Introduction
Gastric cancer (GC) remains one of the most common
types of cancer globally (1). Gastric
carcinogenesis is a multistep and multifactorial process (2) and may be affected by various factors
that participate in each carcinogenic step, including the
activation of oncogenes, the inactivation of tumor suppressor genes
(3) and the effects of epigenetic
modification (4). Each of these
factors may affect the occurrence and development of GC.
Investigating the molecular regulation of GC development is
essential for diagnosis and treatment. Methyl-CpG binding protein 2
(MeCP2) is a methylated binding protein, which may bind to
methylated CpG islands and inhibit the transcription of genes
(5). MeCP2 may also serve an
oncogenic function in GC (6). Long
noncoding RNAs (lncRNAs) are a class of noncoding RNA which are
>200 nucleotides long (7).
Increasingly, studies have revealed that the deregulation of
lncRNAs are involved in a variety of human diseases and serve
important functions in cell proliferation, apoptosis, metastasis
and invasion (8–11). Small integral membrane protein 10 like
2A (linc00086), located at the X chromosome, is required for the
tumor protein p53 transcriptional response (12).
In the present study, the expression of linc00086
was revealed to be downregulated in GC. Bioinformatics analyses
revealed that the linc00086 gene has CpG islands. In order to
further investigate whether linc00086 methylation was responsible
for the downregulated expression of linc00086 in GC, in the present
study GC cells were treated with 5-Aza-2′-deoxycytidine (5-aza-dC)
and it was demonstrated that linc00086 expression was upregulated.
Bisulfite sequencing-polymerase chain reaction (PCR) was used to
detect the methylation effect on linc00086, and analysis of CpG
methylation by bisulfite sequencing-PCR demonstrated that DNA
methylation may regulate the expression of linc00086. As MeCP2 is
involved in gene regulation by binding to methylated promoters, the
expression of MeCP2 was silenced in order to detect the expression
of linc00086. The results revealed that silencing the expression of
MeCP2 may promote the expression of linc00086. These results
suggested that DNA methylation may contribute to silencing the
expression of linc00086 in GC.
Materials and methods
Human tissue samples and cell
lines
The GC cell lines SGC-7901, MKN45, BGC-823, AGS and
the normal gastric cell line GES-1 were sourced from the Key
Laboratory of Environment and Genes Related to Diseases, Xi'an
Jiaotong University, Ministry of Education of China (Xi'an, China).
These cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a
humidified chamber with 5% CO2. A total of 20 GC tissues
(all males; mean age, 59.5 years; age range, 43–74 years) and the
matched normal tissues were sourced from the First Affiliated
Hospital of Xi'an Jiaotong University College of Medicine (Xi'an,
China). No radiotherapy or chemotherapy was conducted prior to
surgery. The present study was approved by the Medical Ethical
Committee of the College of Medicine, Xi'an Jiaotong University.
Written informed consent was provided by all patients.
RNA extraction and reverse
transcription-quantitative (RT-q)PCR analysis
RNA was extracted using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Complementary DNA synthesis was performed
using the Prime-Script RT reagent kit (Takara Biotechnology Co.,
Ltd., Dalian, China) according to the manufacturer's protocol. qPCR
was performed by SYBR Green PCR kit (Takara Biotechnology Co.,
Ltd.) to analyze the expression levels of the genes. β-actin was
used as an endogenous control Thermocycling conditions were as
follows: Initial denaturation at 95°C for 30 sec, followed by 40
cycles at 95°C for 5 sec and at 60°C for 30 sec. The relative
expression of genes was calculated using the 2-ΔΔCq method
(13). The primers used are listed in
Table I.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Name of primer | Sequence (5′-3′) |
|---|
| Linc00086,
forward |
CTCACGGCTTGGATTGCTC |
| Linc00086,
reverse |
AGATGGTAAGGGCGAGGGT |
| Linc00086 CPG |
TTTATTTTGGGAAATGGTTTTA |
| islands, forward | AAG |
| Linc00086 CPG
islands, reverse |
CCCCCAAACCAACATAAATC |
5-aza-dC treatment
SGC-7901 and MKN45 cells were treated with the DNA
methyltransferase inhibitor, 5-aza-dC (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) at differing concentrations (0.0, 2.0, 4.0, 6.0
and 8.0 µM) for 48 h 37°C. Then the RNA was isolated from SGC-7901
or MKN45 cells using TRIzol (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. RT-qPCR was
performed to detect the expression of linc00086 as aforementioned.
The primers used are listed in Table
I.
Bisulfite sequencing PCR
SGC-7901 and MKN45 cells were treated with 2.0 µM
5-aza-dCor equal amounts of DMSO as a control for 48 h at 37°C, and
DNA was extracted and modified using the bisulfite reaction
according to the manufacturer's protocol of the Qiagen
EpiTect® Bisulfite kit (Qiagen, Inc., Valencia, CA,
USA). Primer sequences are listed in Table I. PCR used 2×Taq MasterMix (Dye; CoWin
Biosciences Co., Ltd., Jiangsu, China). The total reaction volume
was 50 µl in a mixture containing 0.4 µg DNA, and the thermocycler
conditions were as follows: Pre-denaturation at 94°C for 2 min,
followed by 35 cycles of denaturation at 94°C for 30 seconds,
annealing at 59°C for 30 seconds, extension at 72°C for 30 seconds,
with a final extension step at 72°C for 2 min. The PCR products of
bisulfite-modified DNA of SGC-7901 cells were purified and cloned
into a T-vector (Takara Biotechnology Co., Ltd.), then sequenced by
Sangon Biotech Co., Ltd. (Shanghai, China). The methylation level
was analyzed using the Quantification Tool for Methylation Analysis
(14).
Small interfering RNA (siRNA)
transfection
A total of 5×105 SGC-7901 or MKN45 cells
were seeded for ~24 h prior to transfection. SiRNAs against MeCP2
(si-MeCP2) with sequences as follows: si-MeCP2-S,
GCUUAAGCAAAGGAAAUCUTT and si-MeCP2-A, AGAUUUCCUUUGCUUAAGCTT
(15) and its respective
siRNA-negative controls (si-control) with sequences as follows:
siRNA-ctrl-S UUCUCCGAACGUGUCACGUTT and siRNA-ctrl-A,
ACGUGACACGUUCGGAGAATT (Shanghai GenePharma Co., Ltd., Shanghai,
China) were transfected into the cells using Lipofectamine 2000
(Invitrogen) according to the manufacturer's protocol. RNA was
extracted 48 h post-transfection, and then RT-qPCR was used to
detect the expression of MeCP2 and linc00086 as aforementioned.
Statistical analysis
All statistical analyses were performed using SPSS
13.0 software (SPSS, Inc., Chicago, IL, USA). Student's t-test was
used to analyze the data. P<0.05 was considered to indicate a
statistically significant difference.
Results
Linc00086 is downregulated in GC
tissue and cell lines
RT-qPCR was performed to detect the expression
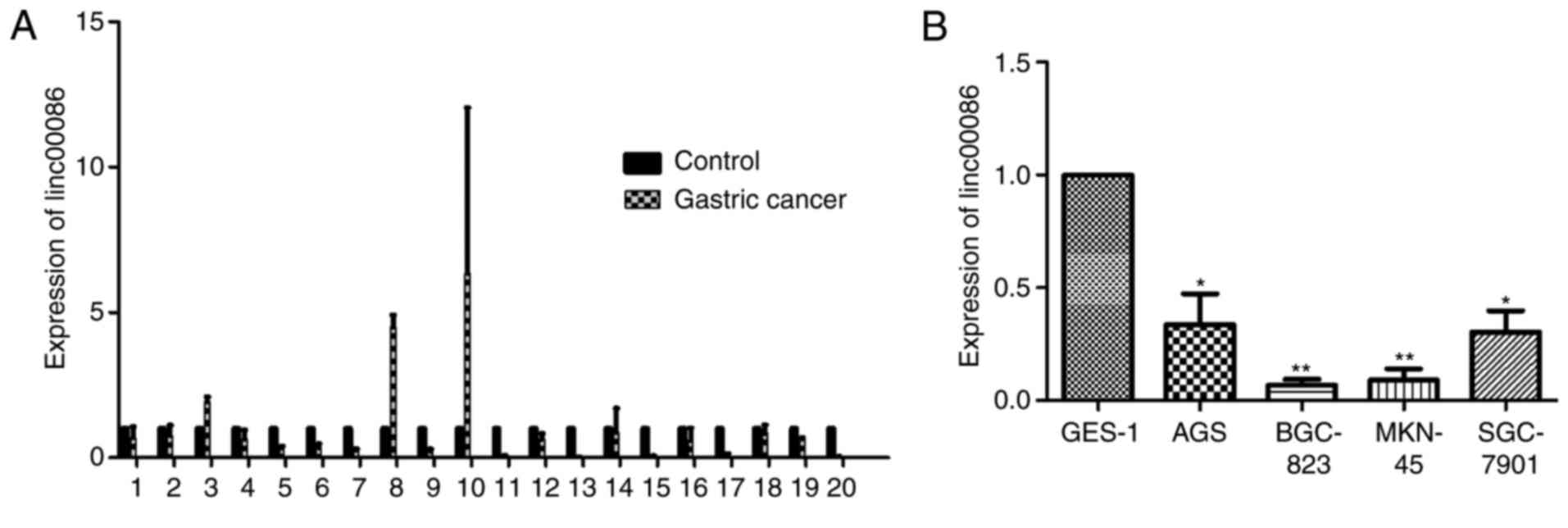
levels of linc00086 in GC tissues and cell lines. The results
revealed that among the 20 paired samples, 17 of them (85%)
exhibited a lower expression of linc00086 in GC tissues compared
with the matched non-tumor gastric tissues (Fig. 1A). Furthermore, when comparing the
expression levels of linc00086 in AGS, BGC-823, MKN-45 and
SGC-7901cells with GES-1 cells, the expression level of linc00086
was significantly downregulated in the GC cell lines, compared with
GES-1 cells (Fig. 1B). The results of
the present study suggest that linc00086 was downregulated in GC
tissue and cell lines.
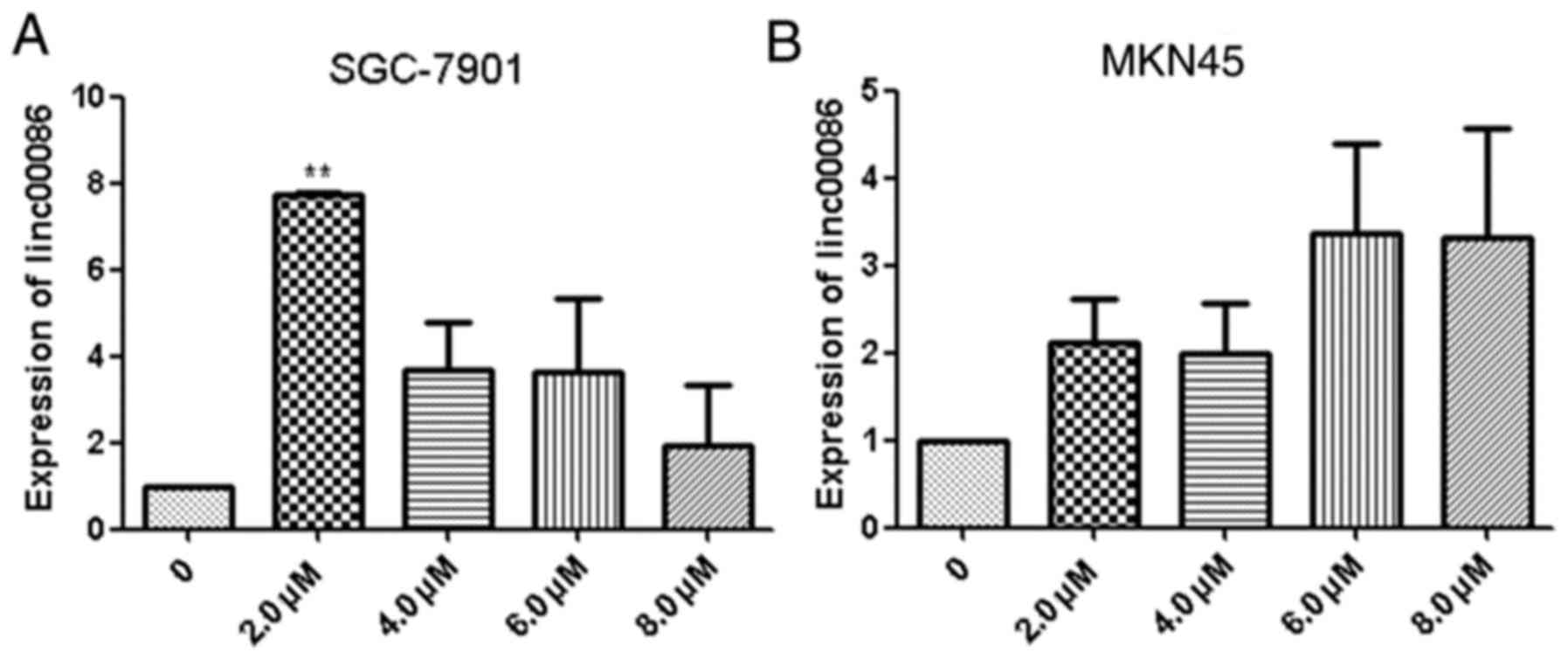
5-aza-dC treatment may increase the
expression of linc00086
The potential for DNA methylation to contribute to
the silencing of the expression of linc00086 in GC was
investigated. SGC-7901 and MKN-45 cells were treated with 0.0, 2.0,
4.0, 6.0 and 8.0 µM 5-aza-dC for 48 h. The results revealed that
treatment with 5-aza-dC may induce the expression of linc00086
(Fig. 2A and B). It was revealed that
treatment with 2.0 µM 5-aza-dC for 48 h was the optimal
concentration.
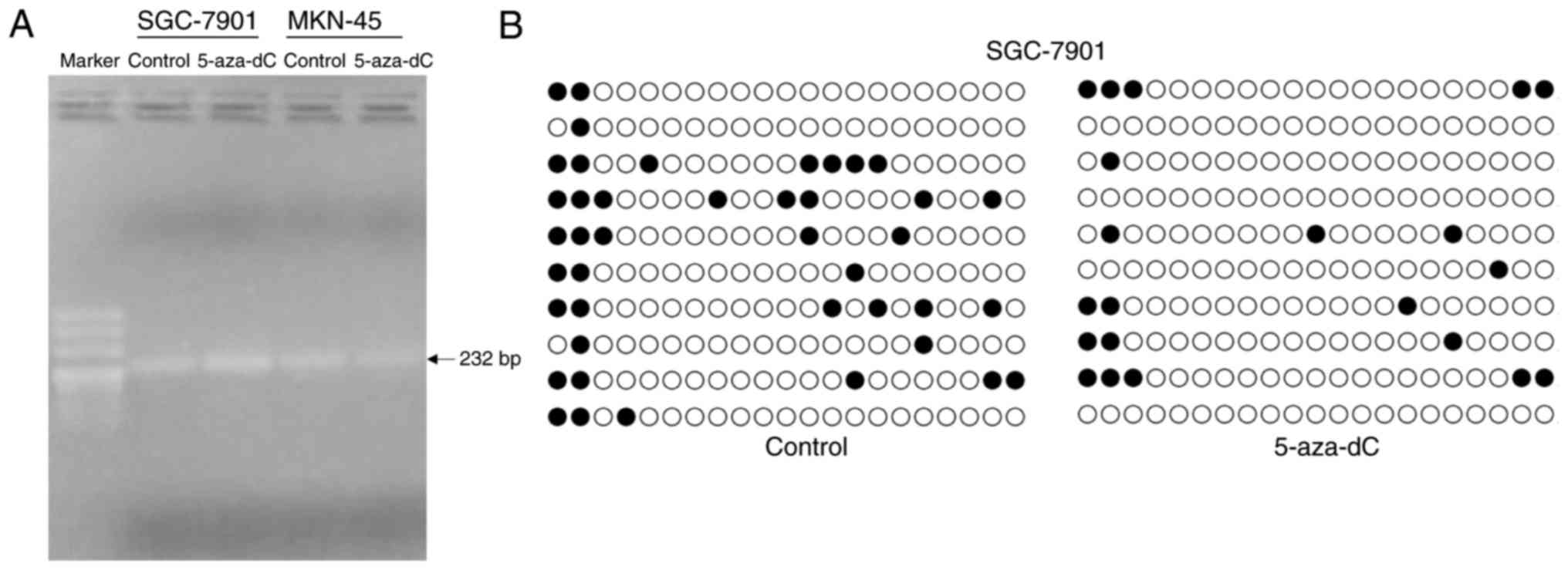
Methylation levels declined following
treatment with 5-aza-dC
SGC-7901 and MKN45 cells were treated with 2.0 µM
5-aza-dCor control, and then DNA was extracted. DNA was modified
using the bisulfite reaction. Primers of bisulfite sequencing-PCR
and bisulfite modified DNA were used to determine the interest
region sequences of the linc00086 CPG islands using PCR (Fig. 3A). Next, the PCR products of
bisulfite-modified DNA of SGC-7901 were cloned into a T-vector and
sequenced. The methylation level of the control group was 20% and
the methylation level of the 5-aza-dC treatment group was 10% in
the SGC-7901 cells. The methylation level of 5-aza-dC treatment
group was decreased compared with the control group (Fig. 3B). The results suggested that the
methylation level declined following treatment with 5-aza-dC.
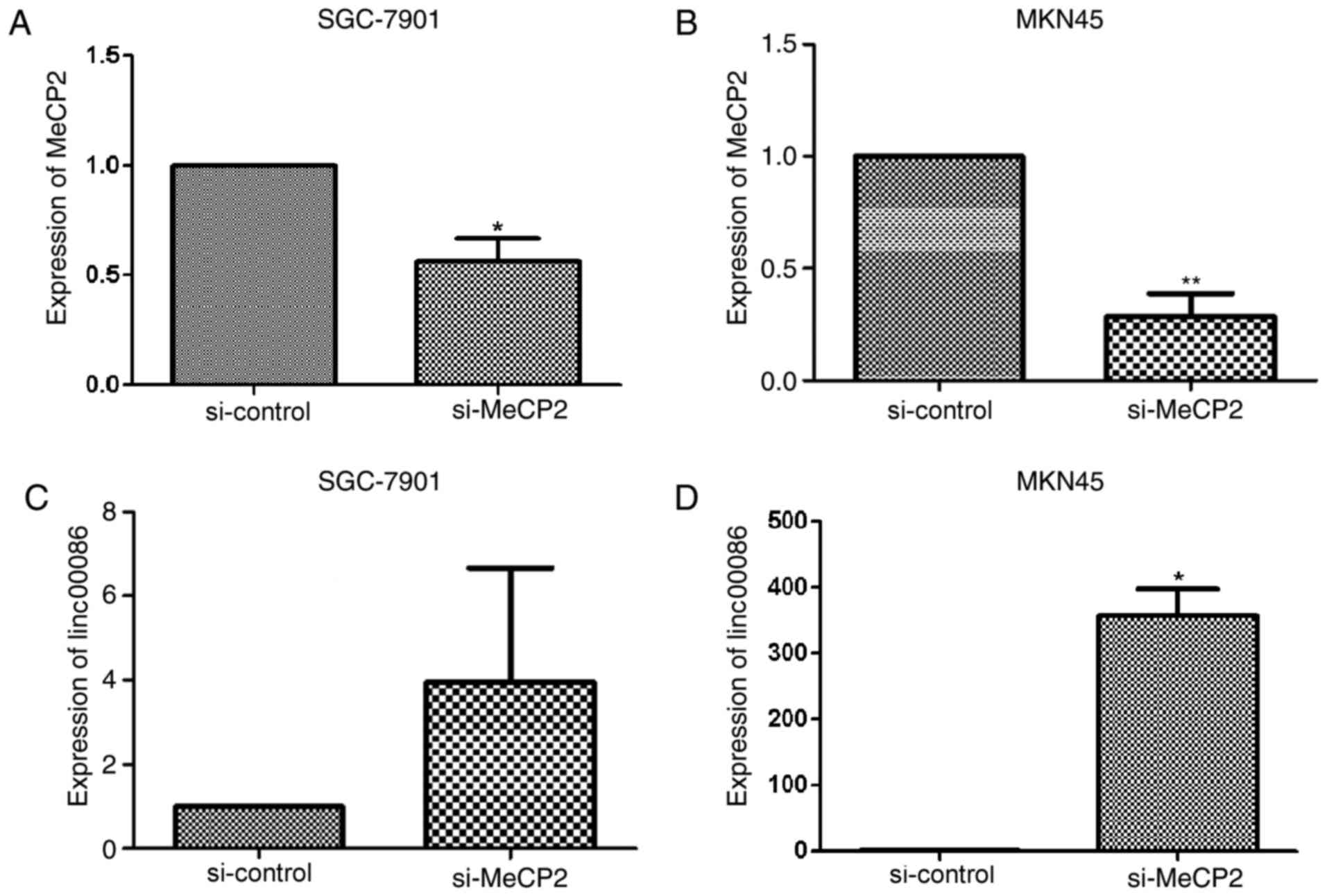
Silencing MeCP2 may induce the
expression of linc00086
SGC-7901 andMKN45 cells were transfected with
si-MeCP2 or si-control. RNA was extracted 48 h post-transfection,
and then expression of MeCP2 and linc00086 was detected. It was
revealed that si-MeCP2 may substantially reduce the expression of
MeCP2 (Fig. 4A and B), and si-MeCP2
may increase the expression of linc00086 in SGC-7901 and MKN45
cells (Fig. 4C and D). The results of
the present study further confirm that the low expression of
linc00086 in GC is regulated by DNA methylation.
Discussion
DNA methylation is an important type of epigenetic
modification, and has been reported to be involved in
tumorigenesis. Previous studies have identified that DNA
methylation changes may result in aberrant gene expression in
cancer (16,17), and a number of them may be an early
molecular marker (18,19). A number of differing factors may
silence a number of tumor suppressors due to DNA methylation,
including miR-122 (20), miR-219-2-3p
(21), miR-203 (22), and lncRNAs SRHC (23) and maternally expressed 3 (non-protein
coding) (24). The focus of the
present study was on the silencing of linc00086 by DNA methylation
in GC.
Previously, studies have revealed the participation
of lncRNAs in various biological processes. Furthermore, a number
of lncRNAs are detectable as biomarkers in the diagnosis of certain
types of cancer, including the following lncRNAs: Urothelial cancer
associated 1 for GC (25), POU class
3 homeobox 3 for esophageal squamous cell carcinoma (26), PVT1 oncogene (non-protein coding) for
cervical cancer (27) and CCDC26 long
non-coding RNA for pancreatic cancer (28).
The present study revealed that linc00086 expression
was lower in GC tissues compared with normal tissues. Accumulating
evidence has revealed that a number of lncRNAs were aberrant in
cancer (29). The association between
linc00086 expression and the survival period of patients with GC
will be further studied through the use of databases of a suitable
sample size. Furthermore, the present study revealed that the
aberrant expression of linc00086 may be regulated by DNA
methylation in GC cell lines. DNA methylation and histone
modification are the two major types of epigenetic modification. In
the present study, bisulfite sequencing-PCR was used to identify
the DNA methylation of linc00086 in SGC-7901 cells in order to
explore the reason behind the low expression of linc00086. The
results of the present study have demonstrated that DNA methylation
may be one of the reasons for the silenced expression of linc00086
in GC. MeCP2 belongs to the family of methyl-CpG-binding proteins
that regulate gene expression by DNA methylation (30). MeCP2 has previously emerged as an
important oncogene in a multitude of types of cancer, and is
involved in cancer progression. It has been reported that MeCP2
expression is increased in hepatocellular carcinoma and promotes
the proliferation of human hepatocellular carcinoma HepG2 cells via
the activation of extracellular signal-regulated kinase 1/2
signaling pathways (31). MeCP2 was
overexpressed in GC and serves an important function in gastric
carcinogenesis (15). MeCP2 may
regulate gene expression by binding methylated CpG islands
(32). The present study revealed
that silencing MeCP2 may induce the expression of linc00086, which
may suggest that the downregulated expression of linc00086 in GC is
associated with DNA methylation. The results of the present study
identified that the aberrant expression of linc00086 may be
regulated by DNA methylation.
Acknowledgements
Not applicable.
Funding
The present study was funded by Research Support
Project of New Teacher of Xi'an Jiaotong University (grant no.
YX1K078).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CH and YS conceived the study. RS, LZ, LW and XS
collected the cancer tissues. YY, ZZ, QJ, YLL, YXL and FW performed
the experiments. CH and YY calculated the data. YXL and YY wrote
the paper.
Ethics approval and consent to
participate
The research protocol was approved by the Medical
Ethical Committee of the College of Medicine, Xi'an Jiaotong
University. Written informed consent was provided by all
patients.
Consent for publication
Written informed consent was obtained from all
patients.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bertuccio P, Chatenoud L, Levi F, Praud D,
Ferlay J, Negri E, Malvezzi M and La Vecchia C: Recent patterns in
gastric cancer: A global overview. Int J Cancer. 125:666–673. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Correa P: Human gastric carcinogenesis: A
multistep and multifactorial process-first American cancer society
award lecture on cancer epidemiology and prevention. Cancer Res.
52:6735–6740. 1992.PubMed/NCBI
|
|
3
|
Tahara E, Yasui W and Yokozaki H: Genetic
alterations in stomach cancer. Nihon Geka Gakkai zasshi.
97:252–256. 1996.(In Japanese). PubMed/NCBI
|
|
4
|
Patel TN, Roy S and Ravi R: Gastric cancer
and related epigenetic alterations. Ecancermedicalscience.
11:7142017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wakefield RI, Smith BO, Nan X, Free A,
Soteriou A, Uhrin D, Bird AP and Barlow PN: The solution structure
of the domain from MeCP2 that binds to methylated DNA. J Mol Biol.
291:1055–1065. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhao L, Liu Y, Tong D, Qin Y, Yang J, Xue
M, Du N, Liu L, Guo B, Hou N, et al: MeCP2 promotes gastric cancer
progression through regulating FOXF1/Wnt5a/β-Catenin and
MYOD1/Caspase-3 signaling pathways. EBioMedicine. 16:87–100. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Annu Rev Biochem. 81:145–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang F, Yuan JH, Wang SB, Yang F, Yuan SX,
Ye C, Yang N, Zhou WP, Li WL, Li W and Sun SH: Oncofetal long
noncoding RNA PVT1 promotes proliferation and stem cell-like
property of hepatocellular carcinoma cells by stabilizing NOP2.
Hepatology. 60:1278–1290. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu ZJ, Li Y, Wu YZ, Wang Y, Nian WQ, Wang
LL, Li LC, Luo HL and Wang DL: Long non-coding RNA CCAT2 promotes
the breast cancer growth and metastasis by regulating TGF-β
signaling pathway. Eur Rev Med Pharmacol Sci. 21:706–714.
2017.PubMed/NCBI
|
|
10
|
Wang D, Wang D, Wang N, Long Z and Ren X:
Long Non-Coding RNA BANCR promotes endometrial cancer cell
proliferation and invasion by regulating MMP2 and MMP1 via ERK/MAPK
signaling pathway. Cell Physiol Biochem. 40:644–656. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang J, Zhang X, Shi J, Cao P, Wan M,
Zhang Q, Wang Y, Kridel SJ, Liu W, Xu J, et al: Fatty acid synthase
is a primary target of MiR-15a and MiR-16-1 in breast cancer.
Oncotarget. 7:78566–78576. 2016.PubMed/NCBI
|
|
12
|
Leveille N, Melo CA, Rooijers K,
Díaz-Lagares A, Melo SA, Korkmaz G, Lopes R, Akbari Moqadam F, Maia
AR, Wijchers PJ, et al: Genome-wide profiling of p53-regulated
enhancer RNAs uncovers a subset of enhancers controlled by a
lncRNA. Nat Commun. 6:65202015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kumaki Y, Oda M and Okano M: QUMA:
Quantification tool for methylation analysis. Nucleic Acids Res.
36:W170–W175. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tong D, Zhao L, He K, Sun H, Cai D, Ni L,
Sun R, Chang S, Song T and Huang C: MECP2 promotes the growth of
gastric cancer cells by suppressing miR-338-mediated
antiproliferative effect. Oncotarget. 7:34845–34859. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nordstrom L, Andersson E, Kuci V,
Gustavsson E, Holm K, Ringnér M, Guldberg P and Ek S: DNA
methylation and histone modifications regulate SOX11 expression in
lymphoid and solid cancer cells. BMC Cancer. 15:2732015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tian Y, Wei W, Li L and Yang R:
Down-Regulation of miR-148a Promotes Metastasis by DNA Methylation
and is associated with prognosis of skin cancer by targeting TGIF2.
Med Sci Monit. 21:3798–3805. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu K, Zhang Y, Zhang C, Zhang Q, Li J,
Xiao F, Li Y, Zhang R, Dou D, Liang J, et al: Methylation of S100A8
is a promising diagnosis and prognostic marker in hepatocellular
carcinoma. Oncotarget. 7:56798–56810. 2016.PubMed/NCBI
|
|
19
|
Pimson C, Ekalaksananan T, Pientong C,
Promthet S, Putthanachote N, Suwanrungruang K and Wiangnon S:
Aberrant methylation of PCDH10 and RASSF1A genes in blood samples
for non-invasive diagnosis and prognostic assessment of gastric
cancer. PeerJ. 4:e21122016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xing TJ, Xu HT, Yu WQ and Jiang DF:
Methylation regulation of liver-specific microRNA-122 expression
and its effects on the proliferation and apoptosis of
hepatocellular carcinoma cells. Genet Mol Res. 12:3588–3597. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lei H, Zou D, Li Z, Luo M, Dong L, Wang B,
Yin H, Ma Y, Liu C, Wang F, et al: MicroRNA-219-2-3p functions as a
tumor suppressor in gastric cancer and is regulated by DNA
methylation. PLoS One. 8:e603692013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Noguchi S, Mori T, Nakagawa T, Itamoto K,
Haraguchi T and Mizuno T: DNA methylation contributes toward
silencing of antioncogenic microRNA-203 in human and canine
melanoma cells. Melanoma Res. 25:390–398. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zheng H, Yang S, Yang Y, Yuan SX, Wu FQ,
Wang LL, Yan HL, Sun SH and Zhou WP: Epigenetically silenced long
noncoding-SRHC promotes proliferation of hepatocellular carcinoma.
J Cancer Res Clin Oncol. 141:1195–1203. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun M, Xia R, Jin F, Xu T, Liu Z, De W and
Liu X: Downregulated long noncoding RNA MEG3 is associated with
poor prognosis and promotes cell proliferation in gastric cancer.
Tumour Biol. 35:1065–1073. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gao J, Cao R and Mu H: Long non-coding RNA
UCA1 may be a novel diagnostic and predictive biomarker in plasma
for early gastric cancer. Int J Clin Exp Pathol. 8:12936–12942.
2015.PubMed/NCBI
|
|
26
|
Tong YS, Wang XW, Zhou XL, Liu ZH, Yang
TX, Shi WH, Xie HW, Lv J, Wu QQ and Cao XF: Identification of the
long non-coding RNA POU3F3 in plasma as a novel biomarker for
diagnosis of esophageal squamous cell carcinoma. Mol Cancer.
14:32015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang JP, Yang XJ, Xiao L and Wang Y: Long
noncoding RNA PVT1 as a novel serum biomarker for detection of
cervical cancer. Eur Rev Med Pharmacol Sci. 20:3980–3986.
2016.PubMed/NCBI
|
|
28
|
Peng W and Jiang A: Long noncoding RNA
CCDC26 as a potential predictor biomarker contributes to
tumorigenesis in pancreatic cancer. Biomed Pharmacother.
83:712–717. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gibb EA, Vucic EA, Enfield KS, Stewart GL,
Lonergan KM, Kennett JY, Becker-Santos DD, MacAulay CE, Lam S,
Brown CJ and Lam WL: Human cancer long non-coding RNA
transcriptomes. PLoS One. 6:e259152011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lyu JW, Yuan B, Cheng TL, Qiu ZL and Zhou
WH: Reciprocal regulation of autism-related genes MeCP2 and PTEN
via microRNAs. Sci Rep. 6:203922016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhao LY, Zhang J, Guo B, Yang J, Han J,
Zhao XG, Wang XF, Liu LY, Li ZF, Song TS and Huang C: MECP2
promotes cell proliferation by activating ERK1/2 and inhibiting p38
activity in human hepatocellular carcinoma HEPG2 cells. Cell Mol
Biol (Noisy-le-grand) Suppl. 59:OL1876–OL1881. 2013.
|
|
32
|
Xu M, Bian S, Li J, He J, Chen H, Ge L,
Jiao Z, Zhang Y, Peng W, Du F, et al: MeCP2 suppresses LIN28A
expression via binding to its methylated-CpG islands in pancreatic
cancer cells. Oncotarget. 7:14476–14485. 2016.PubMed/NCBI
|