Introduction
Esophageal cancer is a type of malignant tumor
originating from the esophageal mucus epithelia or glands. It is
also one of the most commonly-identified malignant GI tract tumor
types in China (1). There are two
main factors that cause esophageal cancer. The first is lifestyle
factors including high-temperature beverages, heavy alcohol
drinking and tobacco smoking. The second is genetic predisposition
in a population (2). The incidence
rate of esophageal squamous cell carcinoma (ESCC) in China is
significantly increased compared with that in the USA (3). As the disease presentation of
early-stage ESCC varies extensively, the majority of patients
exhibit mid-to advanced-stage disease when clinically diagnosed.
The majority of patients are treated predominantly by a combination
of chemo- and radiotherapy instead of surgery (4). Chemotherapeutic drugs that are
recommended by the National Comprehensive Cancer Network in the
United States of America include cisplatin, paclitaxel, irinotecan,
docetaxel, fluorouracil and epirubicin (5). Cisplatin is the primary choice among
these for advanced esophageal cancer in China, due to its high
single-drug efficiency and relatively cheap price (6). However, the single-drug efficiency of
cisplatin in esophageal cancer treatment remains~20%, and the
efficiency of cisplatin-based chemotherapy is not >50% (7). These data indicates that a certain group
of patients may not benefit from the chemotherapy using cisplatin,
but also suffer from the adverse effects of the treatment and the
associated financial burden (8).
Individuals react differently to the same drug,
which is hypothesized to result from distinctions with in
individual genomes. Gene polymorphisms are the primary factor that
causes this variance between individuals (9). The C8092A polymorphism of the Excision
repair cross-complementation group 1 (ERCC1) gene have three
genotypes, wild-type ERCC1-8092 (C/C genotype), the heterozygous
mutation of ERCC1-8092 (C/A genotype) and the homozygous mutation
of ERCC1-8092 (A/A genotype). It has been demonstrated that the
C8092A polymorphism of the ERCC1 gene is significantly associated
with the outcome of platinum treatment in nasopharyngeal and lung
cancer, pleural mesothelioma, breast cancer and ovarian cancer
(10–14). Bradbury et al (15) investigated 150 patients with
esophageal cancer treated with chemoradiation therapy using
platinum, and identified that the patients possessing an ERCC1-8092
A/A genotype or A/C genotype exhibited improved disease-free
survival and overall survival (OS) compared with the patients with
the C/C genotype (P=0.04 and P=0.03, respectively). Our previous
study also indicated that among patients with advanced esophageal
cancer treated by cisplatin and fluorouracil, the A/A or A/C
patient groups demonstrated improved response rates (RR) and
progression-free survival (PFS) compared with the C/C group
(8). These results indicate that A/A
and A/C patients are likely to be more sensitive to platinum
compared with patients with the C/C polymorphism (16). However, whether differentiation of
treatments for esophageal cancer based on the ERCC1 C8092A genotype
may increase the efficiency of chemotherapy and prolong the
survival of patients in China remains unknown. Therefore, in order
to verify this hypothesis, and to optimize the individualized
treatment for patients with advanced esophageal cancer. These
patients of the individualized treatment group were treated based
on their ERCC1 C8092A genotype. The outcomes, including RR, PFS, OS
and adverse events, from the standard and individualized treatment
groups were analyzed.
Materials and methods
Patients
Anhui Provincial Hospital Oncology Department
(Hefei, China), Anhui Provincial Cancer Hospital Oncology
Department (Hefei, China) and Anhui Provincial Cardiovascular
Hospital Oncology Department (Hefei, China) all maintain
prospective databases. Eligible patients exhibited histologically
confirmed advanced squamous cell carcinoma of esophagus and
measurable lesion(s). All patients provided written informed
consent. The present study was approved by the Clinical Research
Ethics Committee of the Anhui Provincial Hospital. Other inclusion
criteria were: Histologically confirmed un-resectable or recurrent
advanced esophageal cancer following surgery; Eastern Cooperative
Oncology Group performance status (ECOG-PS) ≤2, survival not <3
months; clinically measurable lesion(s); last dose of adjuvant
chemotherapy occurred no later than 6 months ago or no history of
chemotherapy; and adequate hematological, hepatic and renal
functions for chemotherapy. Patients were excluded if they had
psychiatric disease, severe cardiological, pulmonary, hepatic or
renal diseases, suffered from colitis gravis or were pregnant. A
total of 140 patients were enrolled in the present study. Patients
were followed up primarily by clinic visits, phone calls and
e-mails until December 2015. Follow-up information included
measurement of lesions, tumor markers (carcinoembryonic antigen and
SCC) and ECOG-PS. Among the patients, genotyping information was
not available for 3 patients, 4 failed to receive the protocol
treatment due to adverse events, 2 were determined to be
non-evaluable and an additional 4 were lost to follow-up.
Therefore, 127 patients were analyzed. Of those, 49 were female
(38.6%) and 78 were male (61.4%); 21 were aged<60 years (16.5%)
and 106 were aged ≥60 years (83.5%). The clinical stages of
esophageal cancer were classified in accordance with the
internationally accepted tumor node-metastasis (TNM) staging system
(7th edition) (17,18). A total of 75 exhibited low
differentiation (59.1%), 37 exhibited mid differentiation (29.1%)
and 15 exhibited high differentiation (11.8%).
Treatment
The standard regimen consisted of cisplatin (25
mg/m2 from days 1 to 3) and paclitaxel (150
mg/m2 on day 1). The length of standard regimen was 3
days. For the individualized treatment group, blood samples were
obtained for the ERCC1-C8092A genotype analysis prior to initiation
of treatment. If the genotype was not C/C, the regimen was the same
as the standard regimen, otherwise the regimen consisted of
fluorouracil (750 mg/m2 from days 1 to 5) and paclitaxel
(150 mg/m2 on day 1) which took 5 days to complete.
These regimens were repeated every 21 days as one cycle. Patients
continued to be treated with the regimen according to the protocol
until progressive disease (PD) was confirmed according to a
contrast computed tomography or magnetic resonance imaging scan. If
PD was confirmed, the regimen was either changed to second-line
chemotherapy or supportive treatment according to the ECOG-PS of
the patients. All patients completed >2 cycles of chemotherapy.
The maximum number of chemotherapy cycles was 6 cycles.
Response evaluation
Response Evaluation Criteria in Solid Tumors version
1.1 (19) was used for efficacy
assessment. Complete response (CR) was defined as the disappearance
of all lesions and no recurrence over a 4-week period. Partial
response (PR) was defined as ≥30% decrease in the tumor sum of
longest diameters (SLD) and PD as ≥20% increase in the SLD or the
occurrence of new lesions. Stable disease (SD) was calculated as
non-PR/PD, and response rate (RR) was calculated using the
following formula: RR=CR + PR. PFS was defined as the time interval
between inclusion and the first progression or mortality from all
causes. OS was defined as the time interval between inclusion and
mortality from all causes. The response evaluation was performed
every 2 weeks. If symptoms that supported PD were observed in the
clinic, the evaluation was performed prior to the date
predetermined within the protocol. If PD was confirmed, the regimen
was either changed or optimized to better support the treatment.
Patients with CR, PR or SD continued to be treated with the regimen
on protocol for a maximum of 6 cycles, until PD was confirmed.
Lesions in all the patients that discontinued chemotherapy were
evaluated every month.
Adverse events and adjustments
The toxicity of the chemotherapy was graded
according to National Cancer Institute Common Terminology Criteria
for Adverse Events version 4.03 (20). Treatment for adverse events was
administered to all grade 1 and 2 toxicities, and the original
regimen was continued when adverse events were alleviated. Regimen
dose was decreased by 25% for all grade 3 and 4 toxicities and
discontinued in case of recurrence and persistence. To increase
patient tolerance of chemotherapy, adjuvant treatment using
anti-emetics, hydration and diuretics drugs was also administered
over the period of chemotherapy. Granulocyte colony-stimulating
factor was administered to grade 3 and grade 4 myelotoxicities 24 h
after chemotherapy.
Genotype analysis of ERCC1 C8092A
Promoter and coding single nucleotide polymorphisms
(SNPs) in ERCC1 C8092A (rs3212986) in the SNP database of the
National Center for Biotechnology Information (BUILD 151;
http://www.ncbi.nlm.nih.gov/SNP) were
sourced. The DNA of eukaryotes in peripheral blood of patients was
obtained using phenol-chloroform extraction and analyzed with
polymerase chain reaction (PCR) amplification. The primers for PCR
was prepared by Sangon Biotech Co., Ltd. (Shanghai, China). The PCR
primers used to amplify the DNA were as follows: ERCC1-8092
Forward, 5′-ACAGTGCCCCAAGAGGAGAT-3′ and reverse,
5′-AGTCTCTGGGGAGGGATTCT-3′. The reaction mixture consisted of: 10X
buffer solution (Sangon Biotech Co., Ltd.) (15
mmol/lMg2+), 2 µl 2.5 mM dNTPs (2.5 mmol/l), 0.5 µl of
each primer (10 mmol/l), 0.2 µl Taq (Sangon Biotech Co., Ltd.)
enzyme (5 U/µl), 50 ng genomic DNA sample and sterile distilled
water (total volume, 50 µl). The thermo cycler conditions were as
follows: 95°C for 5 min; then, 94°C for 15 sec, 60°C for 25 sec and
72°C for 30 sec for 40 cycles, then 72°C for 10 min. The products
were kept at 4°C until use. The products of PCR were sequenced by
Sangon Biotech Co., Ltd., and the gene polymorphisms were analyzed.
The genotypes were identified using MxPro-Mx3000P v4.00 analysis
software (Agilent Technologies, Inc., Santa Clara, CA, USA). The
data were analyzed using the 2−ΔΔCq method (21). The sequence chromatograms were
analyzed using Chromas software v2.0 (Technelysium Pty Ltd., South
Brisbane, Australia) to search for SNPs at the target locus of each
gene.
Statistical analysis
Statistical analyses were performed using SPSS
software version 17.0 (SPSS, Inc., Chicago, IL, USA). Prior to
analysis, the Hardy-Weinberg equation for the equilibrium of allele
distributions was used to statistically evaluate the data along
with the χ2 test. The quantitative data was presented as
frequencies and percentages while quantitative data with a normal
distribution was presented as the mean ± standard deviation.
The distribution of demographic variables was
compared between groups through nonparametric tests. The
Mann-Whitney U test was used to compare the data between two
groups. The Kruskal-Wallis test was used to compare the data
between three groups. The response rate was evaluated along with
the χ2 test or Fisher's exact test for 2×2 contingency
tables.
Independent influential factors of treatment were
assessed using logistic regression analysis, with variables
significant in the univariate analysis were included into a
multivariate model. All testing was 2-sided with significance
determined at P≤0.05.
All survival analyses were performed with the
Kaplan-Meier method. Survival rate was evaluated by the log-rank
test. The significance level α was 0.05, and P<0.05 was
considered to indicate a statistically significant difference.
Results
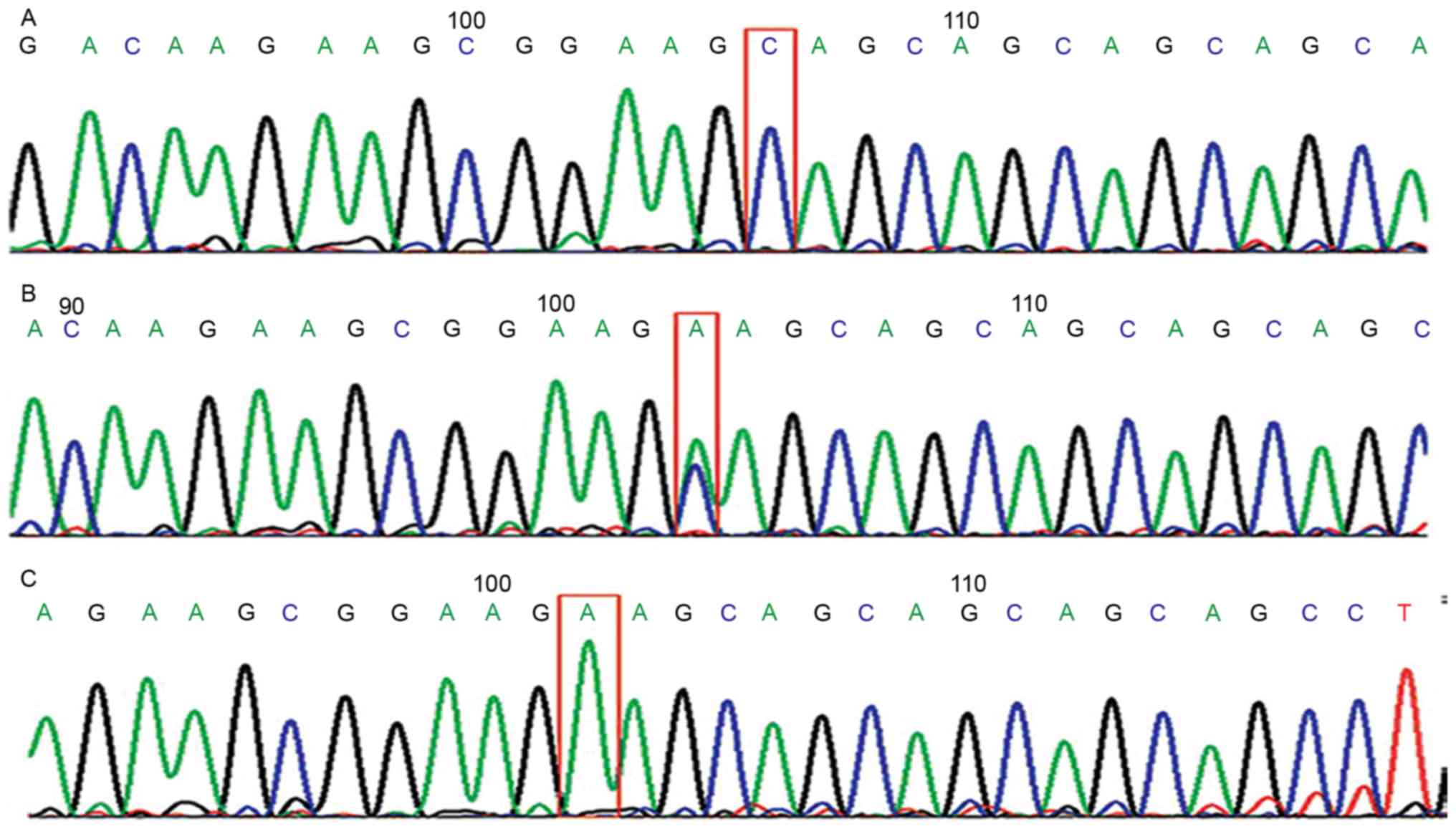
Sequencing of ERCC1 C8092A genes and
genetic equilibrium test
The sequencing result of the ERCC1 C8092A genes from
84 individualized treated patients is presented in Fig. 1. According to the Hardy-Weinberg
Principle, the Hardy-Weinberg equilibrium was reached in the
distribution of genotypes, indicating that the samples were from
the same Mendelian population (P>0.05; Table I).
 | Table I.Genetic equilibrium test in the
individualized treatment group. |
Table I.
Genetic equilibrium test in the
individualized treatment group.
| Genotype | N | χ2 | P-value |
|---|
| ERCC1 C8092A | 84 | 0.7001 | 0.402a |
| A/A | 14 |
| – |
| A/C | 36 |
| – |
| C/C | 34 |
| – |
Clinical characteristics in treatment
groups
The sex, median age, ECOG-PS, TNM staging and
pathological differentiation grading were not significantly
different between the individualized and the standard treatment
groups based on the χ2 or Mann-Whitney U test results
(Table II).
 | Table II.Clinical characteristics in treatment
groups (n=127). |
Table II.
Clinical characteristics in treatment
groups (n=127).
| Clinical
Characteristics | Standard treatment
group (n=43) [n(%)] | Individualized
treatment group (n=84) [n(%)] |
χ2/Z | P-value | A/C & A/A
(n=50) | C/C (n=34) |
χ2/Z | P-value |
|---|
| Sex |
|
|
|
|
|
|
|
|
|
Male | 27 (62.8) | 51 (60.7) | 0.052 | 0.821 | 34 (68.0) | 18 (52.9) | 1.946 | 0.163 |
|
Female | 16 (37.2) | 33 (39.3) | – | – | 16 (32.0) | 16 (47.1) | – | – |
| Age, years | 63.37±4.76 | 63.06±5.16 | 0.332 | 0.741 | 63.14±5.48 | 62.94±4.72 | 0.172 | 0.864 |
| ECOG-PS |
|
|
|
|
|
|
|
|
| 0 | 5
(11.6) | 8 (9.5) | 0.109a | 0.913 | 6
(12.0) | 2 (5.9) | 0.678a | 0.497 |
| 1 | 25 (58.1) | 53 (63.1) | – | – | 31 (62.0) | 22 (64.7) | – | – |
| 2 | 13 (30.2) | 23 (27.4) |
|
| 13 (26.0) | 10 (29.4) | – | – |
| TNM staging |
|
|
|
|
|
|
|
|
|
III | 19 (44.2) | 39 (46.4) | 0.058 | 0.810 | 23 (46.0) | 16 (47.1) | 0.009a | 0.924 |
| IV | 24 (55.8) | 45 (53.6) | – | – | 27 (54.0) | 18 (52.9) | – | – |
| Pathological
differentiation |
|
|
|
|
|
|
|
|
|
Low | 25 (58.1) | 50 (59.5) | 0.265a | 0.791 | 28 (56.0) | 20 (58.5) | 0.407 | 0.684 |
|
Medium | 12 (27.9) | 25 (29.8) | – | – | 15 (30.0) | 11 (32.4) | – | – |
|
High | 6
(14.0) | 9 (10.7) | – | – | 7
(14.0) | 3 (8.8) | – | – |
| Location of
tumor |
|
|
|
|
|
|
|
|
|
Cervical | 2 (4.7) | 8 (9.5) | 0.259 | 0.879 | 6
(12.0) | 3 (8.8) | 0.324 | 0.85 |
|
Middle | 23 (53.4) | 39 (46.4) | – | – | 23 (46.0) | 15 (44.1) | – | – |
|
Lower | 18 (41.9) | 37 (44.1) | – | – | 21 (42.0) | 16 (47.1) | – | – |
| Radiation
therapy |
|
|
|
|
|
|
|
|
|
Yes | 28 (65.1) | 60 (71.4) | 0.533 | 0.466 | 35 (70.0) | 24 (70.6) | 0.003 | 0.954 |
| NA | 15 (34.9) | 24 (28.6) | – | – | 15 (30.0) | 10 (29.4) | – | – |
RR
The RR of the individualized and standard treatment
groups were 53.6 and 34.8%, respectively, which were significantly
different (P=0.046). Within the individualized treatment group, the
RR of the patients with non-C/C genotypes was 52.0%, and the RR of
the patients with the C/C genotype was 55.8%, which were not
significantly different (P=0.726; Table
III). Multivariate analysis indicated that the ECOG-PS,
individualized treatment, high differentiation of tumors and the
TNM staging were all independent prognostic factors of RR (Table IV).
 | Table III.Comparison of RR. |
Table III.
Comparison of RR.
| Patient
grouping | Response (CR+PR),
n | No response
(SD+PD), n | RR (%) | χ2 | P-value |
|---|
| Standard treatment
group | 15 | 28 | 34.8 | 3.095 | 0.046 |
| Individualized
treatment group | 45 | 39 | 53.6 |
|
|
| A/C and A/A | 26 | 24 | 52.0 | 0.123 | 0.726 |
| C/C | 19 | 15 | 55.8 |
|
|
 | Table IV.Univariate and multivariate
analyses. |
Table IV.
Univariate and multivariate
analyses.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variables | OR (95% CI) | P-value | OR (95% CI) | P-value |
|---|
| Sex |
|
|
|
|
|
Male | 1 | – | – | – |
|
Female | 1.044
(0.509–2.142) | 0.906 | – | – |
| Age, years |
|
|
|
|
|
≥60 | 1.236
(0.481–3.179) | 0.660 | – | – |
|
<60 | 1 | – | – | – |
| ECOG-PS |
|
|
|
|
| 0 | 1 | – | 1 | – |
| 1 | 0.369
(0.094–1.443) | 0.152 | 0.486
(0.114–2.078) | 0.331 |
| 2 | 0.072
(0.016,0.335) | 0.001 | 0.146
(0.028–0.771) | 0.023 |
| Treatment |
|
|
|
|
|
Standard | 0.464
(0.217–0.992) | 0.048 | 0.377
(0.152–0.931) | 0.034 |
|
Individualized | 1 | – | 1 | – |
| Treatment |
|
|
|
|
|
Standard | 1.169
(0.482–2.806) | 0.726 | – | – |
|
C/C | 0.495
(0.214–1.142) | 0.099 | – | – |
| A/C
& A/A | 1 | – | – | – |
| Status-OS |
|
|
|
|
| 0 | 1 | – | – | – |
| 1 | 0.894
(0.055–14.612) | 0.937 | – | – |
| Histologic
gradea |
|
|
|
|
| G3 | 1 | – | 1 | – |
| G2 | 2.750
(1.224–6.177) | 0.014 | 1.617
(0.629–4.155) | 0.318 |
| G1 | 9.333
(2.444–35.636) | 0.001 | 6.858
(1.583–29.711) | 0.010 |
| Location of
tumor |
|
|
|
|
|
Cervical | 1 | – | – | – |
|
Middle | 1.586
(0.421–5.980) | 0.496 | – | – |
|
Lower | 1.687
(0.443–6.428) | 0.443 | – | – |
| TNM staging |
|
|
|
|
| 3 | 1 | – | 1 | – |
| 4 | 0.213
(0.101–0.452) | <0.001 | 0.355
(0.144–0.874) | 0.024 |
Adverse events
The adverse events in the two groups were primarily
nausea and vomiting, hair loss, myelotoxicity and neurotoxicity. In
the individualized treatment group, the rate of nausea and vomiting
was 89.3% (75/84), within which the rate of grade 3–4 events was
33.3% (28/84). In the standard treatment group, the rate of nausea
and vomiting was 97.7% (42/43), within which the rate of grade 3–4
events was 65.1% (28/43). The rates of nausea and vomiting in
general from the two groups were significantly different (P=0.001).
In the individualized treatment group, the rate of anemia was 91.6%
(77/84), within which the rate of grade 3 to 4 events was 17.9%
(15/84). In the standard treatment group, the rate of anemia was
95.3% (41/43), within which the rate of grade 3 to 4 events was
32.5% (14/43). These rates of anemia in general from the two groups
were significantly different (P=0.004). In the individualized
treatment group, the rates of hair loss, aleukocytosis and
neurotoxicity were 100.0 (84/84), 94.0 (79/84), and 84.5% (71/84),
respectively. In the standard treatment group, the rates of hair
loss, aleukocytosis and neurotoxicity were 100.0 (43/43), 90.7
(39/43), and 83.7% (36/43), respectively. The rates from the two
groups were not significantly different from each other
(P>0.05). Other adverse events such as diarrhea,
thrombocytopenia, hepatic and renal toxicity were less common, and
the rates from the two groups were not statistically different
(Table V).
 | Table V.Comparison of adverse events. |
Table V.
Comparison of adverse events.
|
|
Grades |
|
|
|---|
|
|
|
|
|
|---|
|
| Individualized
(n=84) | Standard
(n=43) |
|
|
|---|
|
|
|
|
|
|
|---|
| Adverse events | 0 | I | II | III | IV | 0 | I | IIa | III | IV |
Za | P-value |
|---|
| Nauseaand
vomiting | 9 | 19 | 28 | 17 | 11 | 1 | 4 | 10 | 18 | 10 | 3.350 | 0.001 |
| Diarrhea | 69 | 9 | 5 | 0 | 1 | 36 | 4 | 3 | 0 | 0 | 0.217 | 0.829 |
| Hair loss | 0 | 10 | 74 | 0 | 0 | 0 | 6 | 37 | 0 | 0 | 0.324 | 0.746 |
| Aleukocytosis | 5 | 23 | 40 | 12 | 4 | 4 | 11 | 21 | 6 | 1 | 0.438 | 0.662 |
|
Thrombocytopenia | 65 | 12 | 6 | 1 | 0 | 31 | 7 | 3 | 2 | 0 | 0.712 | 0.476 |
| Anemia | 7 | 40 | 22 | 11 | 4 | 2 | 9 | 18 | 12 | 2 | 2.921 | 0.004 |
| Hepatic
toxicity | 66 | 15 | 3 | 0 | 0 | 35 | 6 | 2 | 0 | 0 | 0.323 | 0.747 |
| Renal toxicity | 83 | 1 | 0 | 0 | 0 | 42 | 1 | 0 | 0 | 0 | 0.473 | 0.637 |
| Neurotoxicity | 13 | 26 | 45 | 0 | 0 | 7 | 15 | 21 | 0 | 0 | 0.441 | 0.659 |
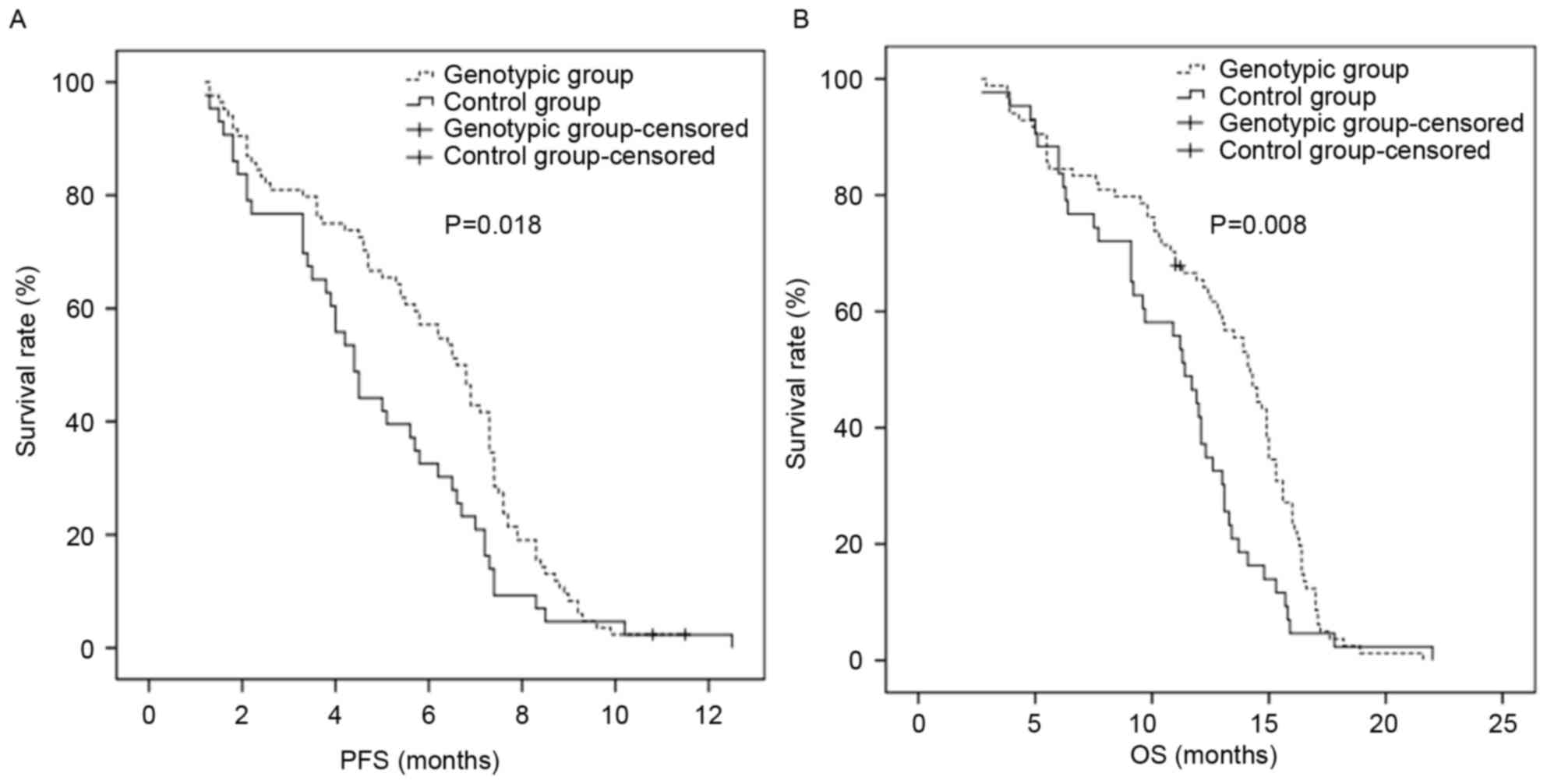
PFS and OS
The PFS in the standard treatment group was 4.4
months [95% confidence interval (CI), 3.8–5.0 months], and the PFS
in the individualized treatment group was 6.6 months (95% CI,
5.8–7.4 months), which were significantly different (P=0.018). The
OS in the standard treatment group was 11.4 months (95% CI,
10.1–12.7 months), and the OS in the individualized treatment group
was 14.2 months (95% CI, 13.2–15.2 months), which were
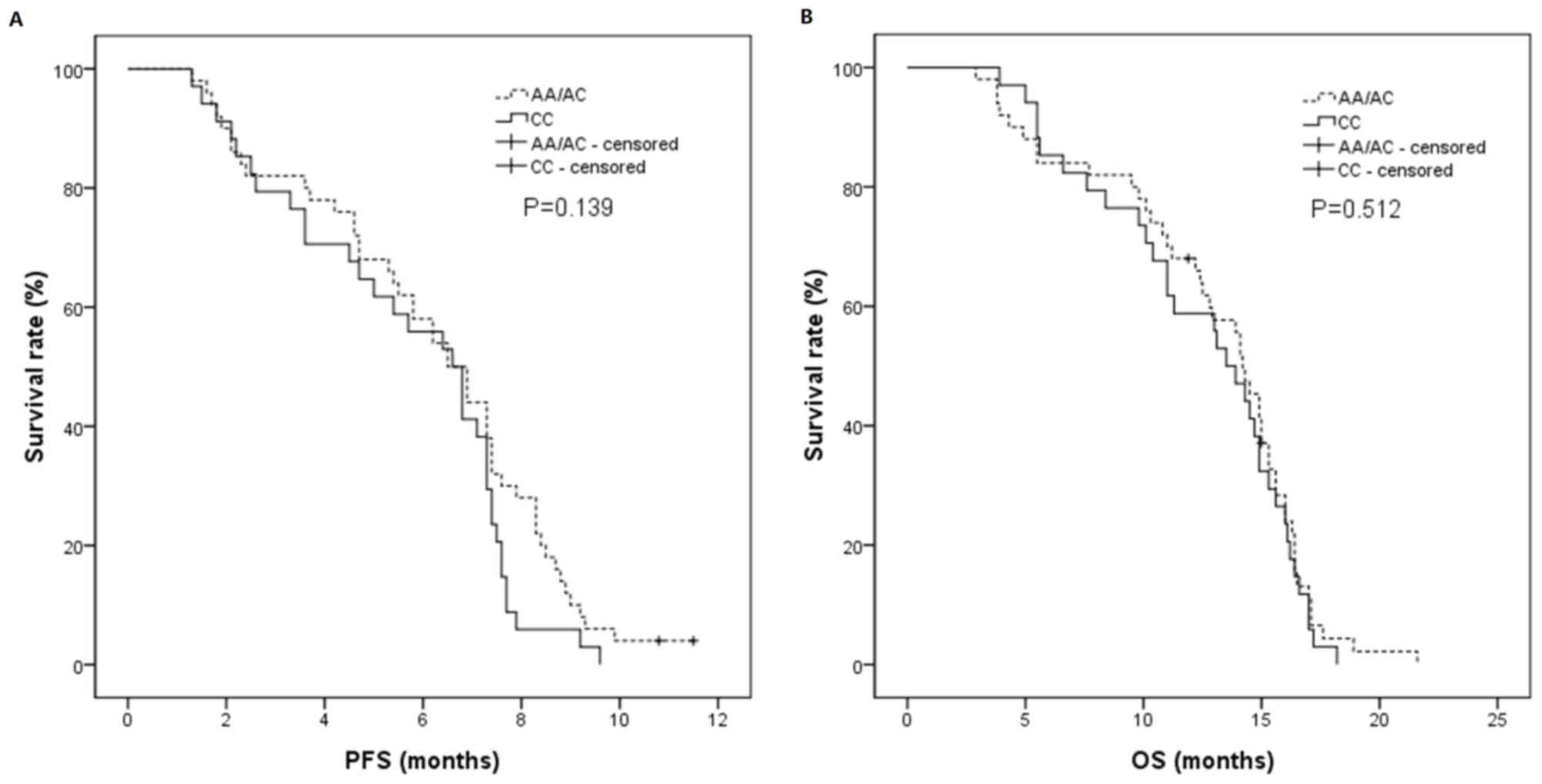
significantly different (P=0.008). In the individualized treatment
group, the PFS in the A/C or A/A groups was 6.5 months (95% CI,
5.4–7.6 months), and the PFS in the C/C group was 6.6 months (95%
CI, 5.3–7.9 months), which were not significantly different
(P=0.139). The OS in A/C or A/A groups was 14.2 months (95% CI,
13.2–15.2 months), and the OS in C/C group was 13.5 months (95% CI,
11.6–15.4 months), which were not significantly different (P=0.512;
Table VI; Figs. 2 and 3).
 | Table VI.Comparison of PFS and OS. |
Table VI.
Comparison of PFS and OS.
| A, Treatment
groups |
|---|
|
|---|
|
| Standard
(n=43) | Individualized
(n=84) |
|
|---|
|
|
|
|
|
|---|
| Survival | Duration,
month | 95% CI | Duration,
month | 95% CI | P-value |
|---|
| Medium PFS |
4.4 | 3.758–5.042 |
6.6 | 5.777–7.423 | 0.018 |
| Medium OS | 11.4 | 10.115–12.685 | 14.2 | 13.228–15.172 | 0.008 |
|
| B,
Genotype-specific groups |
|
|
| A/C & A/A
(n=50) | C/C
(n=34) |
|
|
|
|
|
|
|
Survival | Duration,
month | 95% CI | Duration,
month | 95% CI |
|
|
| Medium PFS |
6.5 | 5.411–7.589 |
6.6 | 5.343–7.857 | 0.139 |
| Medium OS | 14.2 | 13.231–15.169 | 13.5 | 11.643–15.357 | 0.512 |
Discussion
Cisplatin is one of the primary compounds used in
the chemotherapy of esophageal cancer, due to its efficacy and
relatively low price (22). By
forming a platinum-DNA complex in the tumor cell, intra- or
inter-strand cross links are formed, resulting in the termination
of cell cycle at G2/M phase, which consequently triggers apoptosis
in the proliferating cells (23).
However, DNA repairing mechanisms are responsible for the integrity
and stability of the genetic information. On one hand, when
exogenous factors cause changes in the DNA, the cell repairs the
DNA through these mechanisms to prevent additional damage to the
cell. Conversely, when the DNA of tumor cells are damaged in
chemotherapy, the same repairing system is also able to fix the
damaged DNA and ensure that the tumor cells survive the
chemotherapy. Therefore, it is hypothesized that the sensitivity
and resistance to chemotherapy of tumors are associated with DNA
repairing mechanisms. In the human body, there are 6DNA repair
processes: Base excision repair, mismatch repair, homologous
recombination (HR), non-homologous end joining, trans lesion DNA
synthesis and nucleotide excision repair (NER) (24). Among those, NER is primarily
responsible for repairing the DNA adducts induced by polycyclic
aromatic hydrocarbons, ultraviolet light and other exogenous
chemicals (25,26). With respect to platinum-induced DNA
damage in chemotherapy, NER is a multi-functional repairing system.
By excising the DNA adduct and replicating DNA from the
complementary strand, NER retains the integrity of the genome
(27).
ERCC1 is located on the chromosome 19q13.2–13.3. The
whole gene consists of 15 kbp and encodes a protein of 297 amino
acid residues (28). ERCC1 has to
form a heterodimer with DNA repair endonuclease XPF (XPF), which is
responsible for the recognition and incision of the damaged DNA
strand 5′ of the lesion. This step is the rate-determining step in
NER, and also an important step for the regulation of NER (29–31). The
activity of ERCC1 indicates the activity of the repair system of
NER (32). The over expression of
ERCC1 increased the clearance level of platinum-DNA adducts induced
by cisplatin and leads to the resistance of cisplatin in patients.
Previous studies have demonstrated that the ERCC1-C8092A
polymorphism is relevant to the outcome of platinum treatment, and
that the patients with A/A and A/C genotypes are likely to be more
sensitive to platinum treatment compared with the patients with the
C/C genotype (11,33). Bradbury et al (15) investigated 150 patients with
esophageal cancer treated with platinum-based chemo radiation
therapy, and identified that the patients possessing an ERCC1
C8092A genotype of A/A or A/C exhibited improved PFS and OS
compared with the patients with the C/C genotype (P=0.03 and
P=0.04, respectively). Wang et al (33) also demonstrated that in a trial with
256 patients with esophageal cancer treated by cisplatin together
with fluorouracil, patients with the A/A or A/C genotypes exhibited
improved response rate and PFS compared with patients with the C/C
genotype (P<0.01 and P<0.0001, respectively). Therefore, the
present study was designed, according to previous results, to
examine the role of the ERCC1 genotype in the individualized
treatment of advanced esophageal cancer.
The majority of previous studies investigating ERCC1
utilized immunohistochemical analysis, which has certain
disadvantages. Firstly, the tumor tissue for biopsy is small and
the amount of tumor cells available may not be sufficient for
accurate diagnoses following immunohistochemical staining.
Secondly, the analysis in immunohistochemistry is only partially
quantitative, and the result is susceptible to investigator bias.
The present study was based on the expression of ERCC1 at the
molecular level, and therefore was more reliable and accurate. In
addition, the analysis of ERCC1 in previous studies primarily
arises from the analysis of tissue samples, which is complex
(34,35). Schena et al (36) identified that the expression levels of
ERCC1 in tumor tissues and peripheral blood were associatedin
patients with non-small cell lung cancer and patients with head and
neck squamous cell carcinoma.
In the present study, peripheral blood samples were
obtained for ERCC1 C8092A analysis. The patients were randomized
into the individualized and standard treatment groups at a ratio of
2:1, respectively, based on the genotype. The standard regimen was
paclitaxel and cisplatin. In the individualized group, patients
with the non-C/C genotype were treated with paclitaxel and
cisplatin, and patients with the C/C genotype were treated with
paclitaxel and fluorouracil. Paclitaxel is an anticancer drug with
high efficacy. Paclitaxel stabilizes the microtubule polymers and
protects them from disassembly, by inhibiting cell mitosis
(37). Clinical data has demonstrated
that the single-drug efficacy of paclitaxel intreating advanced
esophageal cancer is 32%, suggesting that paclitaxel is relatively
efficient in advanced esophageal cancer treatment (38). The combination of paclitaxel and
cisplatin is one of the most widely-used chemotherapy strategies in
paclitaxel therapies. Zhang et al (39) identified that the efficacy of
paclitaxel and cisplatin treatment in late-stage esophageal cancer
treatment was 48.6% in a Phase II clinical trial. For patients who
are resistant to cisplatin, an alternative choice is paclitaxel
combined with fluorouracil. Yun et al (40) revealed that in recurrent or metastatic
esophageal squamous cell carcinoma, the efficacies of paclitaxel
and capecitabine were 75% in the first-line and 45% in the
second-line treatment. Matsumoto et al (41) and Schnirer et al (42) also demonstrated positive outcomes,
including the RR in the treatment of advanced esophageal cancer
with paclitaxel and fluorouracil. Therefore, the present study
selected the combination of paclitaxel and fluorouracil as the
treatment for patients with the C/C genotype that were not
sensitive to platinum treatment. The RRs of the individualized and
standard treatment groups were 53.6 and 34.8%, respectively. The
difference was statistically significant (χ2=3.095;
P=0.046). The result supported the study hypothesis that
individualized treatment based on ERCC1 genotype increases the RR
of chemotherapy compared with conventional treatment. The adverse
events in the individualized treatment group, including nausea and
vomiting, and anemia, were significantly decreased compared with
the standard treatment group (P=0.001 and P=0.004, respectively).
This indicated that individualized treatment based on the genotype
of patients avoided the side effects induced by cisplatin on those
patients that were not sensitive to cisplatin, increasing the
tolerance of chemotherapy in the patients receiving individualized
treatment.
The PFS in the standard treatment group was 4.4
months (95% CI, 3.8–5.0 months) and 6.6 months (95% CI, 5.8–7.4
months) in the individualized treatment group, which were
significantly different (P=0.018). This result also suggested that
the differentiation of treatments for esophageal cancer based on
the ERCC1 C8092A genotype may benefit the patients during
chemotherapy and prolong the PFS of patients, particularly for
those that were not sensitive to platinum treatment. The OS in the
standard treatment group was 11.4 months (95% CI, 10.1–12.7 months)
and 14.2 months (95% CI, 13.2–15.2 months) in the individualized
treatment group, which were significantly different (P=0.008). This
may be due to the fact that patients in the individualized
treatment group exhibited improved PFS and physical conditions
subsequent to PD and a tolerance for chemotherapy compared with the
standard treatment group. It may also be due to a failure to
control the disease with the second-line treatment following PD in
the standard treatment group. In addition, the differences between
the supportive treatments may also have an effect on the survival
of patients. In the individualized treatment group, the PFS in the
A/C or A/A group was 6.5 months (95% CI, 5.4–7.6 months) and 6.6
months (95% CI, 5.3–7.9 months) in the C/C group, which were not
significantly different (P=0.139). The OS in the A/C or A/A group
was 14.2 months (95% CI, 13.2–15.2 months) and 13.5 months (95% CI,
11.6–15.4 months) in the C/C group, which were not significantly
different (P=0.512). The analysis within the individualized
treatment group indicated that patients with the C/C genotype that
were not sensitive to platinum treatment exhibited improved PFS and
OS compared with AC/AA genotypes when treated based on individual
ERCC1 C8092A genotypes.
The present study only focused on the individualized
treatment for advanced esophageal cancer based on the ERCC1 C8092A
genotype. The population was relatively small, therefore an
increase in the study population and decrease in the distinctions
between the second-line treatments following PD is required to
additionally support the benefit of the individualized treatment.
The present study may provide a molecular basis for the
individualized systemic treatment of advanced esophageal carcinoma.
It may also be valuable to investigate individualized treatment
based on multiple genes simultaneously.
Acknowledgements
The authors would like to thank Dr Xuan-Liang Zhou
(Sangon Biotech Co., Ltd. Shanghai, China) for his technical
assistance with the genotype analysis.
Funding
The present study was supported by the Medical
Scientific Research Foundation of Anhui Province (grant nos.
2010B001 and 13zc012) and by the Science Foundation of Anhui
Province (grant no. 1408085MH179).
Availability of data and materials
All data that were generated or analyzed in the
present study are included in this manuscript.
Authors' contributions
BH and CSJ proposed the study. YWY performed the
majority of the experiments and was a major contributor in writing
the manuscript. YWY analyzed the data and designed the figures. XHH
and YFH contributed to the conception of this study, were involved
in drafting and revising the manuscript and agreed to be
accountable for all aspects of the work. All authors read and
approved the final manuscript for publication.
Ethics approval and consent to
participate
The present study was approved by the Clinical
Research Ethics Committee of the Anhui Provincial Hospital, which
was conducted in accordance with The Declaration of Helsinki.
Participants were fully informed of the procedures, and written
informed consent was obtained from all patients.
Consent for publication
Consent for publication was obtained from all
patients.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hao J and Shao K: The epidemiology,
current status of management, challenge and future strategy for
esophageal cancer in China. China Oncol. 21:501–504. 2011.
|
|
2
|
Domper Arnal MJ, Ferrández Arenas Á and
Lanas Arbeloa Á: Esophageal cancer: Risk factors, screening and
endoscopic treatment in Western and Eastern countries. World J
Gastroenterol. 21:7933–7943. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Okada E, Ukawa S, Nakamura K, Hirata M,
Nagai A, Matsuda K, Ninomiya T, Kiyohara Y, Muto K, Kamatani Y, et
al: Demographic and lifestyle factors and survival among patients
with esophageal and gastric cancer: The Biobank Japan Project. J
Epidemiol. 27:(Suppl):. S29–S35. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xiang K and Fan Q: Progress in medication
treatment of esophageal cancer. World J Gastroenterol.
20:3482–3487. 2012.
|
|
5
|
National Comprehensive Cancer Network
(NCCN): NCCN. Fort Washington, PA; https://www.nccn.org/professionals/physician_gls/pdf/esophageal.pdfDecember
14–2009
|
|
6
|
Nakamura T, Takahashi M, Niigata R,
Yamashita K, Kume M, Hirai M and Yasui H: Changes in blood
concentrations of trace metals in cancer patients receiving
cisplatin-based chemotherapy. Biomed Rep. 5:737–744. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sun H, Qin TJ, Ruan ZP, Wang H and Ma Y:
Clinical study of vinorelbine combined with cisplatin in the
treatment of advanced esophageal cancer. Cancer Res Prevent
Treatment. 33:682–685. 2006.(In Chinese).
|
|
8
|
Chen J, He Y, Hu B, Ji CS, Hu CL and Fan
PS: Prognostic value of the ERCC1 and TS genetic polymorphisms in
advanced esophageal cancer treated with Cisplatin/fluorouracil
chemotherapy. Tumor. 4:314–321. 2010.
|
|
9
|
Ryu H, Song IC, Choi YS, Yun HJ, Jo DY,
Kim JM, Ko YB and Lee HJ: ERCC1 expression status predicts the
response and survival of patients with metastatic or recurrent
cervical cancer treated via platinum-based chemotherapy. Medicine
(Baltimore). 96:e94022017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Palomba G, Atzori F, Budroni M, Ombra M,
Cossu A, Sini M, Pusceddu V, Massidda B, Frau B and Notari F: ERCC1
polymorphisms as prognostic markers in T4 breast cancer patients
treated with platinum-based chemotherapy. J Transl Med. 12:2722014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ting S, Mairinger FD, Hager T, Welter S,
Eberhardt WE, Wohlschlaeger J, Schmid KW and Christoph DC: ERCC1,
MLH1, MSH2, MSH6, and βIII-tubulin: Resistance proteins associated
with response and outcome to platinum-based chemotherapy in
malignant pleural mesothelioma. Clin Lung Cancer. 14:558–567.e3.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen C, Wang F, Wang Z, Li C, Luo H, Liang
Y, An X, Shao J and Li Y: Polymorphisms in ERCC1 C8092A predict
progression-free survival in metastatic/recurrent nasopharyngeal
carcinoma treated with cisplatin-based chemotherapy. Cancer
Chemother Pharmacol. 72:315–322. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Moxley KM, Benbrook DM, Queimado L, Zuna
RE, Thompson D, McCumber M, Premkumar P, Thavathiru E, Hines L and
Moore KN: The role of single nucleotide polymorphisms of the ERCC1
and MMS19 genes in predicting platinum-sensitivity,
progression-free and overall survival in advanced epithelial
ovarian cancer. Gynecol Oncol. 130:377–382. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kalikaki A, Kanaki M, Vassalou H,
Souglakos J, Voutsina A, Georgoulias V and Mavroudis D: DNA repair
gene polymorphisms predict favorable clinical outcome in advanced
non-small-cell lung cancer. Clin Lung Cancer. 10:118–123. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bradbury PA, Marshall AL, Kulke AH, et al:
Prognostic significance of nuclear excision (NER) and base excision
(BER) DNA repair gene polymorphisms in esophageal cancer. J Clin
Oncol (ASCO Annual Meeting). 25:25112007.
|
|
16
|
Shan B, He Y, Chen J, Li XQ, Ji CS, Hu CL
and Hu B: Clinical outcome of advanced esophageal cancer treated
with cisplatin influenced by ERCC1 gene polymorphism in peripheral
blood. Chin J Cancer Prevention Treatment. 18:1447–1450. 2010.
|
|
17
|
Edge SB, Byrd DR, Compton CC, Fritz AG,
Greene FL and Trotti A: AJCC cancer staging manual. 7th edition.
Springer-Verlag; New York, NY; pp. 103–15. 2009
|
|
18
|
Rice TW, Blackstone EH and Rusch VW: 7th
edition of the AJCC cancer staging manual: Esophagus and
esophagogastric junction. Ann Surg Oncol. 17:1721–1724. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Eisenhauer EA, Therasse P, Bogaerts J,
Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S,
Mooney M, et al: New response evaluation criteria in solid tumours:
Revised RECIST guideline (version 1.1). Eur J Cancer. 45:228–247.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
National Cancer Institute (NCI): Common
Terminology Criteria for Adverse Events (CTCAE). Version 4.03.
https://evs.nci.nih.gov/ftp1/CTCAE/CTCAE_4.03/CTCAE_4.03_2010-06-14_QuickReference_5x7.pdfJune
14–2010
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Polee MB, Hop WC, Kok TC, Eskens FA, van
der Burg ME, Splinter TA, Siersema PD, Tilanus HW, Stoter G and van
der Gaast A: Prognostic factors for survival in patients with
advanced oesophageal cancer treated with cisplatin-based
combination chemotherapy. Br J Cancer. 89:2045–2050. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dasari S and Tchounwou PB: Cisplatin in
cancer therapy: Molecular mechanisms of action. Eur J Pharmacol.
740:364–378. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Seiwert TY, Wang X, Heitmann J,
Villegas-Bergazzi V, Sprott K, Finn S, O'Regan E, Farrow AD,
Weichselbaum RR, Lingen MW, et al: DNA repair biomarkers XPF and
phospho-MAPKAP kinase 2 correlate with clinical outcome in advanced
head and neck cancer. PLoS One. 9:e1021122014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Benhamou S and Sarasin A: Variability in
nucleotide excision repair and cancer risk: A review. Mutat Res.
462:149–158. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Benhamou S and Sarasin A: ERCC2/XPD gene
polymorphisms and cancer risk. Mutagenesis. 17:463–469. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gregg SQ, Robinson AR and Niedernhofer LJ:
Physiological consequences of defects in ERCC1-XPF DNA repair
endonuclease. DNA Repair (Amst). 10:781–791. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tantraworasin A, Saeteng S,
Lertprasertsuke N, Arayawudhikul N, Kasemsarn C and Patumanond J:
The prognostic value of ERCC1 and RRM1 gene expression in
completely resected non-small cell lung cancer: Tumor recurrence
and overall survival. Cancer Manag Res. 5:327–336. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Reed E, Dabholkar M, Thornton K, Thompson
C, Yu JJ and Bostick-Bruton F: Evidence for in the appearance of
mRNAs of nucleotide excision repair genes, in human ovarian cancer
tissues. Oncol Rep. 7:1123–1128. 2000.PubMed/NCBI
|
|
30
|
Wood RD: DNA repair in eukaryotes. Annu
Rev Biochem. 65:135–167. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Vogel U, Dybdahl M, Frentz G and Nexo BA:
DNA repair capacity: Inconsistency between effect of
over-expression of five NER genes and the correlation to mRNA
levels in primary lymphocytes. Mutat Res. 461:197–210. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang J, Zhou XQ, Li JY, Cheng JF, Zeng XN,
Li X and Liu P: Prognostic significance of ERCC1 expression in
postoperative patients with gastric cancer. Chin J Cancer Res.
26:323–330. 2014.PubMed/NCBI
|
|
33
|
Wang Y, Chen J, Li X, He Y, Hu B, Ji C and
Xu J: Genetic polymorphisms of ERCC1 and their effects on the
efficacy of cisplatin-based chemotherapy in advanced esophageal
carcinoma. Oncol Rep. 25:1047–1052. 2011.PubMed/NCBI
|
|
34
|
Kim M, Ku JH, Kwak C, Kim HH, Lee E, Keam
B, Kim TM, Heo DS, Lee SH and Moon KC: Predictive and prognostic
value of ribonucleotide reductase regulatory Subunit M1 and
excision repair Cross-complementation group 1 in advanced
urothelial carcinoma (UC) treated with first-line gemcitabine plus
platinum combination chemotherapy. PLoS One. 10:e01333712015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Frischknecht L, Meerang M, Soltermann A,
Stahel R, Moch H, Seifert B, Weder W and Opitz I: Importance of
excision repair cross-complementation group 1 and ribonucleotide
reductase M1 as prognostic biomarkers in malignant pleural
mesothelioma treated with platinum-based induction chemotherapy
followed by surgery. J Thorac Cardiovasc Surg. 149:1539–1546.e1.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Schena M, Guarrera S, Buffoni L, Salvadori
A, Voglino F, Allione A, Pecorari G, Ruffini E, Garzino-Demo P,
Bustreo S, et al: DNA repair gene expression level in peripheral
blood and tumour tissue from non-small cell lung cancer and head
and neck squamous cell cancer patients. DNA Repair (Amst).
11:374–380. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hossain M, Banik NL and Ray SK:
Synergistic anti-cancer mechanisms of curcumin and paclitaxel for
growth inhibition of human brain tumor stem cells and LN18 and
U138MG cells. Neurochem Int. 61:1102–1113. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chao YK, Wu YC, Liu YH, Tseng CK, Chang
HK, Hsieh MJ, Chu Y and Liu HP: Distant nodal metastases from
intrathoracic esophageal squamous cell carcinoma: Characteristics
of long-term survivors after chemoradiotherapy. J Surg Oncol.
102:158–162. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang X, Shen L, Li J, Li Y, Li J and Jin
M: A phase II trial of paclitaxel and cisplatin in patients with
advanced squamous-cell carcinoma of the esophagus. Am J Clin Oncol.
31:29–33. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yun T, Han JY, Lee JS, Choi HL, Kim HY,
Nam BH and Kim HT: Phase II study of weekly paclitaxel and
capecitabine in patients with metastatic or recurrent esophageal
squamous cell carcinoma. BMC Cancer. 11:3852011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Matsumoto H, Kubota H, Higashida M, Yoden
E, Hiratsuka J, Haruma K, Nakamura M and Hirai T: Docetaxel/TS-1
with radiation for unresectable squamous cell carcinoma of the
esophagus-a phase II trial. Anticancer Res. 34:3759–3763.
2014.PubMed/NCBI
|
|
42
|
Schnirer II, Komaki R, Yao JC, Swisher S,
Putnam J, Pisters PW, Roth JA and Ajani JA: Pilot study of
concurrent 5-fluorouracil/paclitaxel plus radiotherapy in patients
with carcinoma of the esophagus and gastroesophageal junction. Am J
Clin Oncol. 24:91–95. 2001. View Article : Google Scholar : PubMed/NCBI
|