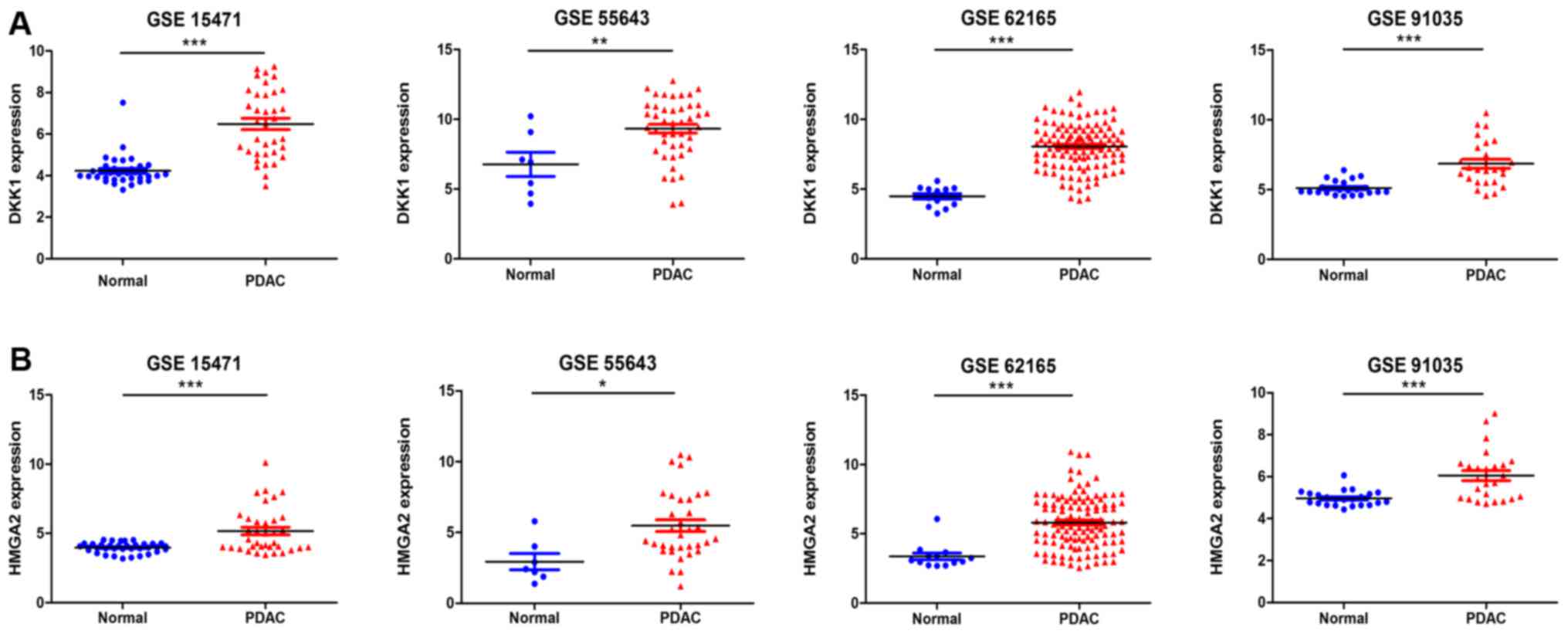
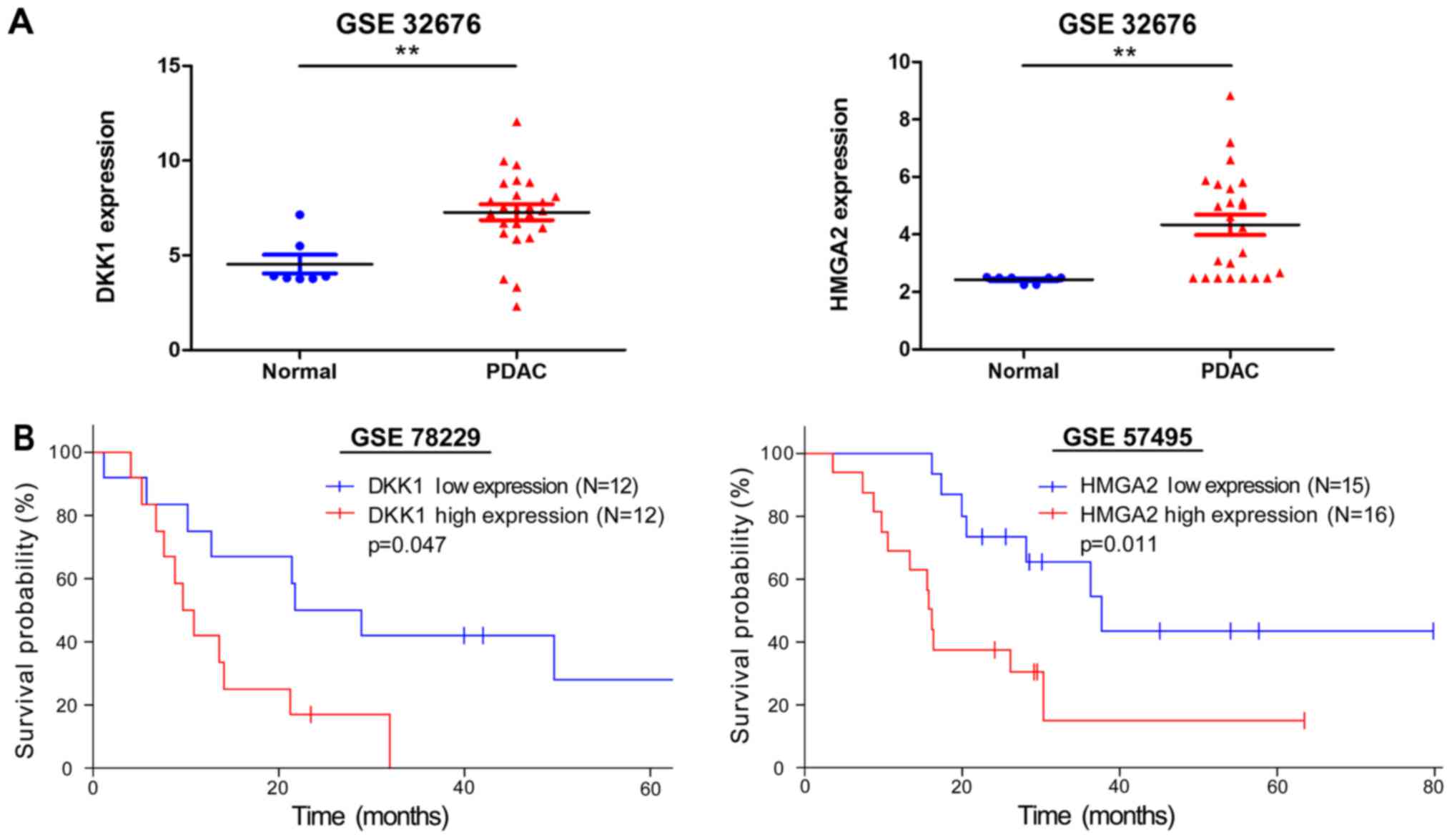
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
DeSantis CE, Lin CC, Mariotto AB, Siegel
RL, Stein KD, Kramer JL, Alteri R, Robbins AS and Jemal A: Cancer
treatment and survivorship statistics, 2014. CA Cancer J Clin.
64:252–271. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kamisawa T, Wood LD, Itoi T and Takaori K:
Pancreatic cancer. Lancet. 388:73–85. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang P, Zhang J, Zhang L, Zhu Z, Fan J,
Chen L, Zhuang L, Luo J, Chen H, Liu L, et al: MicroRNA 23b
regulates autophagy associated with radioresistance of pancreatic
cancer cells. Gastroenterology. 145:1133–1143.e12. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Adamska A, Elaskalani O, Emmanouilidi A,
Kim M, Abdol Razak NB, Metharom P and Falasca M: Molecular and
cellular mechanisms of chemoresistance in pancreatic cancer. Adv
Biol Regul. Nov 22. 2017, (Epub ahead of print). PubMed/NCBI
|
|
6
|
Mazarico JM, Sánchez-Arévalo Lobo VJ,
Favicchio R, Greenhalf W, Costello E, Carrillo-de Santa Pau E,
Marqués M, Lacal JC, Aboagye E and Real FX: Choline Kinase Alpha
(CHKα) as a therapeutic target in pancreatic ductal adenocarcinoma:
Expression, predictive value, and sensitivity to inhibitors. Mol
Cancer Ther. 15:323–333. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Alhasan SF, Haugk B, Ogle LF, Beale GS,
Long A, Burt AD, Tiniakos D, Televantou D, Coxon F, Newell DR, et
al: Sulfatase-2: A prognostic biomarker and candidate therapeutic
target in patients with pancreatic ductal adenocarcinoma. Br J
Cancer. 115:797–804. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41:(Database Issue):.
D991–D995. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cancer Genome Atlas Research Network;
Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA,
Ellrott K, Shmulevich I, Sander C and Stuart JM: The Cancer Genome
Atlas Pan-Cancer analysis project. Nat Genet. 45:1113–1120. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cancer Genome Atlas Research Network.
Electronic address: andrew_aguirre@dfci.harvard.edu; Cancer Genome
Atlas Research Network: Integrated genomic characterization of
pancreatic ductal adenocarcinoma. Cancer Cell. 32:185–203.e13.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000.
|
|
13
|
Kanehisa M, Goto S, Sato Y, Furumichi M
and Tanabe M: KEGG for integration and interpretation of
large-scale molecular data sets. Nucleic Acids Res. 40:(Database
Issue):. D109–D114. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pathan M, Keerthikumar S, Ang CS, Gangoda
L, Quek CY, Williamson NA, Mouradov D, Sieber OM, Simpson RJ, Salim
A, et al: FunRich: An open access standalone functional enrichment
and interaction network analysis tool. Proteomics. 15:2597–2601.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:(Database Issue):. D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Calvo MB, Figueroa A, Pulido EG, Campelo
RG and Aparicio LA: Potential role of sugar transporters in cancer
and their relationship with anticancer therapy. Int J Endocrinol.
2010(pii): 2053572010.PubMed/NCBI
|
|
19
|
Jiang H, Hegde S, Knolhoff BL, Zhu Y,
Herndon JM, Meyer MA, Nywening TM, Hawkins WG, Shapiro IM, Weaver
DT, et al: Targeting focal adhesion kinase renders pancreatic
cancers responsive to checkpoint immunotherapy. Nat Med.
22:851–860. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Morris JP IV, Wang SC and Hebrok M: KRAS,
Hedgehog, Wnt and the twisted developmental biology of pancreatic
ductal adenocarcinoma. Nat Rev Cancer. 10:683–695. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ahn VE, Chu ML, Choi HJ, Tran D, Abo A and
Weis WI: Structural basis of Wnt signaling inhibition by Dickkopf
binding to LRP5/6. Dev Cell. 21:862–873. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qi L, Sun B, Liu Z, Li H, Gao J and Leng
X: Dickkopf-1 inhibits epithelial-mesenchymal transition of colon
cancer cells and contributes to colon cancer suppression. Cancer
Sci. 103:828–835. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sato N, Yamabuki T, Takano A, Koinuma J,
Aragaki M, Masuda K, Ishikawa N, Kohno N, Ito H, Miyamoto M, et al:
Wnt inhibitor Dickkopf-1 as a target for passive cancer
immunotherapy. Cancer Res. 70:5326–5336. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang J, Zhang X, Zhao X, Jiang M, Gu M,
Wang Z and Yue W: DKK1 promotes migration and invasion of non-small
cell lung cancer via β-catenin signaling pathway. Tumour Biol.
39:10104283177038202017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
González-Sancho JM, Aguilera O, García JM,
Pendás-Franco N, Peña C, Cal S, García de Herreros A, Bonilla F and
Muñoz A: The Wnt antagonist DICKKOPF-1 gene is a downstream target
of beta-catenin/TCF and is downregulated in human colon cancer.
Oncogene. 24:1098–1103. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Takahashi N, Fukushima T, Yorita K, Tanaka
H, Chijiiwa K and Kataoka H: Dickkopf-1 is overexpressed in human
pancreatic ductal adenocarcinoma cells and is involved in invasive
growth. Int J Cancer. 126:1611–1620. 2010.PubMed/NCBI
|
|
27
|
Shen Q, Fan J, Yang XR, Tan Y, Zhao W, Xu
Y, Wang N, Niu Y, Wu Z, Zhou J, et al: Serum DKK1 as a protein
biomarker for the diagnosis of hepatocellular carcinoma: A
large-scale, multicentre study. Lancet Oncol. 13:817–826. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kimura H, Fumoto K, Shojima K, Nojima S,
Osugi Y, Tomihara H, Eguchi H, Shintani Y, Endo H, Inoue M, et al:
CKAP4 is a Dickkopf1 receptor and is involved in tumor progression.
J Clin Invest. 126:2689–2705. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hinton J, Callan R, Bodine C, Glasgow W,
Brower S, Jiang SW and Li J: Potential epigenetic biomarkers for
the diagnosis and prognosis of pancreatic ductal adenocarcinomas.
Expert Rev Mol Diagn. 13:431–443. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Are C, Chowdhury S, Ahmad H, Ravipati A,
Song T, Shrikandhe S and Smith L: Predictive global trends in the
incidence and mortality of pancreatic cancer based on geographic
location, socio-economic status, and demographic shift. J Surg
Oncol. 114:736–742. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Perera RM and Bardeesy N: Ready, set, go:
The EGF receptor at the pancreatic cancer starting line. Cancer
Cell. 22:281–282. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li Y, Peng L and Seto E: Histone
deacetylase 10 regulates the cell cycle G2/M phase transition via a
Novel Let-7-HMGA2-Cyclin A2 pathway. Mol Cell Biol. 35:3547–3565.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Watanabe S, Ueda Y, Akaboshi S, Hino Y,
Sekita Y and Nakao M: HMGA2 maintains oncogenic RAS-induced
epithelial-mesenchymal transition in human pancreatic cancer cells.
Am J Pathol. 174:854–868. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Strell C, Norberg KJ, Mezheyeuski A,
Schnittert J, Kuninty PR, Moro CF, Paulsson J, Schultz NA,
Calatayud D, Löhr JM, et al: Stroma-regulated HMGA2 is an
independent prognostic marker in PDAC and AAC. Br J Cancer.
117:65–77. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Haselmann V, Kurz A, Bertsch U, Hübner S,
Olempska-Müller M, Fritsch J, Häsler R, Pickl A, Fritsche H,
Annewanter F, et al: Nuclear death receptor TRAIL-R2 inhibits
maturation of let-7 and promotes proliferation of pancreatic and
other tumor cells. Gastroenterology. 146:278–290. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wei JJ, Wu J, Luan C, Yeldandi A, Lee P,
Keh P and Liu J: HMGA2: A potential biomarker complement to P53 for
detection of early-stage high-grade papillary serous carcinoma in
fallopian tubes. Am J Surg Pathol. 34:18–26. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mahajan A, Liu Z, Gellert L, Zou X, Yang
G, Lee P, Yang X and Wei JJ: HMGA2: A biomarker significantly
overexpressed in high-grade ovarian serous carcinoma. Mod Pathol.
23:673–681. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Italiano A, Bianchini L, Keslair F,
Bonnafous S, Cardot-Leccia N, Coindre JM, Dumollard JM, Hofman P,
Leroux A, Mainguené C, et al: HMGA2 is the partner of MDM2 in
well-differentiated and dedifferentiated liposarcomas whereas CDK4
belongs to a distinct inconsistent amplicon. Int J Cancer.
122:2233–2241. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Iwai A, Marusawa H, Matsuzawa S, Fukushima
T, Hijikata M, Reed JC, Shimotohno K and Chiba T: Siah-1L, a novel
transcript variant belonging to the human Siah family of proteins,
regulates beta-catenin activity in a p53-dependent manner.
Oncogene. 23:7593–7600. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lee KH, Li M, Michalowski AM, Zhang X,
Liao H, Chen L, Xu Y, Wu X and Huang J: A genomewide study
identifies the Wnt signaling pathway as a major target of p53 in
murine embryonic stem cells. Proc Natl Acad Sci USA. 107:69–74.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang J, Shou J and Chen X: Dickkopf-1, an
inhibitor of the Wnt signaling pathway, is induced by p53.
Oncogene. 19:1843–1848. 2000. View Article : Google Scholar : PubMed/NCBI
|