Introduction
According to an epidemiological survey, the
incidence rate of neuroglioma has been significantly increased over
the past 20 years. Neuroglioma, abbreviated as malignant brain
tumors, accounts for 40% or even higher of all primary brain tumors
(1). The death ranking announced by
the World Health Organization (WHO) shows that glioma ranks second
among death causes of tumor patients aged below 34 years and is
ranked third among those of middle-aged and elderly tumor patients
(2). Although the treatment of glioma
patients has obtained a certain effect due to the remarkable
advances in operation and other treatment methods, the average
survival time is only 9–12 months with poor prognoses. Moreover,
the main difficulties for treating glioma are metastasis and
relapse (3).
Micro-ribose nucleic acid (miRNA) is a kind of
non-coding single-stranded RNA with the length of ~19-22
nucleotides (nt) and a main active regulatory factor in biological
bodies at present. miRNA itself does not have the function of
translating proteins, but can adjust the expression of target genes
through the transcription by the complementary pairing with 3′
untranslated region (UTR) base sequences (4). In the adjusting process, miRNA leads to
differential expression of genes, causing degradation and
translation inhibition of messenger RNAs (mRNAs) of migrating and
invading target genes concerned (5).
Literature (6) has reported that
miRNA-130a is relatively highly expressed in glioma tissues, but
few studies exist on the function of glioma. This study
investigated the effects of miRNA-130a on the proliferation,
migration and invasion of glioma cell lines through in vitro
experiments.
Materials and methods
Main experimental materials
Glioma cell lines (U-87MG) and human astrocytes (HA)
1800 were purchased from the Cell Bank of Shanghai Institute of
Biochemistry and Biology, CAS. The U-87MG is different from that of
the original cells but the origin is unknown (7). Fetal bovine serum (FBS) was obtained
from Gibco (Thermo Fisher Scientific, Inc., Waltham, MA, USA) and
high pure miRNA isolation kit was purchased from Roche (Madison,
WI, USA). Both methyl thiazolyl tetrazolium (MTT) and dimethyl
sulphoxide (DMSO) were obtained from Sigma-Aldrich (St. Louis, MO,
USA) and Annexin V-fluorescein isothiocyanate (FITC) was purchased
from Bestbio (Shanghai, China, http://bestbio.bioon.com.cn/). Lipofectamine™ 2000
transfection reagent was purchased from Invitrogen (Thermo Fisher
Scientific, Inc.). miRNA-scramble, miRNA-130a-mimics and
miRNA-130a-inhibitor sequences were synthesized by GenePharma Co.,
Ltd. (Shanghai, China) (Table I).
 | Table I.Primer sequences used in this
study. |
Table I.
Primer sequences used in this
study.
| Items | Primer sequences |
|---|
|
miRNA-130a-inhibitor |
5′AUGCCCUUUUAACAUUGCACUG-3′ |
|
miRNA-130a-mimics |
5′-AUGCCCUUUUAACAUUGCACUG-3′ |
|
miRNA-130a-scramble |
5′-CAGUACUUUUGUGUAGUACAA-3′ |
The study was approved by the Ethics Committee of
The Second Nanning Peoples Hospital. (Nanning, China).
Cell culture
Dulbecco's modified Eagle's medium (DMEM) with 10%
FBS and 1% penicillin streptomycin was used to cultivate U-87MG
cells, which, were then transferred into the thermostatic incubator
with 5% CO2 at 37°C for cultivation, digestion and
passage. Trypsin was used to count cells. The density of cells were
adjusted, and they were successively inoculated into 6-well plates
as per 2×105 cells/well. Afterwards, they were
cultivated in DMEM (containing 10% FBS without 1% penicillin
streptomycin) for 24 h. When the total cells were fused to 80%,
Lipofectamine™ 2000 kit was used for transfection. In this
experiment, four groups were set up: the bare group, the
miRNA-130a-mimics group, the miRNA-130a-inhibitor group and the
miRNA-scramble group. After 6 h of transfection, cells continued to
be cultivated for 24 h.
Expression of miRNA-130a in cells
detected via RT-qPCR
After transfection, the total RNA of cells were
extracted strictly in accordance with the instructions of high pure
miRNA isolation kit, and the concentration of the extracted RNA was
measured by using an ultraviolet spectrophotometer (Hitachi, Tokyo,
Japan). The extracted RNA was reversely transcribed into
complementary deoxyribose nucleic acid (cDNA). Afterwards, RT-qPCR
was conducted for amplification detection under the following
conditions: pre-degeneration at 95°C for 10 min, at 95°C for 15
sec, at 65°C for 30 sec and at 72°C for 30 sec, respectively; 40
cycles in total. U6 was taken as a reference gene. The expression
quantity was calculated by 2−ΔΔCq (6).
Proliferation ability of cells
measured via MTT assay
After 24 h cultivation, cells were added with 40 µl
2 mg/ml MTT solution and continued to be cultivated for 4 h; the
culture solution in wells was sucked by using a pipette, and each
well was added with 100 µl DMSO. Then, they were transferred into
microporous plate oscillators to vibrate for 10 min, and the
crystals in wells were dissolved. Finally, the microplate reader
(Bio-Rad, Hercules, CA, USA) was used to determine the calculation
results.
Invasion ability of cell determined
via Transwell assay
The culture solution after 24 h transfection was
replaced of DMEM without 10% FBS for 24 h cultivation. Trypsin
(0.25%) was used to digest cells, and DMEM without 10% FBS was made
into cell suspension (2×105 cells/ml). The upper chamber
was added with the same cell suspension (200 µl), while the lower
chamber was added with 500 µl DMEM (with 10% FBS). Three groups of
the same wells were set up, additionally. The prepared cells were
put into the thermostatic incubator with 5% CO2 at 37°C
for cultivation (24 h), and 24 h later, Transwell chambers were
taken out. Sterile cotton swabs were used to wipe the Matrigel and
cells in the upper chamber, and then the hematoxylin-eosin (HE)
staining was conducted. Then, 5 fields were randomly selected to
count cells under a low-power microscope (BX-42; Olympus, Tokyo,
Japan) 3 times.
Migration ability of cells measured
via scratch assay
The transfected cells in all groups were inoculated
into 6-well plates for 3 sets of the same wells, in total. When
~90% cells were fused, the scratching was conducted according to
the prepared transverse lines in advance, with 20 µl pipette tip
perpendicular to the 6-well culture plate. Then, phosphate-buffered
saline (PBS) was used to wash cells 3 times, and the cells were
continued to be cultivated in DMEM containing 1% FBS. The above
operations were repeated 3 times.
Detection of cell apoptosis
After cells were transfected for 48 h, 4 groups of
cells were separately transferred into cone-shaped tubes which were
placed on ice. Cells in plates were rinsed by 2 ml PBS, and then,
with PBS removed, tubes were added with 0.5 ml 0.25% tyrisin
without ethylenediamine tetraacetic acid (EDTA) for incubation.
Under a microscope (BX-42; Olympus) it was observed that cells
started to fall off. Afterwards, cells fully fell off the culture
plates by patting, and the resulting cells were re-suspended in
1×PBS (with the density of 1×106 cells/ml). They were
put into clean centrifugal tubes, and then Annexin V-FITC,
apoptosis test fluid was added. After that, they were kept in the
dark at room temperature for 5 min. The supernatant was centrifuged
at 1,200 × g for 5 min, and 1XPBS was added to re-suspend cells
again. Then, 10 µl propidium iodide (PI) was added, and flow
cytometer was used for analysis.
Statistical analysis
Statistical Product and Service Solutions (SPSS)
20.0 software package (IBM Corp., Armonk, NY, USA) was used for the
statistical analysis of the data obtained through the experiment.
The data were compared by using t-test between two groups, and the
differences in data among many groups were analyzed with the
one-way analysis of variance followed by a post hoc test (Least
Significant Difference). P<0.05 was considered to indicate a
statistically significant difference.
Results
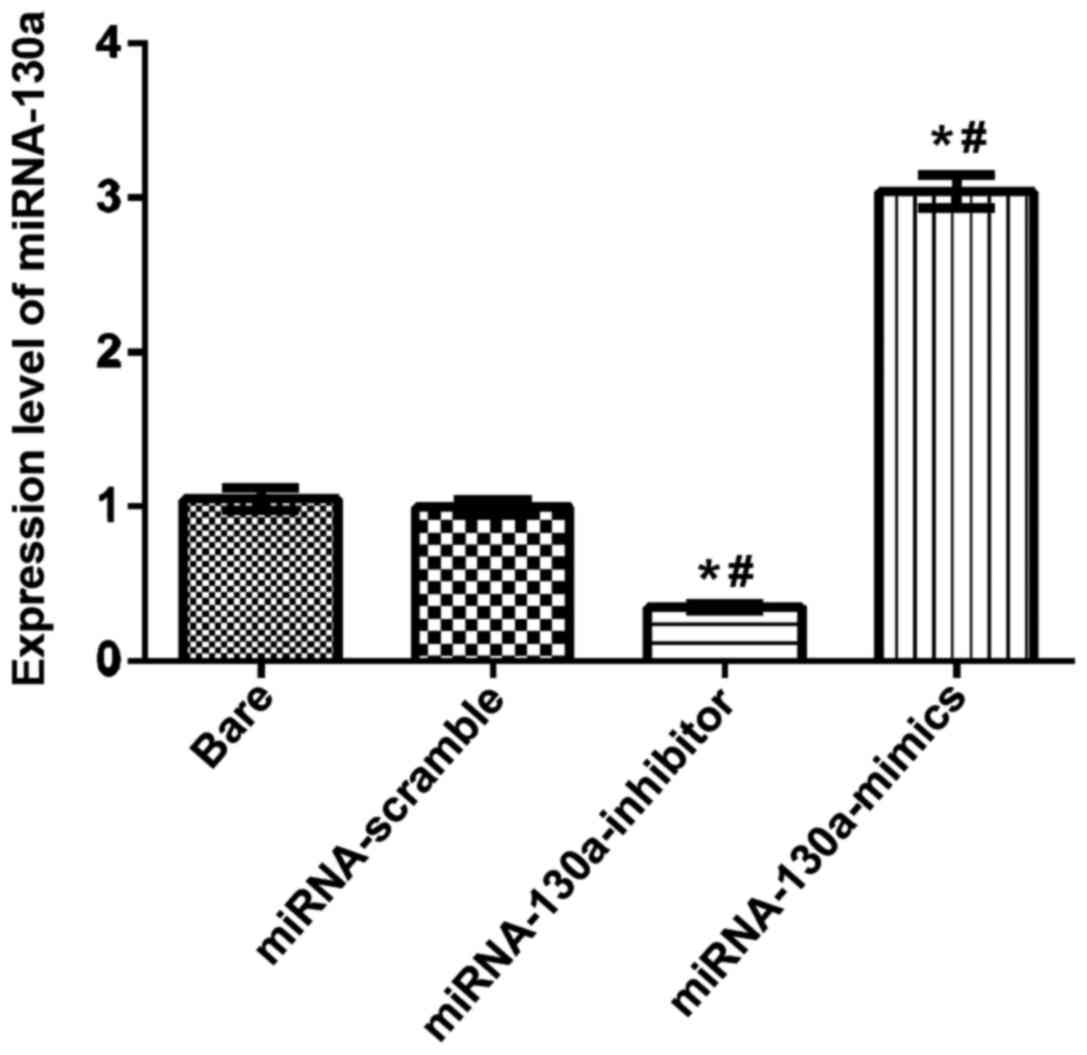
The expression level of miRNA-130a
detected by RT-qPCR
After U-87MG cells were transfected separately as
the bare group, the miRNA-130a-inhibitor, the miRNA-130a-mimics and
the miRNA-scramble groups for 24 h, RT-qPCR detection results
showed that the expression level of miRNA-130a in the
miRNA-130a-mimics group was significantly increased, compared with
those in the other groups; compared with the bare and
miRNA-scramble groups, differences were statistically significant
(P<0.05); the expression level of miRNA-130a in the
miRNA-130a-inhibitor group was obviously downregulated, and
compared with the bare and miRNA-scramble groups, differences were
also statistically significant (P<0.05); the difference between
the bare and miRNA-scramble groups was not statistically
significant (Fig. 1).
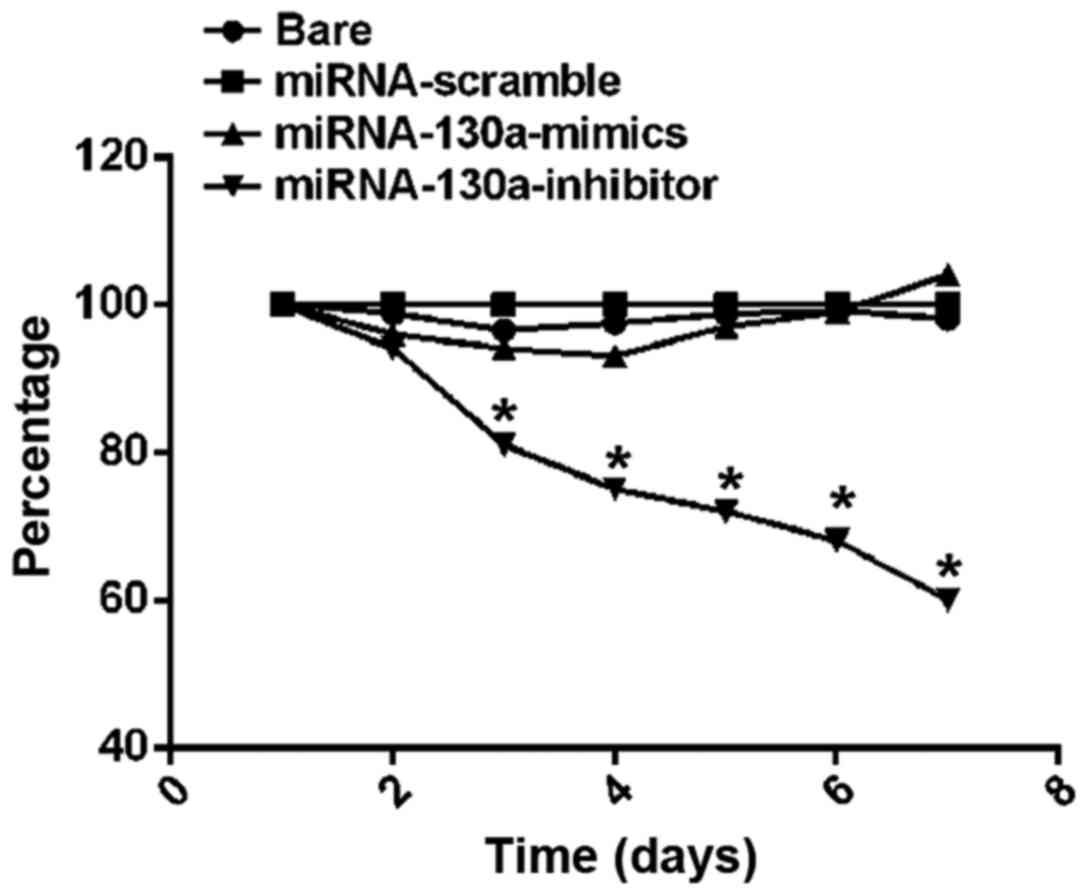
U-87MG cell proliferation activity
tested via MTT assay
MTT assay results on cell proliferation activity
showed that from the 3rd day, the survival rate of U-87MG cells in
the miRNA-130a-inhibitor group began to be significantly decreased
compared with those in the bare, miRNA-scramble and
miRNA-130a-mimics groups, and differences between the groups were
statistically significant (P<0.01); on the 6th day, the cell
survival rate in the miRNA-130a-inhibitor group was the lowest, and
the difference from that in the bare group was statistically
significant (P<0.05) (Fig. 2).
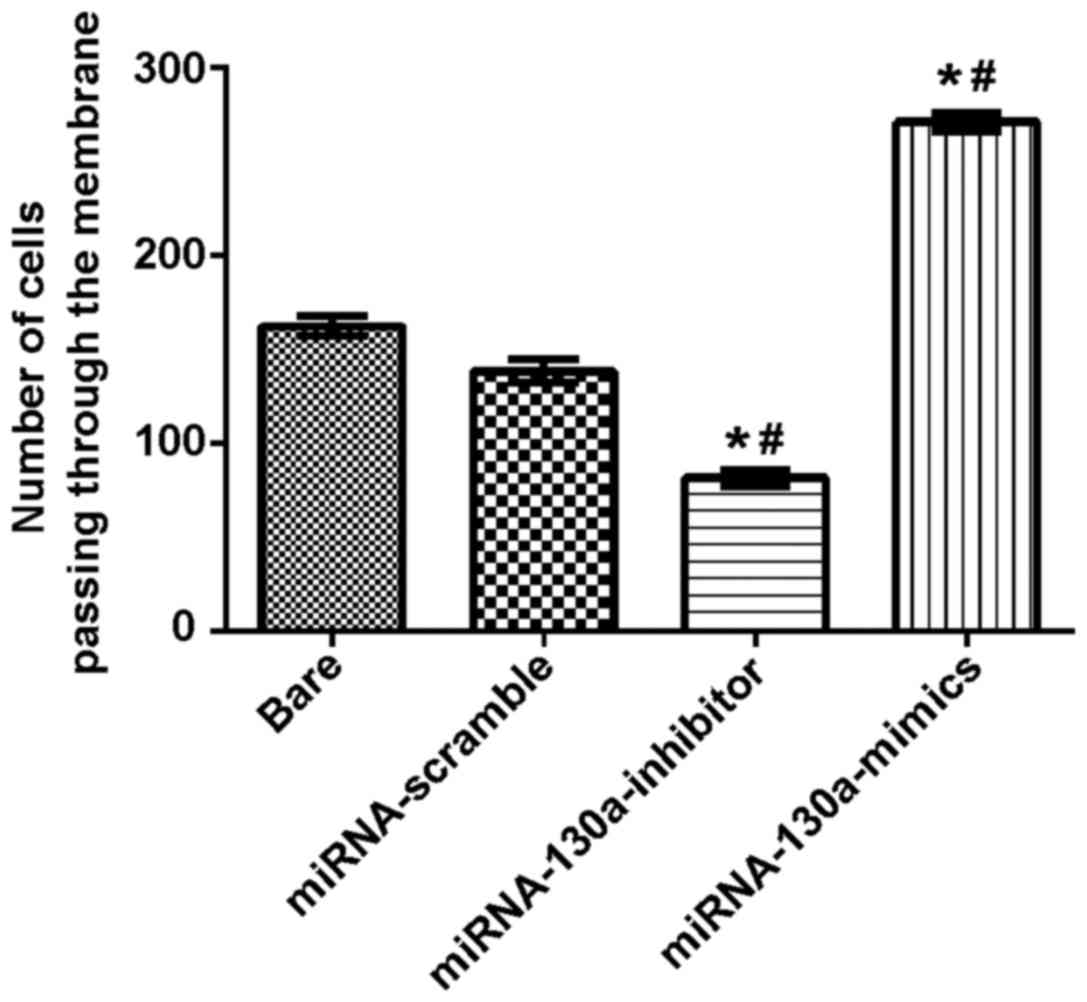
Expression of miRNA-130a and invasion
abilities of U-87MG cells
Transwell chambers were used to test the invading
ability of the miRNA-130a cells, and the results showed that the
number of cells passing through the membrane in the bare,
miRNA-scramble, miRNA-130a-inhibitor and miRNA-130a-mimics groups
were 187.01±8.78, 154.90±8.57, 84.71±4.41 and 285.33±14.35,
respectively. The difference between the miRNA-scramble and bare
groups was not statistically significant; compared with the
miRNA-scramble and bare groups, the miRNA-130a-inhibitor group
showed a significantly decreased invasion ability with obviously
statistical differences (P<0.05), while the miRNA-130a-mimics
group had a remarkably increased invasion ability with
statistically significant differences (P<0.05) (Fig. 3).
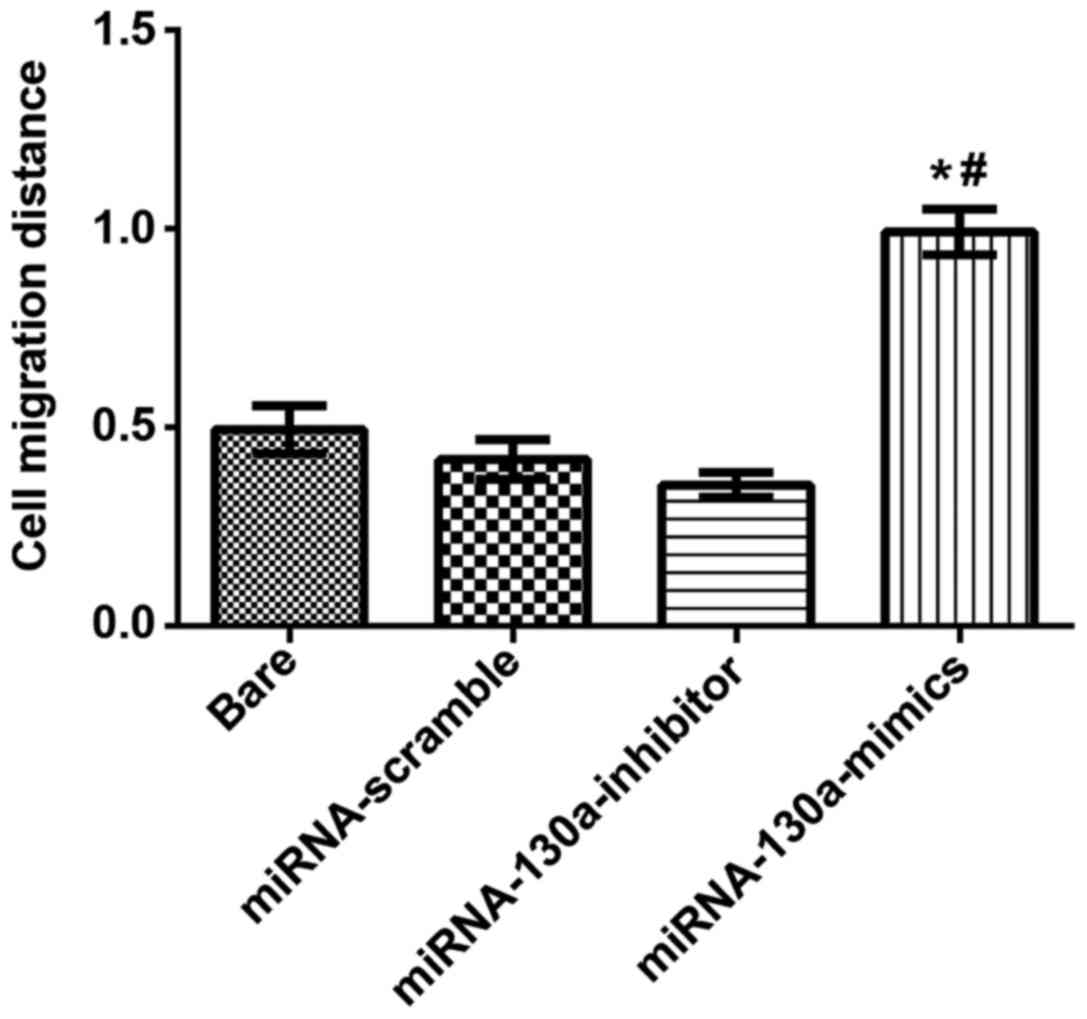
Expression of miRNA-130a and migration
abilities of U-87MG cells
Under an inverted optical microscope (BX-42;
Olympus) the width of scratching wound was observed, and the
results revealed that the cell migration distance in the
miRNA-130a-mimics group was significantly longer than that in the
miRNA-scramble and bare groups with statistically significant
differences (P<0.05); in the miRNA-130a-inhibitor group, the
migration distance was increasingly and significantly shorter than
that of the miRNA-scramble and bare groups with obviously
statistical differences (P<0.05). There were no statistically
significant differences between the bare and miRNA-scramble groups
(Fig. 4).
Apoptosis of glioma cells after
transfection of miRNA-130a
The apoptosis of U-87MG cells varied. After only
miRNA-130a was transfected, the number of apoptotic cells increased
(with the apoptotic cell proportion of 3.56%). Apoptotic cell
proportions in the bare, miRNA-scramble and miRNA-130a-mimics
groups were 1.62, 1.81 and 2.01%, respectively after transfection,
while the apoptosis in the miRNA-130a-inhibitor group (with
apoptotic cell proportion of 13.54%) showed statistical differences
from those in the former three groups (P<0.01).
Discussion
Worldwide tumors jeopardize human health, and the
occurrence and development mechanisms are complicated. According to
the statistical data, the incidence rate of tumors in adults in
2013 is 3.19/100,000 in America, and the number of male patients is
larger than that of female patients. The occurrence of a tumor
shows a positive correlation with age. Elderly people aged 70–80
years are vulnerable to tumors (8,9). The
prognosis is relatively poor, and there are many risk factors, such
as age, place and size of tumor (10). Currently, the main treatment method is
the combination of operation and chemoradiotherapy, and the 2-year
survival rates of patients treated are remarkably increased from
10.4–26.5% (9). However, the
postoperative relapse and metastasis are prone to causing death of
patients. Therefore, it is expected that an effective gene marker
will be found and used to diagnose tumor patients at an early
stage.
In the last 10 years, miRNA, as a new marker, has
gradually attracted people's attention and is abnormally expressed
in multiple tumors. Furthermore, it is involved in the occurrence
and development of tumors, such as proliferation, migration and
invasion and biological functions of many tumors (11–13). The
differential expression of miRNA-130a occurs in many tumors, for
example, its expression is downregulated in leukemia (14) patients. In cell culture in
vitro, miRNA-130a can adjust the survival of cancer cells. In
addition, miRNA-130a is also differentially expressed in diseases,
including lung (15), prostate
(16), breast (17), gallbladder (18) and cervical cancers (19).
Currently, there are few studies on the expression
of miRNA-130a in glioma, and through RT-qPCR, MTT assay, Transwell
migration assay, scratch assay and flow cytometry (FCM), this study
tried to prove the expression of miRNA-130a in glioma cells and
other biological functions. First of all, RT-qPCR was used to
detect the expression of miRNA-130a in the bare, miRNA-130a-mimics,
miRNA-130a-inhibitor and miRNA-scramble groups, and the results
showed that after transfection, the expression quantity in the
miRNA-130a-mimics group was significantly increased (P<0.05),
indicating that miRNA-130a is highly expressed in glioma cells. The
expression in the miRNA-130a-inhibitor group was obviously
decreased, compared with the bare and miRNA-scramble groups
(P<0.05). It is speculated that the functions of glioma cells
including proliferation, invasion, migration and apoptosis can be
inhibited by inhibiting and decreasing the expression of miRNA-130a
in cells. To verify such a speculation, MTT, Transwell migration
and scratch assays were conducted. In the cell proliferation assay,
from the 2nd day, the cell proliferation began to be inhibited in
the miRNA-130a-inhibitor group, and as of the 7th day, it showed
significant differences compared to those in the other 3 groups.
This fact, reveals that the inhibition of miRNA-130a can
effectively reduce the proliferation of cancer cells. Wang et
al (20) reported that miRNA-130a
is highly expressed in non-small cell lung cancer and antagonizes
Gax gene which plays an inhibiting role on proliferation, migration
and angiogenesis of the endothelial cells. This shows that the
expression of the same miRNA is identical in different cancers.
Then, Transwell migration and scratch assays further confirmed this
view. According to the results, the invasion and migration
abilities in the miRNA-130a-mimics group were significantly
improved, compared to those in the other 3 groups (P<0.05).
Among U-87MG cells, the proportions of apoptotic cells in the blank
and negative control groups were 6.88 and 7.54%, respectively.
After the transfection of miRNA-130a, they were increased to 31.84
and 30.31%, respectively. The experiment proved that the expression
of miRNA-130a can weaken the abilities of cell proliferation and
apoptosis. Apoptosis plays a critical role in the occurrence and
development of tumors and is an important method on tumor
treatment.
In conclusion, this study verified that miRNA-130a
is highly expressed in glioma through the experiments. Moreover,
the downregulated miRNA-130a inhibits the proliferation of glioma
cell lines and promotes apoptosis. This indicates that miRNA-130a
can be considered a candidate target for the gene therapy of
glioma.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
KL and HS conceived and designed the study. KL, SZ
and SB were responsible for the collection and analysis of the the
in vitro data. KL, HS and SW interpreted the data and
drafted the manuscript. HS and SZ revised the manuscript critically
for important intellectual content. All authors read and approved
the final study.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
The Second Nanning People's Hospital (Nanning, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sathornsumetee S, Reardon DA, Desjardins
A, Quinn JA, Vredenburgh JJ and Rich JN: Molecularly targeted
therapy for malignant glioma. Cancer. 110:13–24. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Clarke J, Butowski N and Chang S: Recent
advances in therapy for glioblastoma. Arch Neurol. 67:279–283.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Valle-Folgueral JM, Mascarenhas L, Costa
JA, Vieira F, Soares-Fernandes J, Beleza P and Alegria C: Giant
cell glioblastoma: Review of the literature and illustrated case.
Neurocirugia (Astur). 19:343–349. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Iorio MV and Croce CM: MicroRNA
dysregulation in cancer: Diagnostics, monitoring and therapeutics.
A comprehensive review. EMBO Mol Med. 4:143–159. 2012. View Article : Google Scholar
|
|
5
|
Prosdocimo G and Giacca M: Manipulating
the proliferative potential of cardiomyocytes by gene transfer.
Adult Stem Cells. Di Nardo P, Dhingra S and Singla D: 1553. Humana
Press; New York, NY; pp. 41–53. 2017, https://doi.org/10.1007/978-1-4939-6756-8_4
View Article : Google Scholar
|
|
6
|
Qiu S, Lin S, Hu D, Feng Y, Tan Y and Peng
Y: Interactions of miR-323/miR-326/miR-329 and
miR-130a/miR-155/miR-210 as prognostic indicators for clinical
outcome of glioblastoma patients. J Transl Med. 11:102013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Allen M, Bjerke M, Edlund H, Nelander S
and Westermark B: Origin of the U87MG glioma cell line: Good news
and bad news. Sci Transl Med. 8:354re32016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yabroff KR, Harlan L, Zeruto C, Abrams J
and Mann B: Patterns of care and survival for patients with
glioblastoma multiforme diagnosed during 2006. Neuro-oncol.
14:351–359. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ohgaki H, Dessen P, Jourde B, Horstmann S,
Nishikawa T, Di Patre PL, Burkhard C, Schüler D, Probst-Hensch NM,
Maiorka PC, et al: Genetic pathways to glioblastoma: A
population-based study. Cancer Res. 64:6892–6899. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Streiff MB, Ye X, Kickler TS, Desideri S,
Jani J, Fisher J and Grossman SA: A prospective multicenter study
of venous thromboembolism in patients with newly-diagnosed
high-grade glioma: Hazard rate and risk factors. J Neurooncol.
124:299–305. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang Y, Yang P and Wang XF:
Microenvironmental regulation of cancer metastasis by miRNAs.
Trends Cell Biol. 24:153–160. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fujita Y, Yoshioka Y and Ochiya T:
Extracellular vesicle transfer of cancer pathogenic components.
Cancer Sci. 107:385–390. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li BS, Zuo QF, Zhao YL, Xiao B, Zhuang Y,
Mao XH, Wu C, Yang SM, Zeng H, Zou QM, et al: MicroRNA-25 promotes
gastric cancer migration, invasion and proliferation by directly
targeting transducer of ERBB2, 1 and correlates with poor survival.
Oncogene. 34:2556–2565. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kovaleva V, Mora R, Park YJ, Plass C,
Chiramel AI, Bartenschlager R, Döhner H, Stilgenbauer S, Pscherer
A, Lichter P, et al: miRNA-130a targets ATG2B and DICER1 to inhibit
autophagy and trigger killing of chronic lymphocytic leukemia
cells. Cancer Res. 72:1763–1772. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu L, Nie J, Chen L, Dong G, Du X, Wu X,
Tang Y and Han W: The oncogenic role of microRNA-130a/301a/454 in
human colorectal cancer via targeting Smad4 expression. PLoS One.
8:e555322013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fujita Y, Kojima T, Kawakami K, Mizutani
K, Kato T, Deguchi T and Ito M: miR-130a activates apoptotic
signaling through activation of caspase-8 in taxane-resistant
prostate cancer cells. Prostate. 75:1568–1578. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pan Y, Wang R, Zhang F, Chen Y, Lv Q, Long
G and Yang K: MicroRNA-130a inhibits cell proliferation, invasion
and migration in human breast cancer by targeting the RAB5A. Int J
Clin Exp Pathol. 8:384–393. 2015.PubMed/NCBI
|
|
18
|
Ma MZ, Li CX, Zhang Y, Weng MZ, Zhang MD,
Qin YY, Gong W and Quan ZW: Long non-coding RNA HOTAIR, a c-Myc
activated driver of malignancy, negatively regulates miRNA-130a in
gallbladder cancer. Mol Cancer. 13:1562014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
He L, Wang HY, Zhang L, Huang L, Li JD,
Xiong Y, Zhang MY, Jia WH, Yun JP, Luo RZ, et al: Prognostic
significance of low DICER expression regulated by miR-130a in
cervical cancer. Cell Death Dis. 5:e12052014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang XC, Tian LL, Wu HL, Jiang XY, Du LQ,
Zhang H, Wang YY, Wu HY, Li DG, She Y, et al: Expression of
miRNA-130a in nonsmall cell lung cancer. Am J Med Sci. 340:385–388.
2010. View Article : Google Scholar : PubMed/NCBI
|