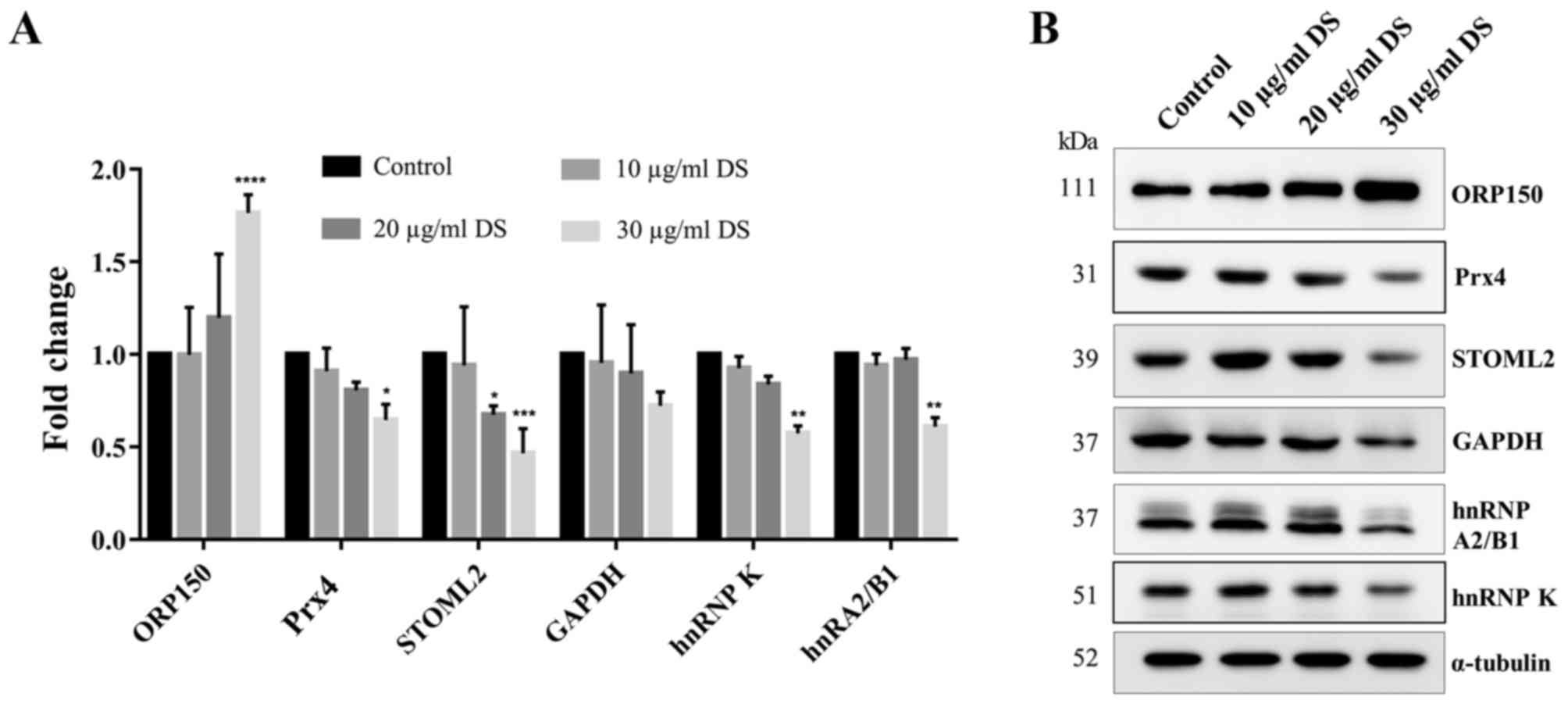 | Figure 5.Verification of the expression of
representative proteins from two-dimensional electrophoresis on the
HCC-S102 cell line treated with D. scandens ethanolic extract.
Cells were treated with different concentrations of D. scandens
ethanolic extract (10, 20 and 30 µg/ml) for 72 h and subjected to
SDS-PAGE (12.5% gel), followed by western blot analysis. (A) Fold
change of and (B) western blot analysis results for ORP150, Prx4,
GAPDH, STOML2, hnRNP A2/B1 and hnRNP K. α-tubulin was used as
loading control. Results are expressed as the mean ± standard
deviation. *P<0.05, **P<0.01, ***P<0.001 and
****P<0.0001 vs. the control. DS, Derris scandens ethanolic
extract; ORP150, oxygen-regulated protein 150; Prx4,
peroxiredoxin-4; STOML2, stomatin-like protein 2; hnRNP,
heterogeneous nuclear ribonucleoprotein. |
Discussion
Cancer is a major health problem, thus developments
of novel cancer treatments are required. It has been reported that
certain plant extracts have the potential to be used as anticancer
drugs by enhancing the immune system, inducing cell
differentiation, inhibiting telomerase activities and inducing
apoptosis in vitro and in vivo (21). In Thailand, certain herbs have been
widely used for a long time. However, the underlying molecular
mechanisms of action of the Thai herbs (e.g., Bridelia ovata
Decne or Dioscorea membranacea Pierre) for cancer treatment
are still being investigated. In the present study, the effect of
D. scandens stem ethanolic extract on the HCC-S102 cell line
were determined.
According to the criteria of cytotoxicity activity
for crude extracts from the American National Cancer Institute
(NCI), a concentration of plant extract ≤30 µg/ml with an
incubation time of between 48 and 72 h is considered to be ‘active’
due to the calculation of the IC50 value (22). The D. scandens ethanolic
extract was investigated for cytotoxic activity in HCC-S102 cells.
The IC50 value following treatment for 72 h was 22.6±1.5
µg/ml, which is of potential interest in terms of the NCI criteria.
One previous study indicated that ethanolic crude extract of D.
scandens exhibited potent cytotoxic activity against the lung
adenocarcinoma cell line (CORL-23) with an IC50 value of
21.04±0.57 µg/ml following treatment for 72 h (23). Furthermore, the cytotoxic activity of
D. scandens ethanolic extract has been investigated using
other lung cancer cell lines (namely, A549 and NCI-H226) compared
with lung normal cells, with the results indicating that D.
scandens ethanolic extract exhibited an increased cytotoxic
activity in lung cancer cell lines compared with normal lung cells
(IC50, 65.04 µg/ml) (24).
D. scandens ethanolic extract has been
revealed to have anti-proliferative effects against colon cancer
cell lines, mediating cell death through necrosis in the SW480
colon cancer cell line (7). Hematulin
et al (5) reported that colon
cancer cells (HT-29) treated with D. scandens ethanolic
extract exhibited increased apoptosis induction compared with cells
treated with a combination of radiation and D. scandens
ethanolic extract. However, cell death from D. scandens
ethanolic extract in combination with radiation was increased
compared with in D. scandens ethanolic extract alone,
suggesting that other modes of cell death were involved in colon
cancer cell lines.
To determine how D. scandens ethanolic
extract exerts cytotoxicity on the HCC-S102 cell line, a cell death
mechanism, apoptosis, was studied using an Annexin V and dead cell
assay, which detects the translocation of phosphatidylserine from
the inner to the outer cell-surface membrane, which occurs during
apoptosis. Using a double stain of Annexin V-fluorescein
isothiocyanate (FITC) and 7-aminoactinomycin D (7-AAD), cells were
classified as living cells or cells at early or late stages of
apoptosis. HCC-S102 cells treated with D. scandens ethanolic
extract were stained with Annexin V-FITC and 7-AAD, and then
quantified using flow cytometric analysis. Results revealed that
cell apoptosis from D. scandens ethanolic treatment was
dose- and time-dependent. At higher doses, treated cells were
killed via the apoptosis pathway following incubation with D.
scandens extract for 48 and 72 h.
PARP is a nuclear DNA-binding protein, which is
typically cleaved by caspase-3 during apoptosis, thus the detection
of PARP cleavage is used as an indicator of apoptosis (25). The results of the present study
revealed that there was a significantly increased intensity of
cleaved PARP protein in treated cells at different doses and times
compared with the control. This corresponds with the results of the
flow cytometric experiments, and confirms that D. scandens
ethanolic extract treatment induced apoptosis in HCC-S102
cells.
Proteomic analysis was used to investigate
differentially expressed proteins between HCC-S102 cells treated
with D. scandens ethanolic extract for 72 h and untreated
control cells. A total of 18 significantly different protein spots
were analyzed using Image Master and identified by LC-MS/MS. Cells
treated with D. scandens extract possessed 6 proteins with
higher expression and 13 proteins with lower expression compared
with untreated controls. A majority of proteins involved in stress
response including Hsp90B1, Hsc70 and ORP150 were upregulated.
Downregulated proteins involved in cell metabolism included RALDH1,
AKR1C, GAPDH, UQCRC1 and STOML2. In addition, TXNL2 and Prx4 which
serve functions as antioxidants were downregulated. Proteins
participating in transcriptional regulation including CBX3 and
hnRNP K were downregulated, as were proteins involved in signal
transduction including hnRNP A2/B1. Among these proteins, ORP150,
Prx4, GAPDH, STOML2, hnRNP A2/B1 and hnRNP K proteins were selected
to validate the differential expression of these proteins between
treated and untreated cells using western blot analysis. In
agreement with the proteomic results, western blot analysis
revealed that ORP150 was upregulated whereas Prx4, GAPDH, STOML2,
hnRNP A2/B1 and hnRNP K proteins were downregulated.
Prx4 is a member of the thiol-specific antioxidant
group of proteins, and is normally expressed in the endoplasmic
reticulum (ER) of cells (15). Its
general function is to serve as an antioxidant by decreasing the
production of reactive oxygen species (ROS). In addition, a
previous study revealed that Prx4 additionally participated in
oxidative protein folding as a molecular chaperone in order to
facilitate protein folding in the ER (26). High expression of Prx4 has been
reported in numerous different tumor types, including lung cancer,
colorectal cancer and high-grade glioma (primary brain malignancy)
(15,27,28). For
example, Prx4 is highly expressed in and associated with the
development of human lung cancer, and has been suggested to be
useful as a good prognostic marker of cancer progression in
early-stage squamous cell carcinoma of lung cancer (15). Additionally, a previous study used
immunohistochemistry and quantitative polymerase chain reaction to
demonstrate that the expression of the Prx4 gene and protein were
higher in colorectal cancer tissues compared with normal tissues
(27). Notably, Kim et al
(28) have previously revealed that
Piperlongumine, a natural plant product, suppressed the expression
of Prx4 and subsequently increased ROS and ER stress levels in
glioma cells. Excess misfolded or unfolded proteins in ER may
induce the ER stress response, which may be detected by the
expression of ORP150.
ORP150 is a notable molecular chaperone in the ER,
which belongs to the heat-shock protein 70 family (29). The ORP150 system is a part of the ER
machinery associated with the folding and assembly of secretory and
membrane proteins in the ER (16).
ORP150 expression was induced by numerous stress conditions
including the accumulation of protein unfolding and the depletion
of glucose or oxygen (16). Under
stress conditions, ORP150 may accumulate in the ER, serving a
cytoprotective function for cells. The upregulation of ORP150 has
been reported in numerous types of cancer including breast cancer
(28), bladder cancer (30) and nasopharyngeal carcinoma (31). Prx4 downregulation concomitantly with
the upregulation of ORP150 indicates an increase in ER stress and
ROS levels in the HCC-S102 cell line. The evidence suggests that,
together, prolonged unfolded protein accumulation and persistent ER
stress in cancer cells may initiate pro-apoptotic signaling
(32). Thus, it may be deduced that
these consequences will occur in HCC-S102 cells when treated with
D. scandens ethanolic extract.
GAPDH and STOML2 are involved in cell metabolism.
GAPDH is a key regulatory enzyme of glycolysis, and serves a
crucial function in several biological processes including the
control of gene expression, DNA replication and repair and
apoptosis (33). There is evidence
that GAPDH expression is increased in various human cancer types,
including ovarian, lung and colon cancer types (17,34,35).
Furthermore, previous studies have revealed that GAPDH knockdown by
small interfering RNA (siRNA) results in a significant decrease in
the proliferative, migratory and invasive abilities of lung
squamous carcinoma cells in vitro (36). STOML2 is a protein on the
mitochondrial inner membrane; however, its functions remain unknown
(37). STOML2 has been implicated in
serving an important function in sustaining the mitochondria
membrane potential and ATP production, and additionally affects
cell activity including cell motility and cell growth in tumor
cells (18). Furthermore, it has been
reported that the downregulation of STOML2 increases the
sensitivity of cancer cells to chemotherapeutic treatments by
inducing cancer cell apoptosis and cell cycle arrest (18). The results of the present study
revealed the downregulation of GAPDH and STOML2, which may be one
of the mechanisms of D. scandens ethanolic extract resulting
in cytotoxicity in HCC-S102 cells.
The hnRNP family are considered to be an important
factor in mRNA processing and biogenesis (38). The present study revealed a decrease
in the expression levels of hnRNP K and hnRNP A2/B1 in the HCC-S102
cell line treated with D. scandens ethanolic extract. hnRNP
K is a member of the hnRNP family. It exerts specific binding for
the c-myc promoter and functions as a transcription factor
(39). Association of hnRNP K with
tumor development in various types of cancer have been reported. A
previous study revealed that hnRNP K may be involved in
hepatocarcinogenesis by enhancing hepatitis B and hepatitis C
replication and cell proliferation (19). In lung cancer, the downregulation of
hnRNP K by siRNA resulted in the inhibition of cell growth and
increased apoptosis in a lung cancer cell line (A549) (40). In addition, there is evidence that
hnRNP K suppresses apoptosis in HCC and nasopharyngeal carcinoma
(41,42).
hnRNP A2/B1 is another member of the hnRNP family
that was revealed to be downregulated in the present study. hnRNP
A2/B1 has several cellular functions including regulating gene
expression at the transcriptional and translational level. hnRNP
A2/B1 has been reported to be overexpressed in tumorigenesis, and
may be a potential biomarker for early detection, particularly in
lung cancer (43). To assess the risk
of human liver cancer, the increased expression and cytoplasmic
localization of hnRNP A2/B1 may be used as a diagnostic biomarker
(20). Furthermore, the inhibition of
hnRNP A2/B1 expression has been demonstrated to increase the
sensitivity of cancer cells to chemotherapy, stimulate apoptosis
and additionally modulate the promotion of pancreatic cancer
(44). hnRNP K and hnRNP A2/B1
expression levels were decreased with the treatment with D.
scandens ethanolic extract in the present study. Therefore,
this may be one of the mechanisms involved in the cytotoxic
activity against HCC-S102 cells.
In conclusion, the present study demonstrated that
the effect of D. scandens ethanolic extract on HCC-S102 cell
line occurs through the apoptotic pathway. The proteomic profiling
of cells treated with D. scandens ethanolic extract
identified numerous protein targets for anticancer activity
including hnRNP K, hnRNP A2/B1, STOML2 and GAPDH. It may be
suggested that the decrease in the survival of HCC-S102 cells by
alterations in these proteins may potentiate the lethal effect of
D. scandens-induced Prx4 downregulation and excess ER stress
as indicated by ORP150 upregulation. Consequently, the present
results indicate the association of the apoptosis mechanism with
cell cytotoxicity upon treatment of D. scandens ethanolic
extract. However, further studies on the function of each protein
are necessary in order to elucidate the mode of action of D.
scandens ethanolic extract.
Acknowledgements
The present study was supported by the Chulabhorn
Graduate Institute (grant no. 10/2556) and the Chulabhorn Research
Institute (grant no. BC2008-02).
References
|
1
|
Somboon K, Siramolpiwat S and Vilaichone
RK: Epidemiology and survival of hepatocellular carcinoma in the
central region of Thailand. Asian Pac J Cancer Prev. 15:3567–3570.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mittal S and El-Serag HB: Epidemiology of
HCC: Consider the population. J Clin Gastroenterol. 47:S2–S6. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Safarzadeh E, Shotorbani SS and Baradaran
B: Herbal medicine as inducers of apoptosis in cancer treatment.
Adv Pharm Bull. 4(Suppl 1): S421–S427. 2014.
|
|
4
|
Hussain H, Al-Harrasi A, Krohn K, Kouam
SF, Abbas G, Shah A, Raees MA, Ullah R, Aziz S and Schulz B:
Phytochemical investigation and antimicrobial activity of Derris
scandens. J King Saud Univ Sci. 27:375–378. 2015. View Article : Google Scholar
|
|
5
|
Hematulin A, Ingkaninan K, Limpeanchob N
and Sagan D: Ethanolic extract from Derris scandens Benth mediates
radiosensitzation via two distinct modes of cell death in human
colon cancer HT-29 cells. Asian Pac J Cancer Prev. 15:1871–1877.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Laupattarakasem P, Sripa B and
Laupattarakasem W: Antimigration of cancer cells by Derris scandens
on Cholangiocarcinoma cells. Srinagarind Med J. 22:339–345.
2010.
|
|
7
|
Kaewkon K, Khamprasert N and Limpeanchob
N: Derris scandens benth extract induces necrosis rather than
apoptosis of SW480 colon cancer cells. Thai J Pharmacol.
33:118–121. 2011.
|
|
8
|
Li Y, Geng X and Zhang W: Application of
proteomics to the study of hepatocellular carcinoma and some
related diseases. Chin Jof Clin Oncol. 2:903–906. 2005. View Article : Google Scholar
|
|
9
|
Lee SY, Kim GT, Roh SH, Song JS, Kim HJ,
Hong SS, Kwon SW and Park JH: Proteomic analysis of the anti-cancer
effect of 20 S-ginsenoside Rg3 in human colon cancer cell lines.
Biosci Biotechnol Biochem. 73:811–816. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Srisomsap C, Sawangareetrakul P,
Subhasitanont P, Chokchaichamnankit D, Chiablaem K, Bhudhisawasdi
V, Wongkham S and Svasti J: Proteomic studies of cholangiocarcinoma
and hepatocellular carcinoma cell secretomes. J Biomed Biotechnol.
2010:4371432010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Laohathai K and Bhamarapravati N:
Culturing of human hepatocellular carcinoma. A simple and
reproducible method. Am J Pathol. 118:203–208. 1985.
|
|
12
|
Chiablaem K, Lirdprapamongkol K,
Keeratichamroen S, Surarit R and Svasti J: Curcumin suppresses
vasculogenic mimicry capacity of hepatocellular carcinoma cells
through STAT3 and PI3K/AKT inhibition. Anticancer Res.
34:1857–1864. 2014.PubMed/NCBI
|
|
13
|
Bradford MM: A rapid and sensitive method
for the quantitation of microgram quantities of protein utilizing
the principle of protein-dye binding. Anal Biochem. 72:248–254.
1976. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Khongmanee A, Lirdprapamongkol K, Tit-oon
P, Chokchaichamnankit D, Svasti J and Srisomsap C: Proteomic
analysis reveals important role of 14–3-3σ in anoikis resistance of
cholangiocarcinoma cells. Proteomics. 13:3157–3166. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hwang JA, Song JS, Yu DY, Kim HR, Park HJ,
Park YS, Kim WS and Choi CM: Peroxiredoxin 4 as an independent
prognostic marker for survival in patients with early-stage lung
squamous cell carcinoma. Int J Clin Exp Pathol. 8:6627–6635.
2015.PubMed/NCBI
|
|
16
|
Krętowski R, Stypułkowska A and
Cechowska-Pasko M: Low-glucose medium induces ORP150 expression and
exerts inhibitory effect on apoptosis and senescence of human
breast MCF7 cells. Acta Biochim Pol. 60:167–173. 2013.PubMed/NCBI
|
|
17
|
Tang Z, Yuan S, Hu Y, Zhang H, Wu W, Zeng
Z, Yang J, Yun J, Xu R and Huang P: Over-expression of GAPDH in
human colorectal carcinoma as a preferred target of 3-bromopyruvate
propyl ester. J Bioenerg Biomembr. 44:117–125. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang Y, Cao W, Yu Z and Liu Z:
Downregulation of a mitochondria associated protein SLP-2 inhibits
tumor cell motility, proliferation and enhances cell sensitivity to
chemotherapeutic reagents. Cancer Biol Ther. 8:1651–1658. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hsieh TY, Matsumoto M, Chou HC, Schneider
R, Hwang SB, Lee AS and Lai MM: Hepatitis C virus core protein
interacts with heterogeneous nuclear ribonucleoprotein K. J Biol
Chem. 273:17651–17659. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cui H, Wu F, Sun Y, Fan G and Wang Q:
Up-regulation and subcellular localization of hnRNP A2/B1 in the
development of hepatocellular carcinoma. BMC cancer. 10:3562010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tavakoli J, Miar S, Zadehzare MM and
Akbari H: Evaluation of effectiveness of herbal medication in
cancer care: A review study. Iran J Cancer Prev. 5:144–156.
2012.PubMed/NCBI
|
|
22
|
Malek SN, Phang CW, Ibrahim H, Norhanom AW
and Sim KS: Phytochemical and cytotoxic investigations of Alpinia
mutica rhizomes. Molecules. 16:583–589. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Saetung A, Itharat A, Dechsukum C,
Wattanapiromsakul C, Keawpradub N and Ratanasuwan P: Cytotoxic
activity of Thai medicinal plants for cancer treatment.
Songklanakarin J Sci Technol. 27(Suppl 2): S469–S478. 2005.
|
|
24
|
Prommee N, Prajuabjinda O and Itharat A:
In vitro cytotoxic, antioxidant and antimicrobial activities of
Derris Scandens. Proceedings of the First Conference on Graduate
Student Network of Thailand. Pathumthani, Bangkok; 2012
|
|
25
|
Li Y, Kong D, Bao B, Ahmad A and Sarkar
FH: Induction of cancer cell death by isoflavone: The role of
multiple signaling pathways. Nutrients. 3:877–896. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zito E, Melo EP, Yang Y, Wahlander Å,
Neubert TA and Ron D: Oxidative protein folding by an endoplasmic
reticulum-localized peroxiredoxin. Mol Cell. 40:787–797. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yi N, Xiao MB, Ni WK, Jiang F, Lu CH and
Ni RZ: High expression of peroxiredoxin 4 affects the survival time
of colorectal cancer patients, but is not an independent
unfavorable prognostic factor. Mol Clin Oncol. 2:767–772. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim TH, Song J, Kim SH, Parikh AK, Mo X,
Palanichamy K, Kaur B, Yu J, Yoon SO, Nakano I and Kwon CH:
Piperlongumine treatment inactivates peroxiredoxin 4, exacerbates
endoplasmic reticulum stress, and preferentially kills high-grade
glioma cells. Neuro Oncol. 16:1354–1364. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Stojadinovic A, Hooke JA, Shriver CD,
Nissan A, Kovatich AJ, Kao TC, Ponniah S, Peoples GE and Moroni M:
HYOU1/Orp150 expression in breast cancer. Med Sci Monit.
13:BR231–BR239. 2007.PubMed/NCBI
|
|
30
|
Asahi H, Koshida K, Hori O, Ogawa S and
Namiki M: Immunohistochemical detection of the 150-kDa
oxygen-regulated protein in bladder cancer. BJU Int. 90:462–466.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhou Y, Liao Q, Li X, Wang H, Wei F, Chen
J, Yang J, Zeng Z, Guo X, Chen P, et al: HYOU1, regulated by
LPLUNC1, is up-regulated in nasopharyngeal carcinoma and associated
with poor prognosis. J Cancer. 7:367–376. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Schönthal AH: Pharmacological targeting of
endoplasmic reticulum stress signaling in cancer. Biochem
Pharmacol. 85:653–666. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang JY, Zhang F, Hong CQ, Giuliano AE,
Cui XJ, Zhou GJ, Zhang GJ and Cui YK: Critical protein GAPDH and
its regulatory mechanisms in cancer cells. Cancer Biol Med.
12:10–22. 2015.PubMed/NCBI
|
|
34
|
Hjerpe E, Egyhazi Brage S, Carlson J,
Frostvik Stolt M, Schedvins K, Johansson H, Shoshan M and
Avall-Lundqvist E: Metabolic markers GAPDH, PKM2, ATP5B and
BEC-index in advanced serous ovarian cancer. BMC Clin Pathol.
13:302013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Puzone R, Savarino G, Salvi S, Dal Bello
MG, Barletta G, Genova C, Rijavec E, Sini C, Esposito AI, Ratto GB,
et al: Glyceraldehyde-3-phosphate dehydrogenase gene over
expression correlates with poor prognosis in non small cell lung
cancer patients. Mol Cancer. 12:972013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hao L, Zhou X, Liu S, Sun M, Song Y, Du S,
Sun B, Guo C, Gong L, Hu J, et al: Elevated GAPDH expression is
associated with the proliferation and invasion of lung and
esophageal squamous cell carcinomas. Proteomics. 15:3087–3100.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xiao B, Xie Z, Guo L, Wu J and Zhang H:
Stomatin-like protein 2 expression is associated with clinical
survival in patients with cervical cancer. Int J Clin Exp Pathol.
8:1804–1809. 2015.PubMed/NCBI
|
|
38
|
Geuens T, Bouhy D and Timmerman V: The
hnRNP family: Insights into their role in health and disease. Hum
Genet. 135:851–867. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lu J and Gao FH: Role and molecular
mechanism of heterogeneous nuclear ribonucleoprotein K in tumor
development and progression. Biomed Rep. 4:657–663. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tang F, Li W, Chen Y, Wang D, Han J and
Liu D: Downregulation of hnRNP K by RNAi inhibits growth of human
lung carcinoma cells. Oncol Lett. 7:1073–1077. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xiao Z, Ko HL, Goh EH, Wang B and Ren EC:
hnRNP K suppresses apoptosis independent of p53 status by
maintaining high levels of endogenous caspase inhibitors.
Carcinogenesis. 34:1458–1467. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen LC, Chung I, Hsueh C, Tsang NM, Chi
LM, Liang Y, Chen CC, Wang LJ and Chang YS: The antiapoptotic
protein, FLIP, is regulated by heterogeneous nuclear
ribonucleoprotein K and correlates with poor overall survival of
nasopharyngeal carcinoma patients. Cell Death Differ. 17:1463–1473.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Qu XH, Liu JL, Zhong XW, Li X and Zhang
QG: Insights into the roles of hnRNP A2/B1 and AXL in non-small
cell lung cancer. Oncol Lett. 10:1677–1685. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gu WJ and Liu HL: Induction of pancreatic
cancer cell apoptosis, invasion, migration, and enhancement of
chemotherapy sensitivity of gemcitabine, 5-FU, and oxaliplatin by
hnRNP A2/B1 siRNA. Anticancer Drugs. 24:566–576. 2013.PubMed/NCBI
|