Introduction
Glioblastoma multiforme is the most common type of
malignant primary brain tumor in adults (1). Mean progression-free survival is just
over 6 months; treatment with surgical resection, chemotherapy and
radiation is invariably followed by tumor recurrence (2). Despite the improvement in outcomes with
this combined chemoradiotherapy approach, few patients survive
beyond 5 years (3). Therefore, future
studies on the molecular mechanism of the disease may provide the
theoretical basis to identify new targets for effective
therapies.
Cytochrome P450, family 27, subfamily A, polypeptide
1 (CYP27A1), also termed CTX\CP27\CYP27 is a ubiquitously expressed
mitochondrial enzyme belonging to the cytochrome P450 family
(4). CYP27A1 catalyzes the
hydroxylation of cholesterol at C-27 to form 27-hydroxycholesterol
and cholestenoic acid (5). A single
serum measurement of 27-hydroxycholesterol can reliably estimate
average levels over a one-year period (6). The role of CYP27A1 in bile acid
synthesis in the liver is well established; it catalyzes the
initial, rate-limiting step in the alternative bile acid synthetic
pathway and the intermediate step in the classic bile acid
synthetic pathway (7,8). CYP27A1 is strongly expressed in
macrophages within human benign and malignant mammary tissue
(4). Breast cancer cell intrinsic
expression of CYP27A1 protein is associated with tumor grade
(4). However, its roles remain
unclear in glioblastoma.
MicroRNAs (miRNAs/miRs) are a class of small RNA
found in a diverse range of eukaryotes, including animals, plants
and DNA viruses, which range in size between 19 and 24 nucleotides
(nts) (9). Aberrant miRNA expression
has been linked to glioblastoma (10). miR-204, a direct negative regulator of
ezrin gene expression, inhibits glioma cell migration and invasion
(11). However, studies continue to
emerge with an improved understanding on the mechanism of miR-204
as a tumor suppressive gene in glioblastoma. In the present study,
it was observed that CYP27A1 overexpression promoted proliferation,
while silencing of CYP27A1 inhibited proliferation in glioblastoma
cells, without affecting migration and invasion. It was revealed
that CYP27A1 was upregulated in glioblastoma tissues, indicating
that CYP27A1 may be an oncogene. The downregulation of specific
miRNAs may contribute to the upregulation of oncogenes in
glioblastoma (12). A common strategy
was used to predict target miRNAs of CPY27A1 using the miRanda
algorithm. It was revealed that miR-211 and miR-204 could target
the 3′untranslated region (UTR) of CPY27A1 mRNA. Overexpressing
miR-204 inhibited CPY27A1 expression in glioblastoma cells. Lastly,
the present study demonstrated that miR-204 was downregulated in
glioblastoma and its overexpression inhibited proliferation,
migration and invasion in glioblastoma cells. Therefore, miR-204
functions as a tumor suppressor gene, at least partly by
suppressing CYP27A1 expression in glioblastoma.
Materials and methods
Glioblastoma tissues, cells,
CYP27A1-expressing plasmids and short hairpin (sh) CYP27A1
plasmids
A total of 7 Chinese women patients with
glioblastoma were recruited from the Department of Neurosurgery,
Yishui Central Hospital (Shandong, China). The mean age was 56
years (range, 31–78 years). All tissues were examined
histologically, and pathologists confirmed the diagnosis. The
present study was approved by the medical ethics committee of
Yishui Central Hospital (Shandong, China). Written informed consent
was obtained from each individual. The human glioblastoma U87MG
cell line was purchased from the Cell Bank of the Chinese Academy
of Sciences (Beijing, China). CYP27A1-expressing plasmids/pcDNA3.1
(pcDNA3.1) and shCYP27A1/scramble were purchased from Tiangen
Biotech Co., Ltd. (Shanghai, China).
Protein extraction and western blot
analysis
Following transfection, cells and tissues were lysed
for 48 h using RIPA Lysis and Extraction Buffer (Beijing Solarbio
Science & Technology Co., Ltd., Beijing, China), and the
protein concentration was measured using Pierce™ BCA Protein Assay
kit (Thermo Fisher Scientific, Inc.). Following heating at 100°C
for 10 min in the presence of a loading buffer, equal amounts of
protein lysates (50 µg) were separated using 10% SDS-PAGE (Bio-West
Inc., Logan, UT, USA) at 100 V for 1 h, and transferred onto
Invitrogen nitrocellulose membranes (Thermo Fisher Scientific,
Inc.) at 120 V for 1 h. Following blocking with 5% skimmed milk
(diluted with PBS), the membranes were incubated overnight at 4°C
with the following primary antibodies: CYP27A1 (ab126785; 1:500;
Abcam, Cambridge, MA, USA), c-myc (ab32072; 1:500; Abcam), RB
(ab181616; 1:500; Abcam), Ki-67 (ab15580; 1:500; Abcam), CDK2
(ab32147; 1:500; Abcam), p21 (ab109520; 1:500; Abcam), p53
(ab32049; 1:500; Abcam), PDCD4 (ab51495; 1:500; Abcam), SOX2
(ab92494; 1:500; Abcam), β-actin (ab8227; 1:500; Abcam).
Subsequently, the membranes were incubated with secondary goat
monoclonal (RMG01) to rabbit IgG Fab region (Biotinylated;
ab222772; 1:10,000; Abcam) at room temperature for 2 h, and
proteins were detected using enhanced chemiluminescence (Pierce™
ECL Western Blotting Substrate; Thermo Fisher Scientific,
Inc.).
MTT assay
A total of 5×103 cells were seeded onto
96-well plates and transfected with CYP27A1 expressing plasmids or
empty vector at 37°C for 24 h.
Subsequent to transfection, MTT (5 mg/ml;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was added to the
wells (20 µl/well) and incubated for 4 h at 37°C, and then the
supernatants were removed and 2% dimethyl sulfoxide was added to
each well. The absorbance of each sample was measured using a UV
spectrophotometer at the wavelength of 490 nm.
Bromodeoxyuridine (BrdU) labeling and
immunofluorescence
Cells grown on coverslips (Thermo Fisher Scientific,
Inc., Waltham, MA, USA) were incubated with BrdU at 37°C for 1 h
and stained with anti-BrdU antibody (1:200; cat. no. ab220076;
Abcam) according to the manufacturer's protocol. For
immunofluorescence analysis, cells were plated on glass coverslips.
Following transfection, cells were exposed to anti-CYP27A1 antibody
(1:250; ab198970; Abcam) overnight at 4°C and goat anti-rabbit
secondary antibody (1:500; ab150079; Abcam). Coverslips were
counterstained with DAPI (Molecular Probes; Thermo Fisher
Scientific, Inc.) to detect cell nuclei. Microscopic analysis was
performed with a confocal laser-scanning microscope (Leica
Microsystems, Bensheim, Germany). Fluorescence intensities were
measured in three randomly selected viewing areas for 200~300
cells/coverslip and analyzed using ImageJ (version 1.37v; National
Institutes of Health, Bethesda, MA, USA).
Migration and invasion assay
Cell suspension (total cells, 3×104) in
medium without fetal bovine serum (FBS) was plated into the top
chamber with 8-µm pore sized filter inserts of Transwell plates
(24-well; Costar, Cambridge, MA, USA). For the Transwell migration
assay, the top surface of filter membranes was not coated with
Matrigel, while the top surface of filter membranes was coated with
Matrigel matrix (BD Biosciences, San Jose, CA, USA) in the Matrigel
invasion assay. Medium containing 10% FBS (Sigma-Aldrich; Merck
KGaA) was added to the bottom chambers in the two assays. The cells
were incubated for 24 h at 37°C, and then the non-migrated or
non-invaded cells on top surface of the membrane were removed with
a cotton swab, the cells under the filter membrane were fixed with
4% paraformaldehyde for 10 min at room temperature and stained with
0.2% crystal violet for 10 min at room temperature (Sigma-Aldrich;
Merck KGaA).
Methods of bioinformatics
The analysis of potential microRNA target sites was
performed using the commonly used miRanda prediction algorithm
(http://www.microrna.org/).
Reverse transcription-polymerase chain
reaction (RT-PCR)
Extraction of total RNA and detection of the mature
form of miRNAs was performed with the mirVanami RNA Isolation kit
(Ambion; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Template DNA was obtained from Dr Xin Shao
(Department of Medicine, Jiujiang College, Jiujiang, China). Taq
DNA polymerase was purchased from Promega Corporation (Madison, WI,
USA). β-actin was used as an experimental control gene. SYBR green
(Takara Biotechnology Co., Ltd., Dalian, China) was used as a
fluorophore. Quantification was performed using the
2−ΔΔCq method (13).
Luciferase (Luc) assays
Total RNA was isolated from the U87MG cells using
TRIzol reagent (cat no. 101472; Invitrogen; Thermo Fisher
Scientific, Inc.). cDNA was synthesized from 1 µg of total RNA in a
20 µl reverse transcription system followed by PCR amplification in
a 50 µl PCR system performed using an RT-PCR kit (cat no. A3500;
Promega Corporation). The 3′UTR of CYP27A1 was amplified [10X
buffer 3 ul, MgCl2 (25 mM) 3 ul, dNTP (25 mM) 0.36 ul,
forward primer (10 µM) 1 ul, reverse primer (10 µM) 1 µl, Taq
enzyme (5 U/µl) 0.3 µl, ddH2O 19.34 µl and cDNA 2 µl;
95°C for 2 min; 95°C for 30 sec; 55°C for 60 sec; 72°C for 30 sec]
using cDNA from U87MG cells, cloned into pRL-TK (Promega
Corporation), checked for orientation, sequenced and termed
Luc-CYP27A1-wild-type (WT). For reporter assays, U87MG cells were
transiently transfected with WT-reporter plasmid and precursor
(pre)-miR-204 using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). Reporter assays were performed 36 h
post-transfection using the dual-luciferase assay-system (Promega
Corporation), normalized for transfection efficiency by
co-transfected Renilla-luciferase.
Northern blot analysis
Northern blot analysis of miRNAs was performed as
described previously (14). Probes
were labeled with (γ-32P) ATP complementary to miR-204 and U6 small
nuclear RNA.
Statistical analysis
All experimental data are presented as the mean ±
standard error, with the number of independent experiments (n=3),
and were analyzed using Student's t-test. P<0.05 was considered
to indicate a statistically significant difference. Analysis was
performed using GraphPad Prism software (version 5.0; GraphPad
Software, Inc., La Jolla, CA, USA).
Results
CYP27A1 promotes proliferation of
glioblastoma cells
To investigate whether CYP27A1 could promote the
proliferation of glioblastoma U87MG cells, firstly using western
blot analysis, it was examined whether CYP27A1-expressing plasmids
could stably express CYP27A1 protein in U87MG cells. The results
demonstrated that CYP27A1 protein could be significantly increased
by CYP27A1-expressing plasmids in the cells (Fig. 1A). In order to identify the effect of
CYP27A1 on proliferation, an MTT assay was performed. The results
demonstrated that overexpression of CYP27A1 significantly
upregulated the proliferation rate of U87MG cells following
transfection (Fig. 1B). To show the
effects of CYP27A1 on proliferation, a BrdU incorporation assay was
performed to analyze its effects on DNA synthesis. The results
confirmed that CYP27A1 significantly promoted DNA synthesis in the
cells, and representative micrographs and quantification of BrdU
incorporating-cells following transfection with CYP27A1 or empty
vector (mock) are shown (Fig. 1C).
Subsequently, western blot analysis was performed to identify
whether proliferation-associated markers were affected by CYP27A1
in the cells. The results of western blot analysis revealed that
c-myc, sex determining region Y-box 2 (SOX2), Ki-67 and
cyclin-dependent kinase 2 were promoted and p21, p53and programmed
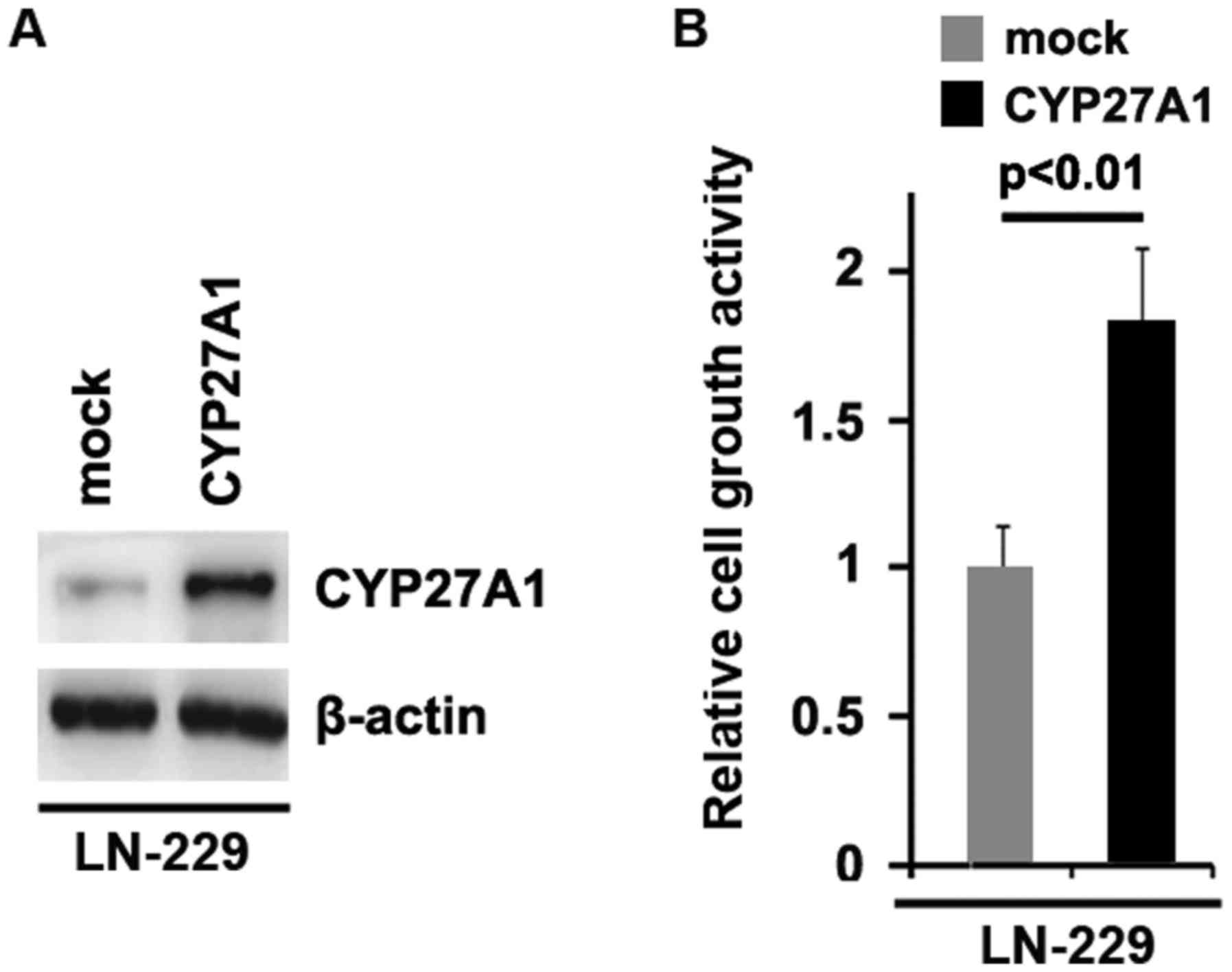
cell death protein 4 (PDCD4) were inhibited by CYP27A1 (Fig. 1D). Meanwhile, it was observed that
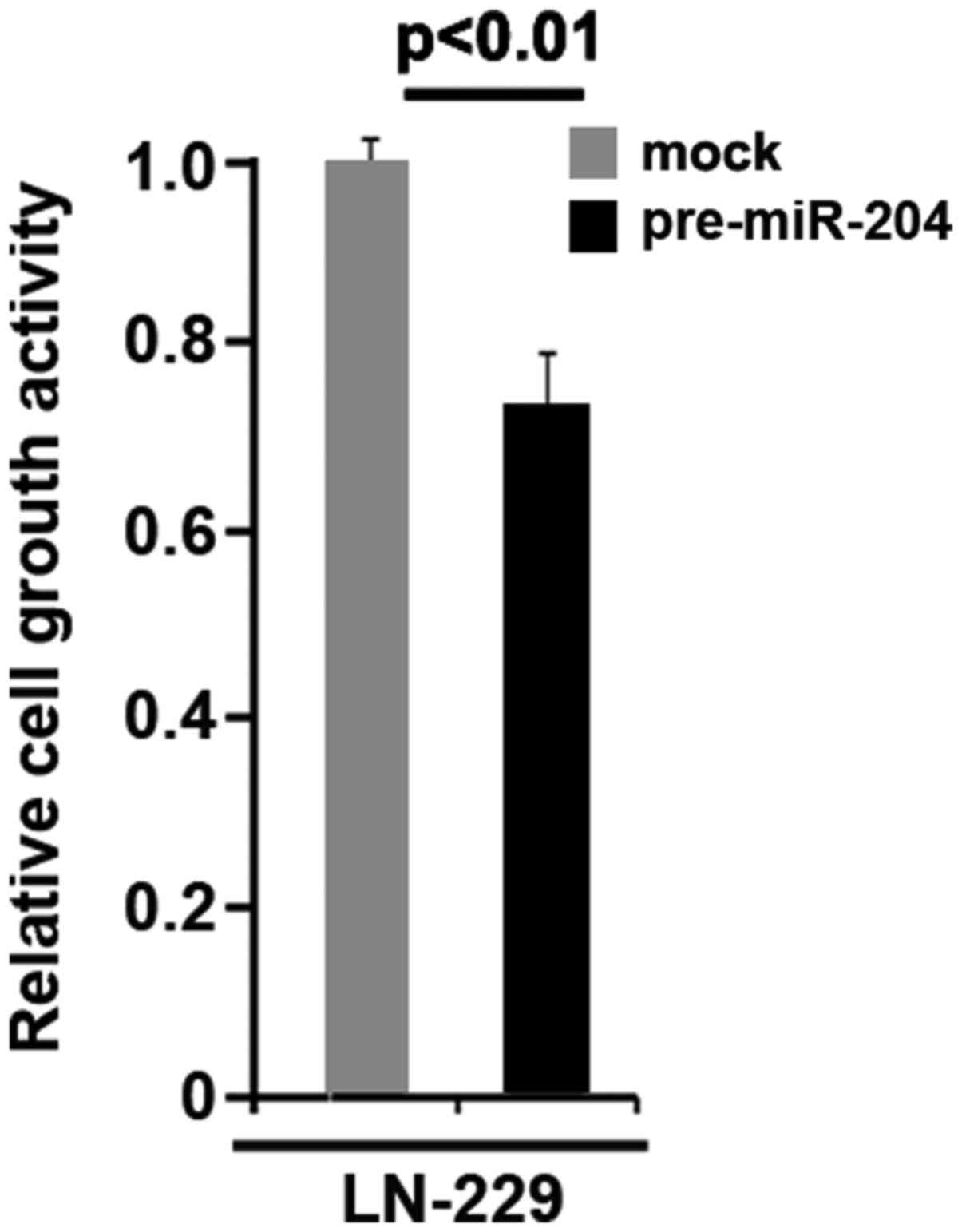
CYP27A1 protein was increased by CYP27A1 expression plasmids
(Fig. 2A) and overexpression of
CYP27A1 promoted proliferation in LN-229 glioblastoma cells
(Fig. 2B). These data supported that
CYP27A1 promoted proliferation in glioblastoma cells.
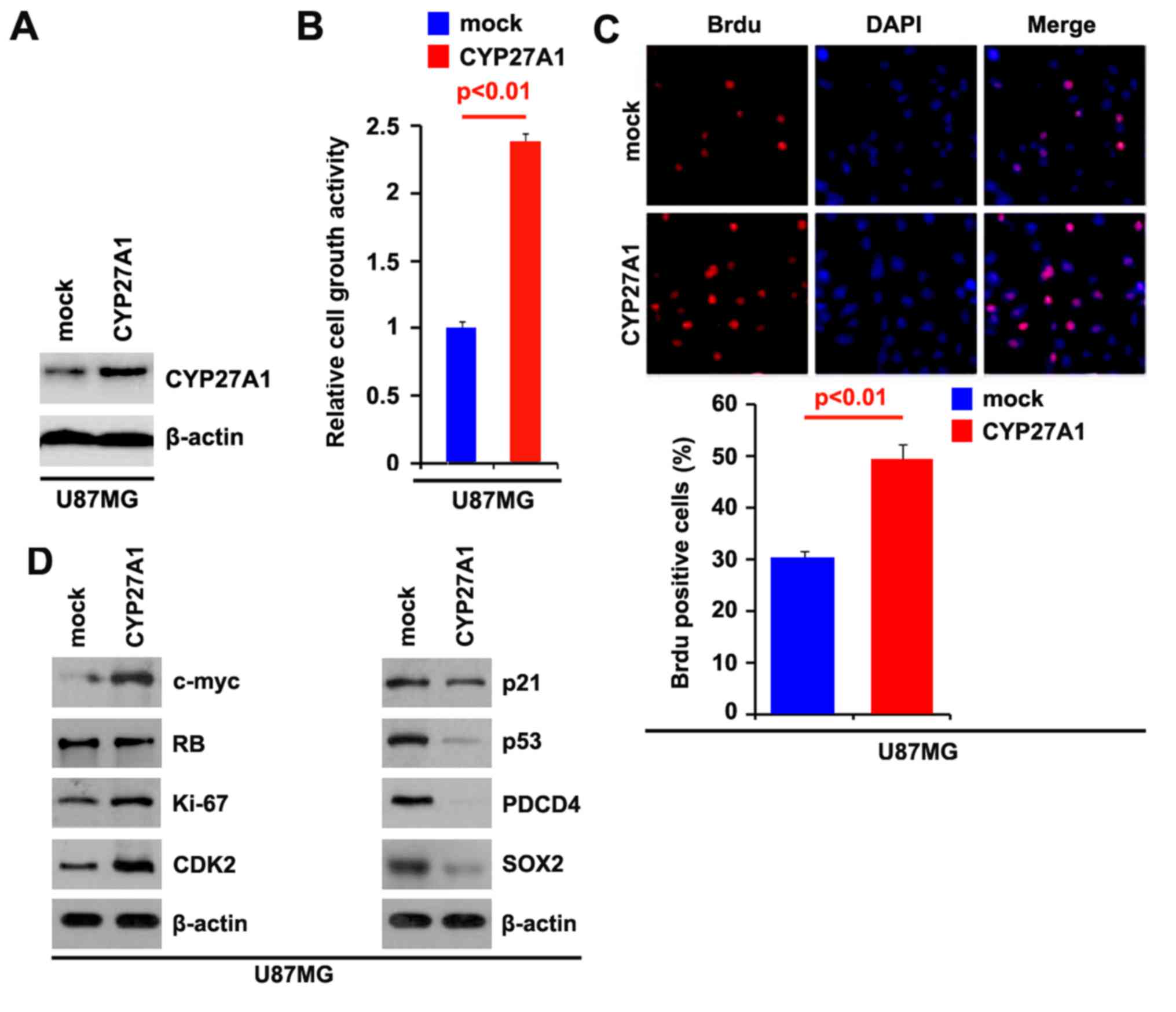 | Figure 1.CYP27A1 promotes proliferation in
glioblastoma U87MG cells. (A) Western blot analysis for CYP27A1 in
U87MG cells. U87MG cells were transfected with CYP27A1-expressing
plasmids or the empty vector (mock). β-actin was used as the
loading control. n=3. (B) MTT assay for U87MG cells. U87MG cells
were transfected with CYP27A1-expressing plasmids or the empty
vector (mock) and cellular viability was then measured at the
indicated time points using an MTT assay. n=3. (C) BrdU
incorporation assay for U87MG cells. Representative micrographs and
quantification of BrdU incorporating-cells following transfection
with CYP27A1-expressing plasmids or the empty vector (mock). n=3.
(D) Western blot analysis for c-myc, RB, Ki67, CDK2, p21, p53,
PDCD4 and SOX2 in U87MG cells transfected with CYP27A1-expressing
plasmids or the empty vector (mock). β-actin was used as the
loading control. n=3. Error bars indicate standard error of the
mean. CYP27A1, cytochrome P450, family 27, subfamily A, polypeptide
1; BrdU, bromodeoxyuridine; RB, retinoblastoma; CDK2,
cyclin-dependent kinase 2; PDCD4, programmed cell death protein 4;
SOX2, sex determining region Y-box 2. |
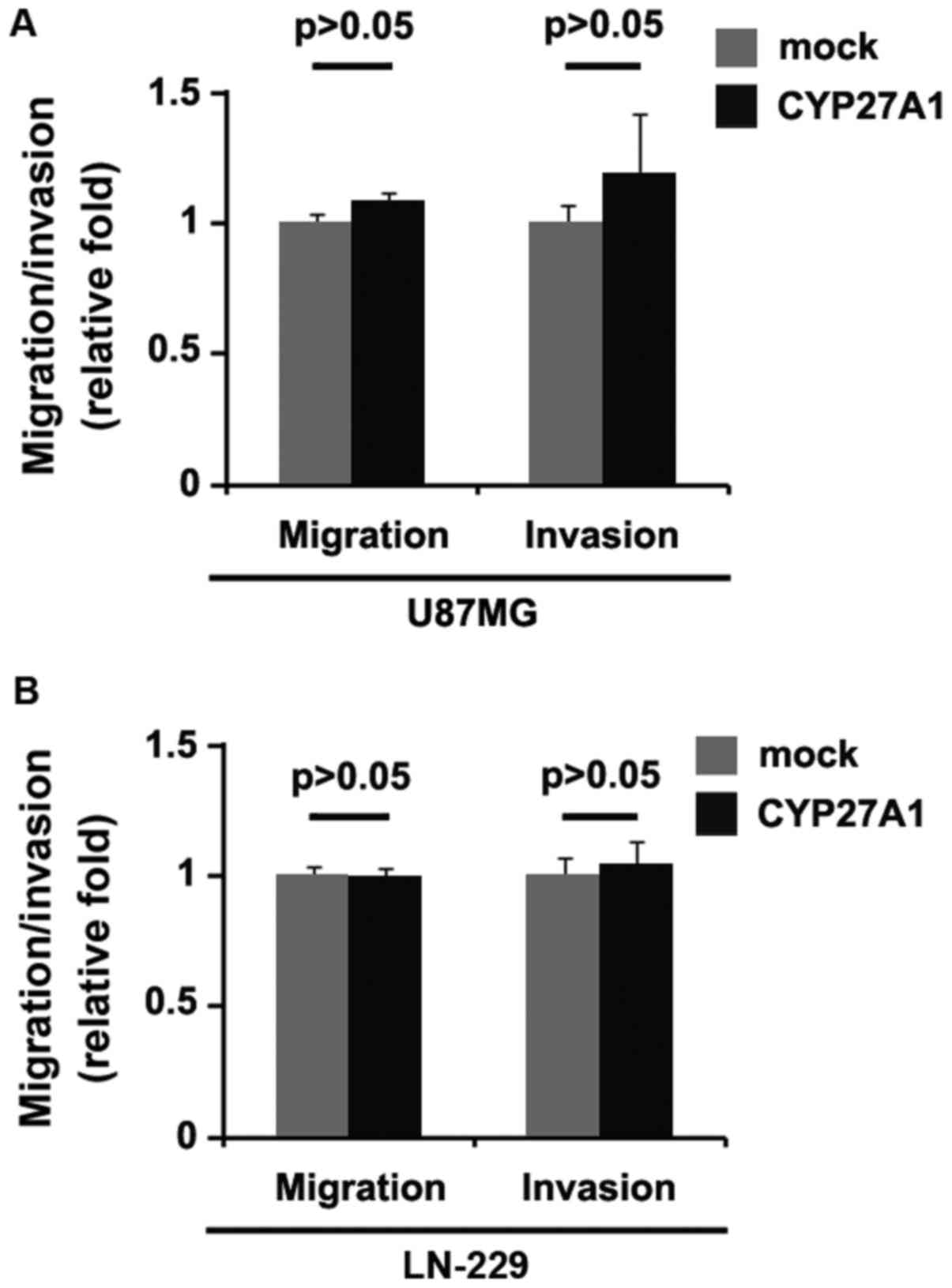
CYP27A1 overexpression does not affect
migration and invasion in glioblastoma cells
Considering that CYP27A1 evidently promoted U87MG
cellular proliferation, it was then determined whether CYP27A1 has
an impact on the migration and invasion of glioblastoma cells. The
migration and invasion assay results demonstrated that the
overexpression of CYP27A1 did not affect migration and invasion in
U87MG and LN-229 cells (Fig. 3A and
B).
Silencing CYP27A1 inhibits
proliferation in glioblastoma cells
It was demonstrated that CYP27A1 overexpression
promoted proliferation in U87MG and LN-229 cells. To provide
additional evidence that CYP27A1 is involved in the proliferation
of glioblastoma cells, the effects of an inhibitor of CYP27A1,
shCYP27A1, were studied. Following stable transfection, CYP27A1
expression was detected by western blot analysis. The results
demonstrated that exogenous shCYP27A1 significantly downregulated
CYP27A1 expression in U87MG cells (Fig.
4A). An MTT assay was performed to detect the proliferation of
U87MG cells transfected with shCYP27A1 and scramble. The results
demonstrated that shCYP27A1 inhibited proliferation in U87MG cells
compared with scramble-transfected groups, and that the inhibition
was dose-dependent (Fig. 4B). To
demonstrate the effects of silencing CYP27A1 on cellular
proliferation, BrdU incorporation assay was performed to detect DNA
synthesis in the cells. The results confirmed that shCYP27A1
significantly suppressed DNA synthesis in the cells and
representative micrographs and quantification of BrdU
incorporating-cells following transfection with shCYP27A1 or
scramble are presented (Fig. 4C).
Subsequently, western blot analysis was performed to identify
whether proliferation-associated markers were affected by shCYP27A1
in the cells. The results of western blot analysis demonstrated
that the proliferating cell nuclear antigen was downregulated and
p21, as well as p53, were upregulated by silencing CYP27A1
(Fig. 4D).
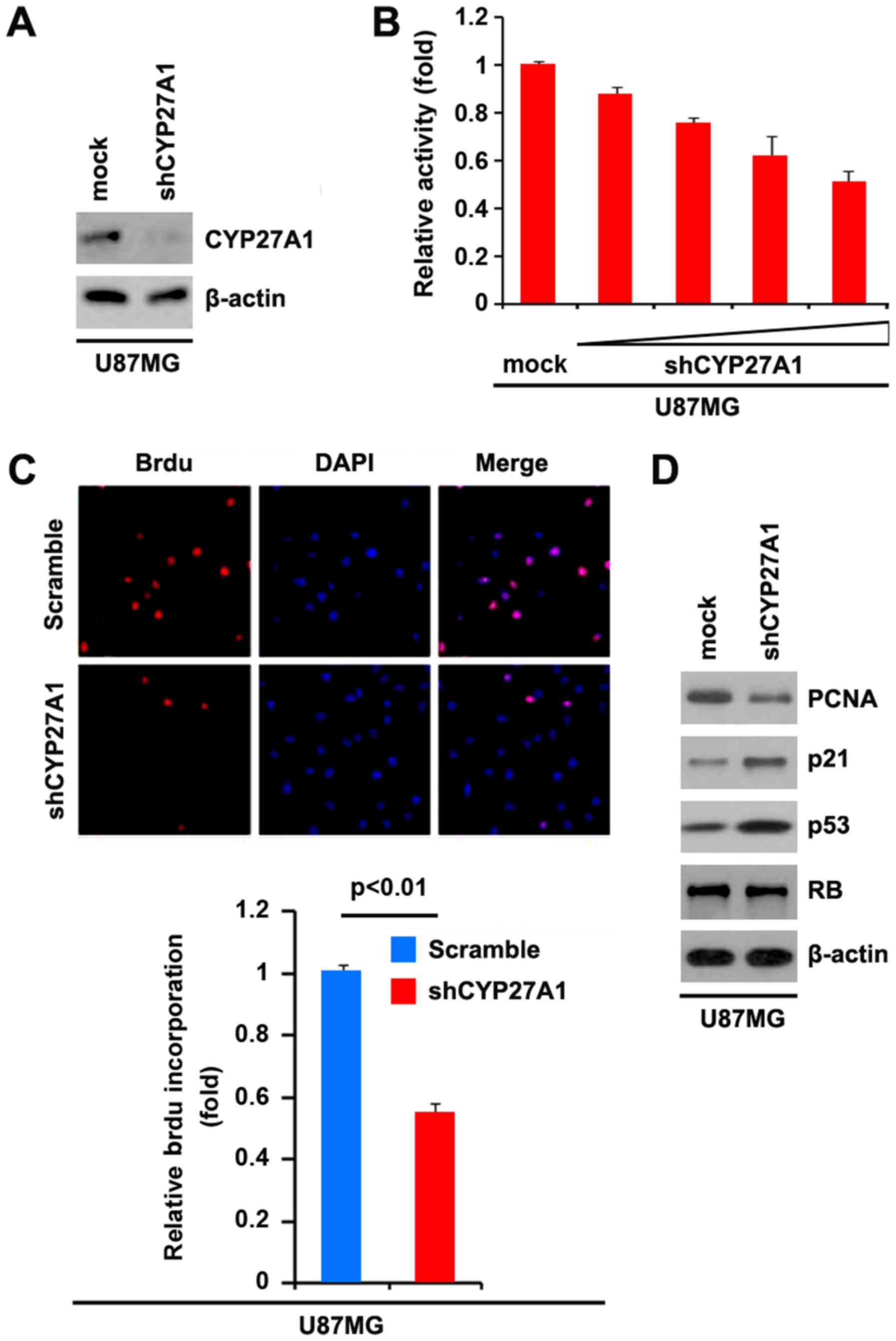 | Figure 4.Silencing CYP27A1 inhibits
proliferation in glioblastoma U87MG cells. (A) Western blot
analysis for CYP27A1 in U87MG cells infected with shCYP27A1 or
scramble. β-actin was used as the loading control. n=3. (B) MTT
assay for U87MG cells transfected with shCYP27A1 or scramble. n=3.
(C) BrdUincorporation assay for U87MG cells. Representative
micrographs and quantification of BrdU incorporating cells
following transfection with shCYP27A1 or scramble. n=3. (D) Western
blot analysis for PCNA, p21, p53 and RB in U87MG cells transfected
with shCYP27A1 or scramble. β-actin was used as the loading
control. n=3. Error bars indicate standard error of the mean.
CYP27A1, cytochrome P450, family 27, subfamily A, polypeptide 1;
BrdU, bromodeoxyuridine; sh, short hairpin; PCNA, proliferating
cell nuclear antigen; RB, retinoblastoma. |
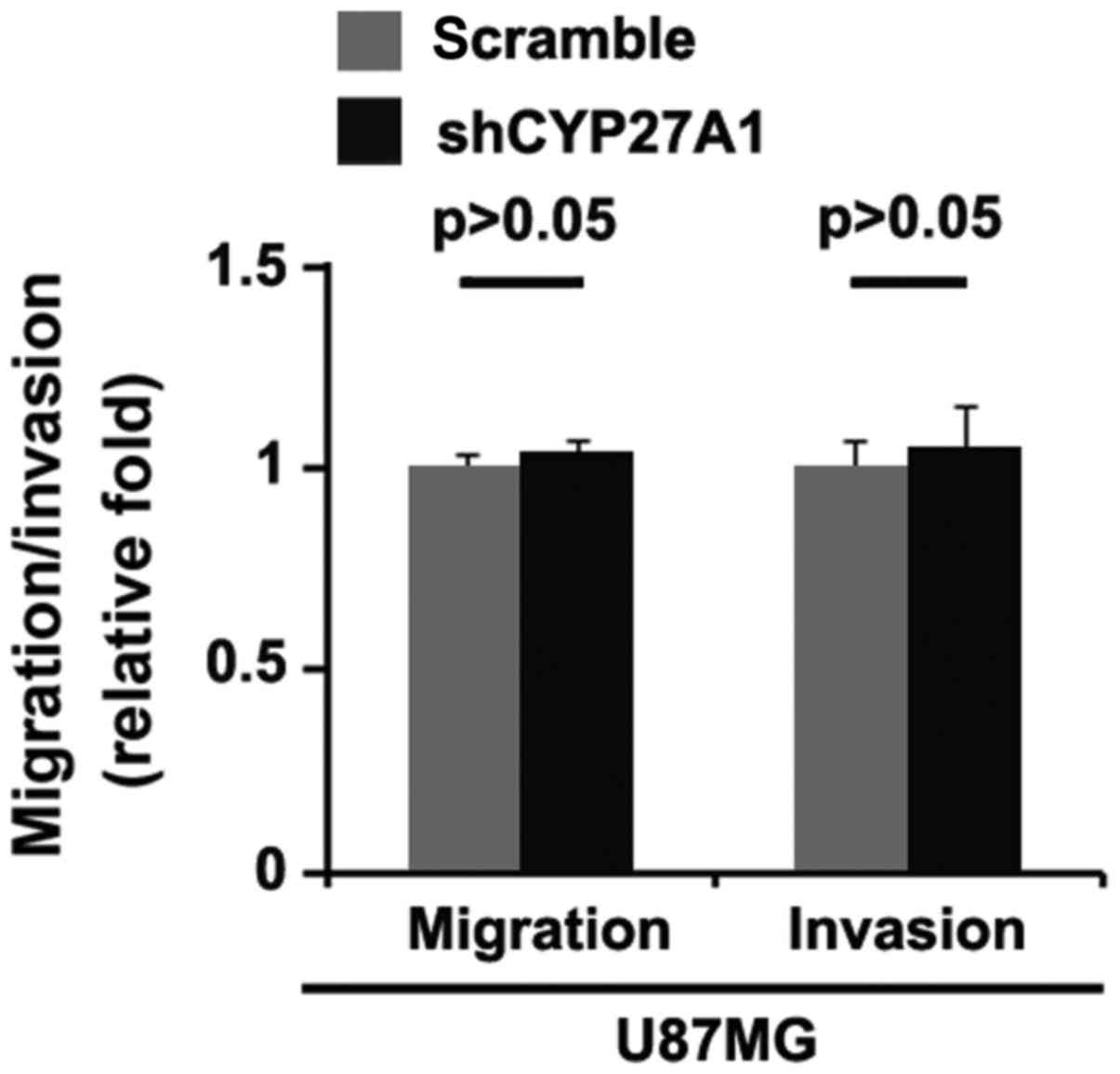
Silencing CYP27A1 does not affect
migration and invasion in glioblastoma cells
It was then determined whether silencing CYP27A1
would have any impact on migration and invasion in U87MG cells. The
migration and invasion assays revealed that silencing CYP27A1 did
not affect migration and invasion of them (Fig. 5).
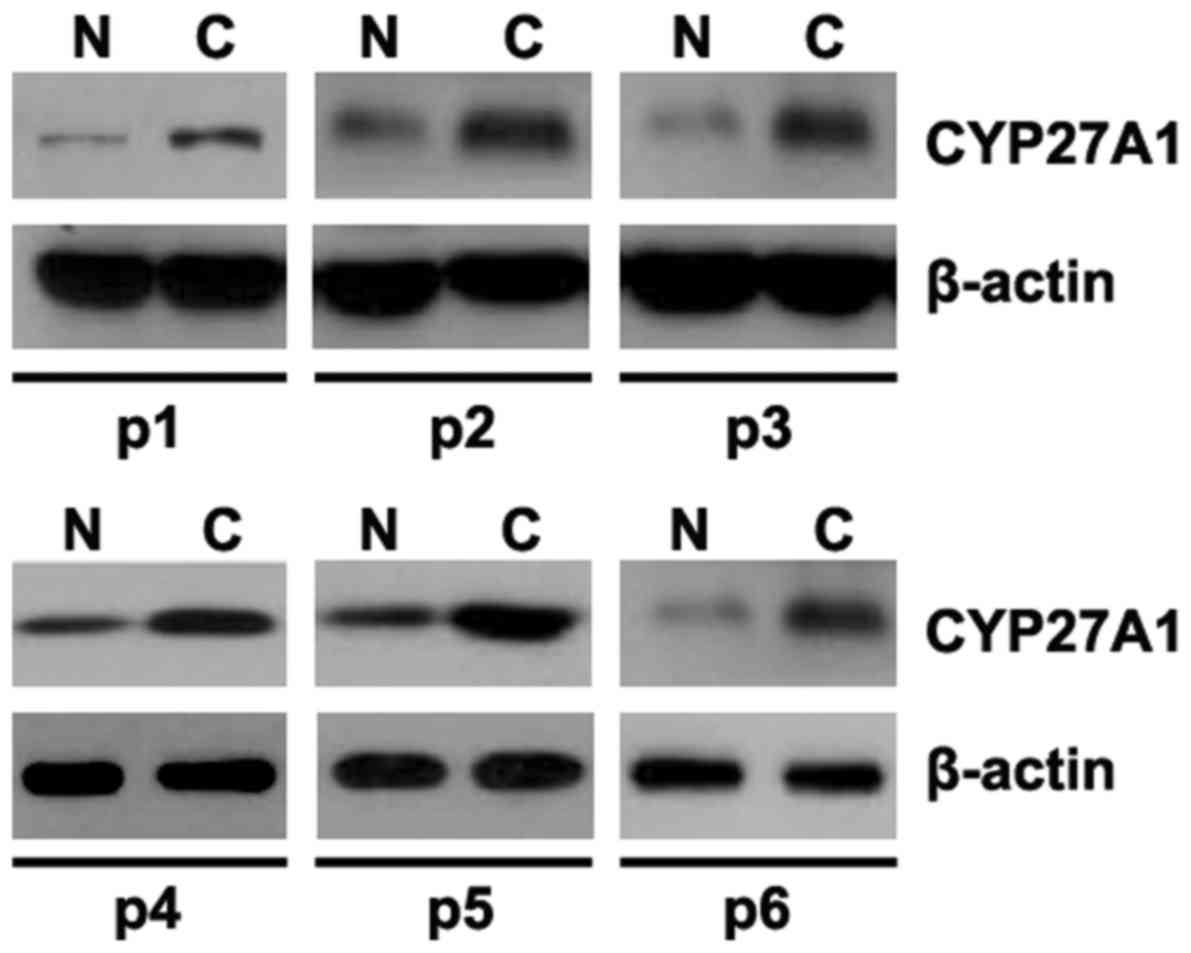
Aberrant expression of CYP27A1 in
glioblastoma tissues
In order to identify CYP27A1 protein expression in
glioblastoma tissues, western blot analysis was performed to detect
CYP27A1 protein between glioblastoma tissues and adjacent normal
tissues. It was revealed that CYP27A1 was increased in cancer
tissues of 6 patients, compared with adjacent normal tissues
(Fig. 6).
CYP27A1 is a target of miR-204 in
glioblastoma U87MG cells
Having demonstrated that CYP27A1 expression is
specifically upregulated in glioblastoma and it can promote
proliferation in glioblastoma cells, the mechanisms promoting
CYP27A1 expression in the disease were then studied. miRNAs are a
new class of small (~22 nt) noncoding RNAs, which negatively
regulate protein-coding gene expression by targeting mRNA
degradation or translation inhibition (15). Downregulation of specific miRNA may
contribute to oncogene overexpression (16). Thus, it was reasoned whether CYP27A1
was upregulated by defect of specific miRNA. To confirm this
reason, a commonly used prediction algorithm, miRanda (http://www.microrna.org/), was used to analyze the
3′UTR of CYP27A1.
The algorithm predicted that miR-204 and miR-221
could target the 3′UTR of CYP27A1 and the predicted target is shown
in Fig. 7A. To study the biological
function of miR-204, it was investigated whether miR-204 expression
could be increased by pre-miR-204 in U87MG cells. RT-PCR was
performed to detect miR-204 expression in the cells transfected
with pre-miR-204 and the results demonstrated that pre-miR-204
could significantly upregulate miR-204 expression in U87MG cells
(Fig. 7B). In order to identify that
CYP27A1 could be downregulated by miR-204, immunofluorescence and
western blot analyses were performed to study whether miR-204 could
affect CYP27A1 protein expression. Immunofluorescence analyses
demonstrated that CYP27A1 was significantly downregulated in U87MG
cells transfected with pre-miR-204 and representative micrographs,
and the quantification of immunofluorescence is shown in Fig. 7C. Consistent with immunofluorescence
analysis, the results of western blot analysis demonstrated that
CYP27A1 protein was significantly downregulated by miR-204 in U87MG
cells (Fig. 7D).
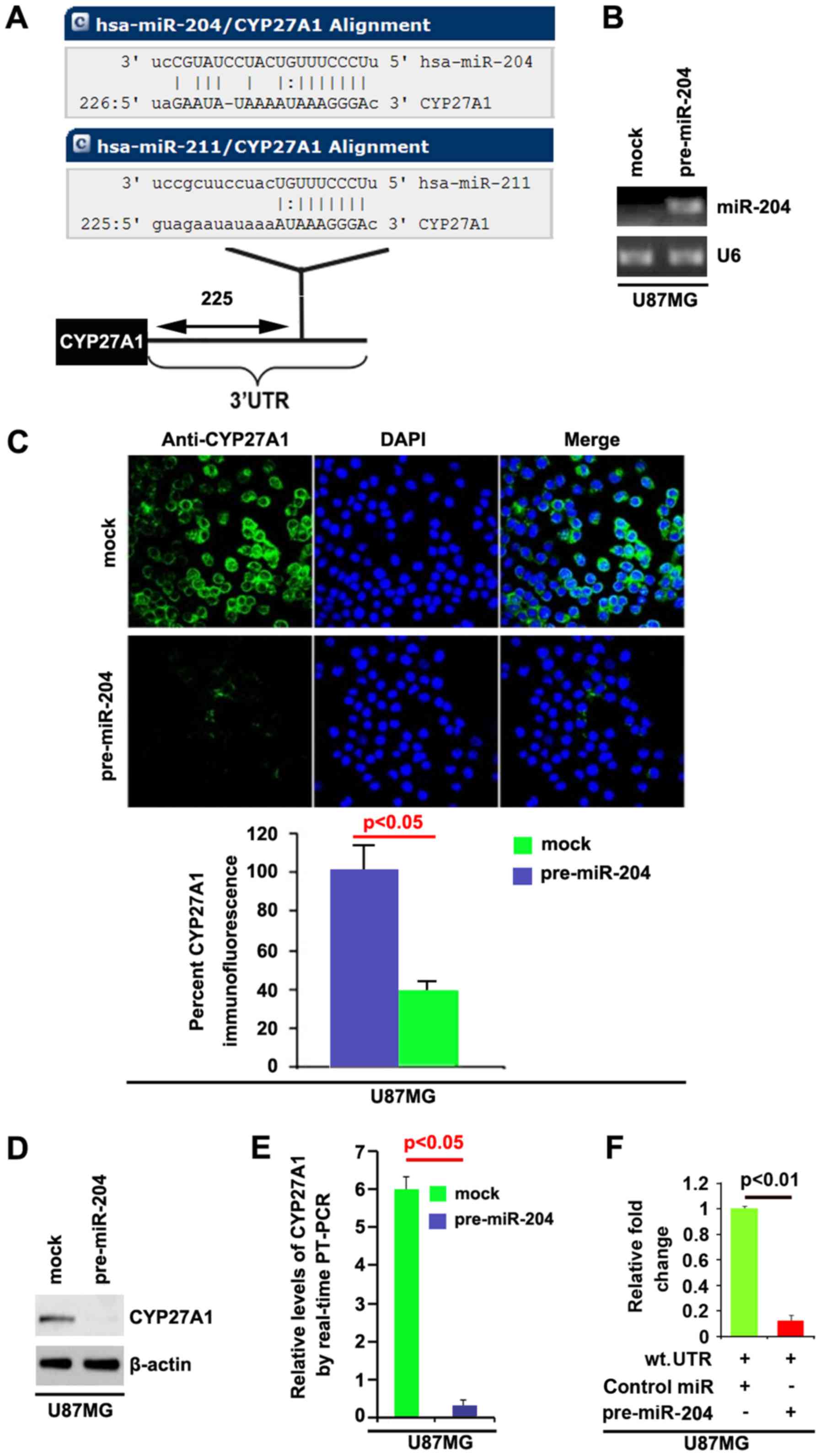 | Figure 7.CYP27A1 is a target of miR-204 in
glioblastoma U87MG cells. (A) Diagram demonstrating that CYP27A1 is
a target gene of miR-204 and miR-211, predicted by miRanda. (B)
Detection of miR-204 by RT-PCR in U87MG cells transfected with
precursor-miR-204 or the control miR (mock). (C) Immunofluorescence
analyses of U87MG cells transfected with pre-miR-204 or the control
miR (mock). Upper panel shows microscopic images of
immunofluorescence staining of one representative experiment
(magnification, ×100). Bottom panel shows graphic presentation of
mean fluorescence intensities of three independent experiments. (D)
Western blot analysis for CYP27A1 in U87MG cells. U87MG cells were
transfected with pre-miR-204 or control-miR (mock). β-actin was
used as the loading control. (E) RT-PCR for CYP27A1 in U87MG cells.
U87MG cells were transfected with pre-miR-204 or control-miR
(mock). GAPDH was a loading control. (F) Reporter assay, with
cotransfection of 500 ng WT-reporter and 50 nM control-miR, or
pre-miR-204 as indicated. 36 h after transfection, cells were
harvested for luciferase reporter assay. + represents existing
inserted fragments; - represents no existing inserted fragments;
n=3. Error bars indicate standard error of the mean. CYP27A1,
cytochrome P450, family 27, subfamily A, polypeptide 1; miR,
microRNA; RT-PCR, reverse transcription-polymerase chain reaction;
WT, wild-type; has, Homo sapiens; UTR, untranslated
region. |
In addition, RT-PCR was also performed to detect
CYP27A1 mRNA in the cells transfected with pre-miR-204. The results
demonstrated that miR-204 downregulated CYP27A1 mRNA level in U87MG
cells (Fig. 7E). To demonstrate the
direct regulation of CYP27A1 by miR-204, luciferase reporters with
the targeting sequences of wild-type (CYP27A1-wt-luc) were used to
detect whether miR-204 targets 3′UTR of CYP27A1 mRNA. A luciferase
assay was performed. The results demonstrated that miR-204
significantly inhibited CYP27A1-WT-luc plasmids in the cells
(Fig. 7F). Meanwhile, we observed
that miR-204 inhibits CYP27A1 expression in LN-229 glioblastoma
cells by targeting 3′UTR of CYP27A1 mRNA (Fig. 8). The results confirmed that miR-204
negatively regulates protein-coding gene CYP27A1 expression by
targeting its 3′UTR in glioblastoma cells.
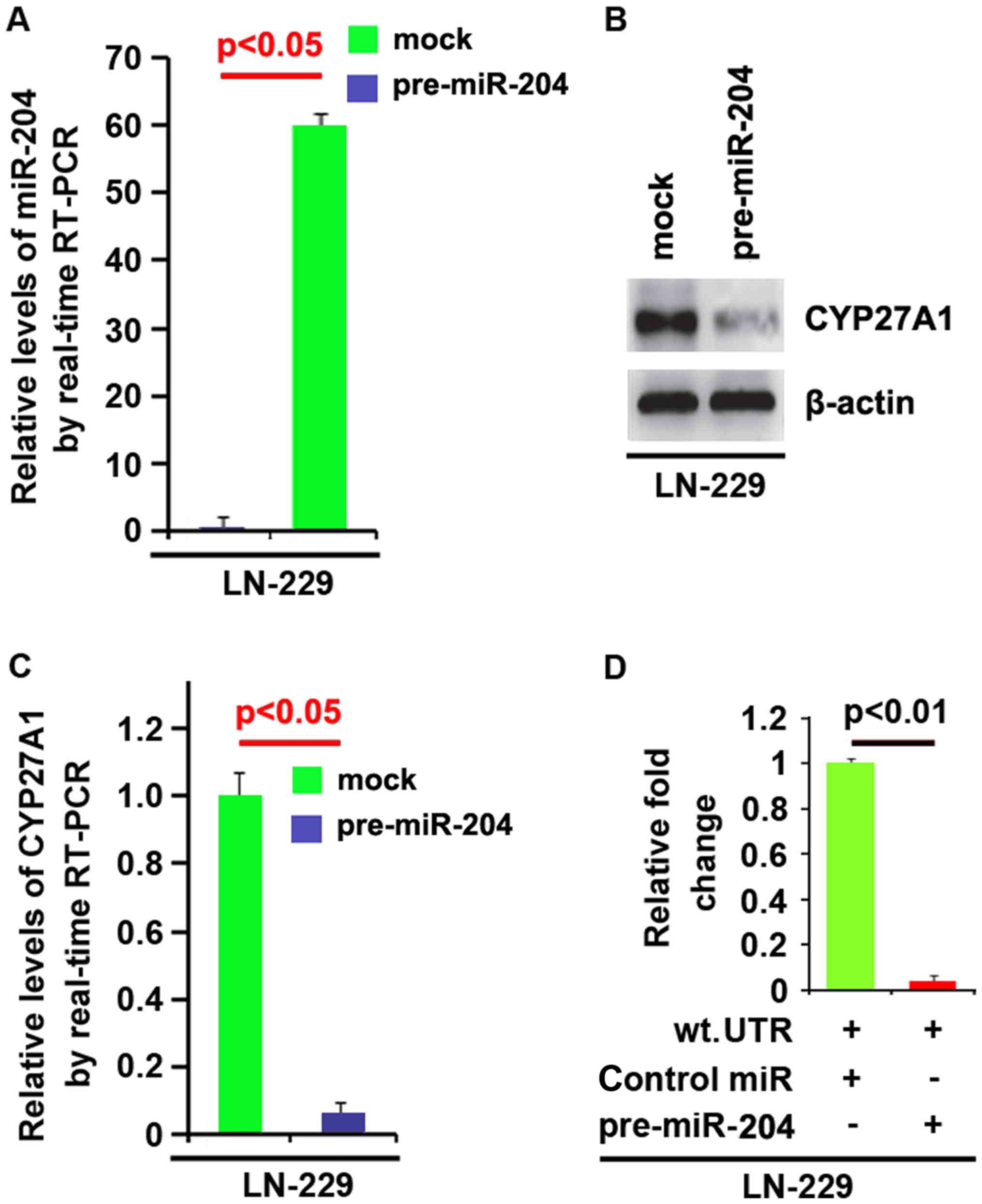 | Figure 8.CYP27A1 is a target of miR-204 in
glioblastoma LN-229 cells. (A) Detection of miR-204 by RT-PCR in
LN-229 cells transfected with pre-miR-204 (precursors miRNA) or the
control miR (mock). (B) Western blot analysis for CYP27A1 in LN-229
cells. LN-229 cells were transfected with pre-miR-204 or
control-miR (mock). β-actin was used as the loading control. (C)
RT-PCR for CYP27A1 in LN-229 cells. LN-229 cells were transfected
with pre-miR-204 or control-miR (mock). GAPDH was a loading
control. (D) Reporter assay, with cotransfection of 500 ng
WT-reporter and 50 nM control-miR, or pre-miR-204 as indicated. 36
h after transfection, cells were harvested for luciferase reporter
assay. + represents existing inserted fragments; - represents no
existing inserted fragments; n=3. Error bars indicate standard
error of the mean. CYP27A1, cytochrome P450, family 27, subfamily
A, polypeptide 1; miR, microRNA; RT-PCR, reverse
transcription-polymerase chain reaction; WT, wild-type. |
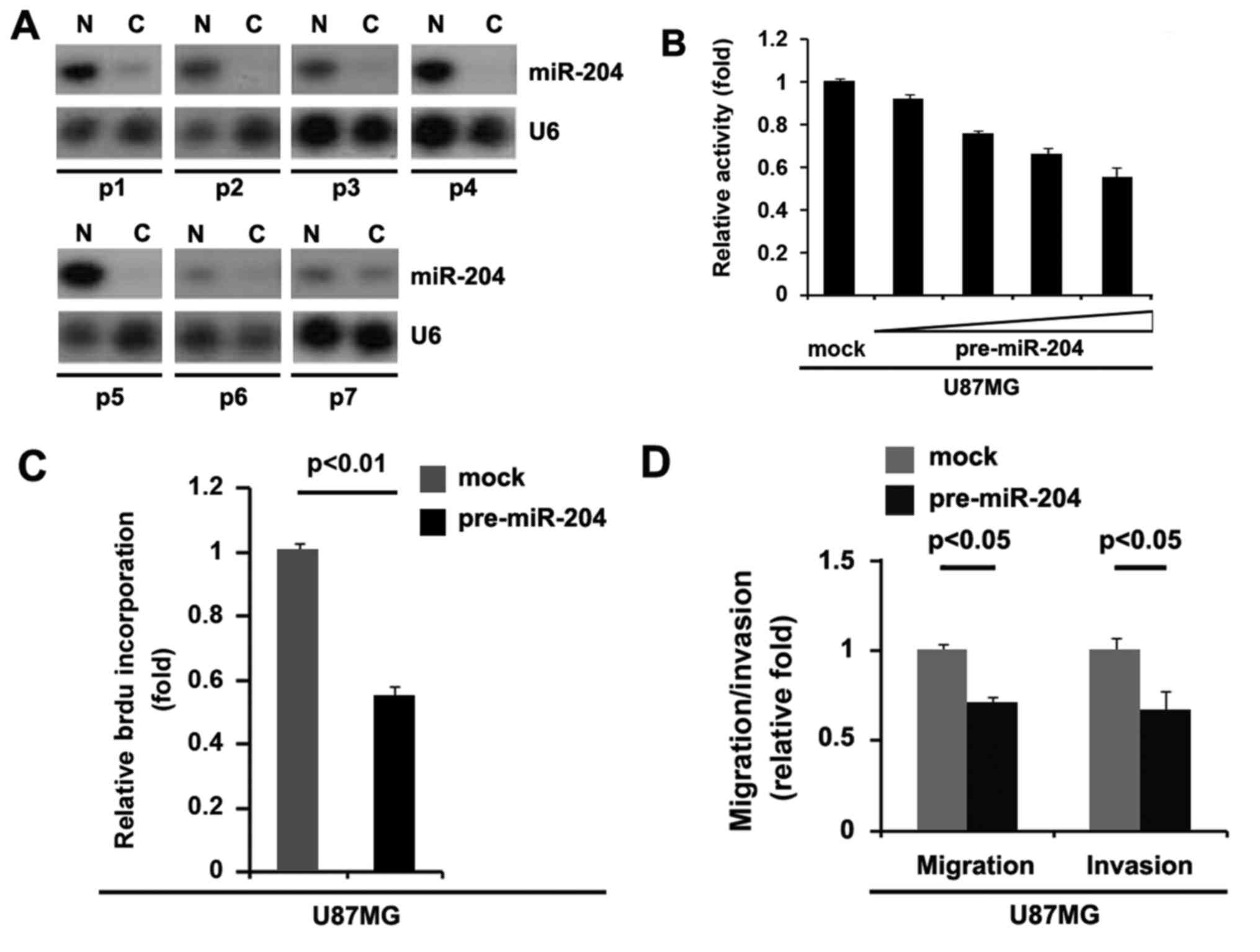
miR-204 is downregulated in
glioblastoma and its overexpression inhibits proliferation,
migration and invasion in glioblastoma cells
To assess the expression of miR-204 in glioblastoma
tissues, northern blot analysis was conducted in 7 pairs of
glioblastoma tissues and matched adjacent normal tissue samples.
The expression of miR-204 was consistently lower in the
glioblastoma tissues compared with normal tissues (Fig. 9A). In an attempt to identify the role
of miR-204 in regulating proliferation of U87MG cells, the cells
were transfected with pre-miR-204. Following stable transfection,
the proliferation rates of U87MG cells were tested by an MTT assay.
The results demonstrated that overexpression of miR-204
significantly inhibited the proliferation rate of U87MG cells and
that the inhibition of cellular proliferation was dose-dependent
(Fig. 9B). This was revealed by BrdU
incorporation analysis showing that transfection with miR-204
resulted in decreased DNA synthesis activity per viable cell in
U87MG cells (Fig. 9C). Considering
that miR-204 evidently inhibited the proliferation of U87MG cells,
the present study then sought to determine whether miR-204 would
have any impact on migration and invasion in U87MG cells. The
migration and invasion assay demonstrated that the overexpression
of miR-204 not only inhibited migration of U87MG cells, but also
suppressed their invasion (Fig. 9D).
Moreover, we observed that miR-204 inhibits proliferation in LN-229
glioblastoma cells (Fig. 10).
Discussion
Increased CYP27A1 expression was detected in
aberrant crypt foci, and adenomatous polyps, as well as the
expression of CYP27A1 protein is associated with tumor grade in
breast cancer (4,14). In the present study, it was identified
that overexpression of CYP27A1 promoted proliferation in
glioblastoma cells and silencing it inhibited proliferation,
indicating that it may be an oncogene. However, its overexpression
and silencing it did not affect migration and invasion in
glioblastoma cells. PDCD4, a known tumor suppressor gene, has been
identified as a functional target of miR-21 (15,16).
Silencing of SOX2 in glioblastoma tumor-initiating cells causes the
cessation of proliferation and loss of tumorigenicity (17). It was observed that CYP27A1
overexpression significantly downregulated PDCD4 and upregulated
the SOX2 protein in glioblastoma cells, indicating that CYP27A1 may
perform an important role in the regulation of tumor-initiating
cells.
In addition, CYP27A1 was found to be upregulated in
glioblastoma tissues. However, only collected 6 pairs of
glioblastoma tissues and adjacent normal tissues were collected.
Thus, future studies should use a bigger sample size.
Previous studies have reported that miR-204 is a
tumor suppressor miRNA: miR-204 can suppress head and neck tumor
metastasis (18); higher tumor grade
of human clear cell renal cell carcinomas was associated with a
concomitant decrease in miR-204 (19); loss of miR-204 expression enhances
glioma migration and stem cell-like phenotype (20); miR-204 targets B-cell lymphoma-2
expression and enhances responsiveness of gastric cancer (21); and miR-204 downregulates sirtuin 1
(SIRT1) and reverts SIRT1-induced epithelial-mesenchymal
transition, anoikis resistance and invasion in gastric cancer cells
(22). Dysregulation of miR-204
mediates migration and invasion of endometrial cancer by regulating
fork head box C1 (23). miR-204
increases sensitivity of neuroblastoma cells to cisplatin and is
associated with a favorable clinical outcome (24). Consistent with these previous studies
(18–24), it was identified that miR-204 was not
only downregulated in glioblastoma, and inhibited proliferation,
migration and invasion in glioblastoma cells. The present study
also revealed that overexpressing miR-204 inhibited CYP27A1
expression in glioblastoma cells, indicating that the
downregulation of miR-204 is associated with the upregulation of
CYP27A1 in the disease. It was concluded that miR-204 inhibited
proliferation by suppressing CYP27A1 expression. However, it was
observed that CYP27A1 did not affect migration and invasion in
glioblastoma cells, although miR-204 inhibited migration and
invasion. This observation indicated that there are other target
genes regulated by miR-204, which may regulate migration and
invasion in glioblastoma cells.
Recently, the U-87 MG cell line from ATCC was
reported to be contaminated or misidentified (25). It has been proposed as a glioblastoma
cell line whose origin is unknown (25). However, the U-87 MG cell line is still
widely used for glioblastoma research (25). In the present study, U87MG and LN-229
cells were used. The results were same from the 2 cell lines. Thus,
the contamination or misidentification does not affect the
conclusion presented.
Elucidating the mechanism by whichmiR-204 inhibits
proliferation by suppressing CYP27A1 may help to improve the
understanding of the molecular mechanism of proliferation in
glioblastoma. Thus, restoration of miR-204 may represent a
promising therapeutic way to inhibit CYP27A1-mediated proliferation
regulation. However, the roles of CYP27A1 require confirmation
in vivo.
Acknowledgements
Not applicable.
Funding
The present study was supported by Yishui Central
Hospital and Linyi People's Hospital.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JX and LQT conceived the study, collected the
experimental data and wrote a draft of the manuscript. LMZ, DKS,
XFL, and PX contributed to the experimental work and data analysis.
All authors edited and approved the final version of the
manuscript.
Ethics approval and consent to
participate
Ethics approval was obtained from ethics committee
of Yishui Central Hospital. All subjects provided written informed
consent at the time of enrollment.
Consent for publication
Consent for publication was obtained from each
patient.
Competing interests
The authors declare that they have no competing
financial interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Das S and Marsden PA: Angiogenesis in
glioblastoma. N Engl J Med. 369:1561–1563. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stupp R, Hegi ME, Mason WP, van den Bent
MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B,
Belanger K, et al: Effects of radiotherapy with concomitant and
adjuvant temozolomide versus radiotherapy alone on survival in
glioblastoma in a randomised phase III study: 5-year analysis of
the EORTC-NCIC trial. Lancet Oncol. 10:459–466. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nelson ER, Wardell SE, Jasper JS, Park S,
Suchindran S, Howe MK, Carver NJ, Pillai RV, Sullivan PM, Sondhi V,
et al: 27-Hydroxycholesterol links hypercholesterolemia and breast
cancer pathophysiology. Science. 342:1094–1098. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Norlin M, von Bahr S, Björkhem I and
Wikvall K: On the substrate specificity of human CYP27A1:
Implications for bile acid and cholestanol formation. J Lipid Res.
44:1515–1522. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lu DL, Sookthai D, Le Cornet C, Katzke VA,
Johnson TS, Kaaks R and Fortner RT: Reproducibility of serum
oxysterols and lanosterol among postmenopausal women: Results from
EPIC-Heidelberg. Clin Biochem. 52:117–122. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Björkhem I: Mechanism of degradation of
the steroid side chain in the formation of bile acids. J Lipid Res.
33:455–471. 1992.PubMed/NCBI
|
|
8
|
Russell DW and Setchell KD: Bile acid
biosynthesis. Biochemistry. 31:4737–4749. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993.
|
|
10
|
Chan JA, Krichevsky AM and Kosik KS:
MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells.
Cancer Res. 65:6029–6033. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mao J, Zhang M, Zhong M, Zhang Y and Lv K:
MicroRNA-204, a direct negative regulator of ezrin gene expression,
inhibits glioma cell migration and invasion. Mol Cell Biochem.
396:117–128. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kefas B, Godlewski J, Comeau L, Li Y,
Abounader R, Hawkinson M, Lee J, Fine H, Chiocca EA, Lawler S and
Purow B: microRNA-7 inhibits the epidermal growth factor receptor
and the Akt pathway and is down-regulated in glioblastoma. Cancer
Res. 68:3566–3572. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Matusiak D and Benya RV: CYP27A1 and CYP24
expression as a function of malignant transformation in the colon.
J Histochem Cytochem. 55:1257–1264. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lu Z, Liu M, Stribinskis V, Klinge CM,
Ramos KS, Colburn NH and Li Y: MicroRNA-21 promotes cell
transformation by targeting the programmed cell death 4 gene.
Oncogene. 27:4373–4379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhu S, Wu H, Wu F, Nie D, Sheng S and Mo
YY: MicroRNA-21 targets tumor suppressor genes in invasion and
metastasis. Cell Res. 18:350–359. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gangemi RM, Griffero F, Marubbi D, Perera
M, Capra MC, Malatesta P, Ravetti GL, Zona GL, Daga A and Corte G:
SOX2 silencing in glioblastoma tumor-initiating cells causes stop
of proliferation and loss of tumorigenicity. Stem Cells. 27:40–48.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lee Y, Yang X, Huang Y, Fan H, Zhang Q, Wu
Y, Li J, Hasina R, Cheng C, Lingen MW, et al: Network modeling
identifies molecular functions targeted by miR-204 to suppress head
and neck tumor metastasis. PLoS Comput Biol. 6:e10007302010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mikhaylova O, Stratton Y, Hall D, Kellner
E, Ehmer B, Drew AF, Gallo CA, Plas DR, Biesiada J, Meller J and
Czyzyk-Krzeska MF: VHL-regulated MiR-204 suppresses tumor growth
through inhibition of LC3B-mediated autophagy in renal clear cell
carcinoma. Cancer Cell. 21:532–546. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ying Z, Li Y, Wu J, Zhu X, Yang Y, Tian H,
Li W, Hu B, Cheng SY and Li M: Loss of miR-204 expression enhances
glioma migration and stem cell-like phenotype. Cancer Res.
73:990–999. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sacconi A, Biagioni F, Canu V, Mori F, Di
Benedetto A, Lorenzon L, Ercolani C, Di Agostino S, Cambria AM,
Germoni S, et al: miR-204 targets Bcl-2 expression and enhances
responsiveness of gastric cancer. Cell Death Dis. 3:e4232012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang L, Wang X and Chen P: miR-204 down
regulates SIRT1 and reverts SIRT1-induced epithelial-mesenchymal
transition, anoikis resistance and invasion in gastric cancer
cells. BMC Cancer. 13:2902013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chung TK, Lau TS, Cheung TH, Yim SF, Lo
KW, Siu NS, Chan LK, Yu MY, Kwong J, Doran G, et al: Dysregulation
of microRNA-204 mediates migration and invasion of endometrial
cancer by regulating FOXC1. Int J Cancer. 130:1036–1045. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ryan J, Tivnan A, Fay J, Bryan K, Meehan
M, Creevey L, Lynch J, Bray IM, O'Meara A, Tracey L, et al:
MicroRNA-204 increases sensitivity of neuroblastoma cells to
cisplatin and is associated with a favourable clinical outcome. Br
J Cancer. 107:967–976. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Allen M, Bjerke M, Edlund H, Nelander S
and Westermark B: Origin of the U87MG glioma cell line: Good news
and bad news. Sci Transl Med. 8:354re32016. View Article : Google Scholar : PubMed/NCBI
|