Introduction
Breast cancer is the main cause of cancer-associated
mortality in women globally (1).
Significant progress has been made in diagnostic techniques and
targeted therapies, however, the prognosis of breast cancer remains
poor (1). Therefore, it is important
to identify the underlying molecular mechanisms in breast cancer.
Various studies indicated that the activated Wnt/β-catenin
signaling pathway serves an essential role in tumorigenesis
(2,3)
and ~40% cases of breast cancer exhibit increased expression levels
of β-catenin (4). It has been
demonstrated that the Wnt/β-catenin signaling pathway is regulated
by cytosolic β-catenin, which may translocate into the nucleus to
interact with transcriptional factors of the T-cell
factor/lymphoid-enhancing factor (TCF/LEF) family resulting in
activation of Wnt target genes (2–6). It is
known that the downstream targets of β-catenin/TCF, including
cyclin D1 and c-Myc, are vital regulators of cell proliferation and
apoptosis and are associated with several types of cancer including
mammary gland carcinogenesis, thyroid carcinogenesis and prostate
cancer (7–9). Additionally, Wnt binds to a frizzled
receptor in existence of the co-receptor, low density
lipoprotein-related protein 5/6, thus inhibiting β-catenin
degradation (4). Furthermore,
cytoplasmic β-catenin forms a complex with axis inhibition protein,
adenomatosis polyposis coli, casein kinase 1 (CK1) and glycogen
synthase kinase-3β (GSK-3β) when Wnt is absent. Notably,
cytoplasmic β-catenin is phosphorylated by CK1 and GSK-3β at the
N-terminal region, leading to degradation of β-catenin through the
ubiquitination proteasome pathway (10). Consequently, the Wnt signaling pathway
is a potential therapeutic target in breast cancer (2–4).
Regulator of differentiation (ROD)1, also termed
polypyrimidine tract binding protein 3, was initially regarded as
an inhibitor of cell differentiation (11). Overexpression of ROD1 was demonstrated
to block phorbol ester and sodium butyrate-induced differentiation
of K562 cells (11). Subsequently, it
has been revealed that ROD1 is a member of the heterogeneous
nuclear ribonucleoproteins family and participates in alternative
splicing of pre-mRNA (12), a
post-transcriptional regulation for gene expression (13). Additionally, abnormal splicing holds a
vital role in various types of cancer (14). As a RNA-binding protein, ROD1 binds to
the post-transcriptional RNA of >104 genes associated
with the differentiation and proliferation of cancer cells detected
by RNA immunoprecipitation-sequencing (15). Notably, ROD1 may promote proliferation
but inhibit the differentiation of human gastric cancer MKN45 cells
(16). However, it remains unknown
whether ROD1 expression is aberrant in breast cancer, and whether
it serves an essential role in the proliferation and invasion of
breast cancer cells.
In the present study, ROD1 expression was analyzed
by western blotting and reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) in normal and cancer tissues.
The effects of ROD1 on invasion of breast cancer cells as well as
tumor growth were investigated in a xenograft model. Finally, the
aim of this study was to investigate the molecular mechanism of
ROD1 in breast cancer cell invasion.
Materials and methods
Cell culture
The human breast cancer cells (MCF-7 and
MDA-MB-231), normal cells (hMC and MCF10A) and 293A cells were
purchased from ATCC (American Type Culture Collection; Manassas,
VA, USA). These cells were maintained in RPMI-1640 medium (Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10%
fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.),
100 U/ml penicillin and 100 mg/ml streptomycin which were obtained
from Gibco; Thermo Fisher Scientific, Inc. Subconfluent cells were
treated with adriamycin (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) diluted into the medium to a final concentration of 1.5
mg/ml. All cells were placed in a humidified incubator containing
5% CO2 and 95% air at 37°C.
Cell proliferation
Cell proliferation rate was examined by Cell
Counting Kit-8 (CCK-8) (Sigma-Aldrich; Merck KGaA), according to
the manufacturer's protocol at the given time points (0, 12, 24, 36
and 48 h). In brief, MDA-MB-231 cells were seeded in 96-well plates
at a density of ~2×103 cells/well and maintained in
RPMI-1640 medium. Subsequently, the testing reagent (10 µl) was
added to each well at 37°C for 2 h. The optical absorption (450 nm)
of each well was detected using a microplate reader. The
adenoviruses were diluted with RPMI-1640 medium at a final
concentration of 2×102 for each well. These cells were
transfected with adenovirueses, Ad-vector, Ad-ROD1 and Ad-shROD1
(Hanbio Biotechnology Co., Ltd., Shanghai, China) for 12 h at 37°C.
Then, the adenovirus containing medium was removed and fresh medium
was added to the wells. Following transfection for another 0, 12,
24, 36 and 48 h, the transfected MDA-MB-231 cell proliferation was
determined by CCK-8 assay. The group ‘con’ (Fig. 1) represents the negative control, with
cells that did not undergo transfection. The experiment was
repeated three times.
Cell invasion
In order to measure the invasiveness of MDA-MB-231
cells, these were plated in Transwell plates (8.0 µm pore size;
Corning Inc., NY, USA) coated with Matrigel (Sigma-Aldrich; Merck
KGaA). Briefly, 5×104 cells/well were placed in the
upper chamber in RPMI-1640 with no serum; the lower chamber was
filled with RPMI-1640 (supplemented with 10% FBS). Following
incubation at 37°C for 48 h, the cells in lower chamber were fixed
with 4% paraformaldehyde at 4°C for 1 h, and stained with 0.1%
crystal violet at 4°C for 1 h, extracted with 33% acetic acid and
the absorbance was measured at a wavelength of 570 nm using a
microplate reader. Paraformaldehyde, crystal violet and acetic acid
were obtained from Sigma-Aldrich; Merck KGaA. The experiment was
repeated three times.
RNA isolation, cDNA synthesis and
RT-qPCR
Total RNA was isolated from cells or tissues using
TRIzol reagent (Thermo Fisher Scientific, Inc.). Subsequently, cDNA
synthesis was performed using the Superscript III RT kit (Thermo
Fisher Scientific, Inc.). RT-qPCR reactions were carried out using
the SYBR Green PCR Master Mix (Thermo Fisher Scientific, Inc.) in
an ABI 7500 thermal cycler (Thermo Fisher Scientific, Inc.). GAPDH
was used as an internal control. The primers used were as follows:
GAPDH, sense, 5′-CACCATCTTCCAGGAGCGAG-3′ and antisense,
5′-GCAGGAGGCATTGCTGAT-3′; ROD1, sense,
5′-CACCTTTCTCTCTCCCCAAGAAACT-3′ and antisense,
5′-TTGCTGTCATTCCCATTAGCTGT-3′ (Invitrogen; Thermo Fisher
Scientific, Inc.). The thermocycling conditions were as follows:
Initial denaturation of 94°C for 5 min, followed by 40 cycles of
94°C for 30 sec and 58°C for 30 sec, and final extension of 72°C
for 15 sec. Each reaction was conducted in duplicate. Relative mRNA
expression of ROD1 gene was evaluated using the 2−∆∆Cq
method (17).
Construction of recombinant adenoviral
vectors (Ad)-ROD1 and Ad-short hairpin(sh)ROD1
The primers for human ROD1 gene were as follows:
sense, 5′-ATGCCTTTCTCTCTCCCCAAGAAACT-3′ and antisense,
5′-TCAGATTGTAGATTTTGAGAAGGAA-3′ (Invitrogen; Thermo Fisher
Scientific, Inc.), and restriction digestion enzymes XbaI
and KpnI (Thermo Fisher Scientific, Inc.) were used for
cloning this gene. The PCR-amplified fragments were used for
subcloning into the pAdEasy/Track-cytomegalovirus (CMV) vector
(Stratagene; Agilent Technologies, Inc., Santa Clara, CA, USA). The
targeting sequence of shROD1 was 5′-GCCCTGTGCTTCGAATAAT-3′
(Invitrogen; Thermo Fisher Scientific, Inc.). The interfering RNA
of ROD1 was subcloned into the pAdEasy/Track-CMV vector with
HindIII and KpnI (Thermo Fisher Scientific, Inc.).
DNA polymerase, restrictive enzymes and T4 ligase were purchased
from Thermo Fisher Scientific, Inc. The ligation reaction mixture
was prepared as follows: 100–500 ng Linear DNA, 1–2 µg
phosphorylated linkers, 2 µl 10X T4 DNA Ligase buffer, 2 µl 50% PEG
4000 solution, 2 U T4 DNA Ligase, with nuclease-free water to 20
µl. The agents were mixed thoroughly and incubated for 1 h at 22°C
and then the reaction was subjected to heat-inactivation at 65°C
for 10 min or at 70°C for 5 min. Finally, the products were used
for transfecting competent bacteria. Additionally, shScramble
adenoviral vectors were not used in this study, and instead the
Track-CMV empty vector was used as the control. The plasmids were
transfected into 293A packaging cells using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) to generate recombinant adenoviruses.
Subsequently, viral particles were purified using cesium chloride
density gradient ultracentrifugation (120,000 × g for 20 h at 4°C)
(18). The titer of virus was
examined by RT-qPCR (19,20). A negative control virus (Ad-Vector)
was used.
Luciferase reporter assay
To determine luciferase of TOPFlash, a total of
5×104 MDA-MB-231 cells/well were plated in 24-well
plates. TOPFlash plasmid (Add gene, Inc., Cambridge, MA, USA) is a
firefly luciferase reporter with wild type TCF/LEF binding sites
(21–24). FOPFlash plasmid with mutant TCF/LEF
binding sites often functions as a control for background
luminescence (21–24). The ratio of the luciferase activity of
TOPflash against that of FOP flash was evaluated. A total of 0.5 µg
TOPFlash or FOPFlash plasmid was transfected/well. The un
transfected cells were used as a control. Firefly luciferase
activity was normalized for transfection efficiency using the
corresponding Renilla luciferase activity. Following 12 h
incubation with Ad-ROD1 or Ad-shROD1, the luciferase activity was
determined using a Dual Luciferase® Reporter Assay kit
(Promega Corporation, Madison, WI, USA), according to the
manufacturer's protocol.
Western blot analysis and
co-immunoprecipitation
Tissues and cells were lysed using a
radioimmunoprecipitation assay (RIPA) buffer (cat no. P1003;
Beyotime Institute of Biotechnology, Haimen, China) followed by
centrifugation (15,000 × g for 30 min at 4°C). In addition, nuclear
and cytoplasmic extraction was performed according to the
manufacturer's protocol (NE-PER® Nuclear and Cytoplasmic
extraction reagents; Pierce Biotechnology; Thermo Fisher
Scientific, Inc.). The protein concentration was determined by BCA
Protein Quantification Kit (Thermo Fisher Scientific, Inc.). A
total of 40 mg protein for western blot analysis was separated in
10% TruPAGE™ precast gels (Sigma-Aldrich; Merck KGaA) and
transferred to a polyvinylidene difluoride (PVDF) membrane
(Sigma-Aldrich; Merck KGaA). Subsequently, the membrane was blocked
with 5% skim milk (Sangon Biotech Co., Ltd., Shanghai, China) and
incubated with primary antibodies at 4°C overnight and incubated
with horseradish peroxidase (HRP)-conjugated goat anti-mouse and
HRP-conjugated goat anti-rabbit secondary antibodies for 2 h at
room temperature (BS12478 or BS13278; 1:5,000; Bio world
Technology, Inc., St. Louis Park, MN, USA). GAPDH was used as an
internal control. In the present study, the primary antibodies used
were as follows: anti-ROD1 (14027-1-AP; 1:1,000; ProteinTech Group,
Inc., Chicago, IL, USA), anti-GAPDH (SAB2701826; 1:1,000 dilution;
Sigma-Aldrich; Merck KGaA), anti-β-catenin (9562; 1:1,000; Cell
Signaling Technology, Inc., Danvers, MA, USA), anti-β-catenin
(Y333; 11574, 1:1,000; AbSci, Vancouver, WA, USA), anti-lumin B
(13435; 1:1,000; Cell Signaling Technology, Inc.), anti-c-Myc
(9402; 1:1,000; Cell Signaling Technology, Inc.). Finally, the
bands were detected using an enhanced chemiluminescence (ECL)
Reagent Plus kit (Cell Signaling Technology, Inc.). The
corresponding semi-quantitative analysis was based on optical
density using Image J software (version 2.1.4.6; National
Institutes of Health, Bethesda, MD, USA).
For the co-immunoprecipitation, cells or tissues
were extracted using RIPA buffer (cat no. P1003; Beyotime Institute
of Biotechnology). Supernatants were obtained by centrifugation
(15,000 × g, 15 min, 4°C) and incubated with the indicated
antibodies (anti-ROD1) for 6 h at 4°C followed by
immunoprecipitation with 30 ml protein A/G agarose (Thermo Fisher
Scientific, Inc.). The precipitates were completely washed with PBS
and evaluated by western blotting, as aforementioned.
Knockdown of β-catenin by small
interfering RNA (siRNA) transfection
Knockdown of β-catenin by siRNA transfection was
conducted in MDA-MB-231 cells using Lipofectamine® 2000
transfection reagent (Thermo Fisher Scientific, Inc.). The sequence
of siRNA against β-catenin was 5′-UGGAUUUGUACCAUUCUUCUG-3′.
Subsequent to transfection at 37°C for 6 h, the cells were
maintained in fresh RPMI-1640 media for another 24 h. Then, the
cells were incubated with Ad-vector, Ad-ROD1 (2×102
plaque forming unit/well in 6-well plate, labeled as Ad-ROD1 L;
2×103 plaque forming unit/well, labeled as Ad-ROD1 H) at
37°C for 24 h. Finally, these cells were harvested for
proliferation analysis and western blot analysis.
Immunohistochemistry
Breast cancer tissues and normal mammary tissues
were fixed in 10% formalin buffer at 4°C for 24 h. The fresh
samples were immediately stored in liquid nitrogen (~-200°C). The
10 µm sections were subjected to dewaxing in xylene at room
temperature for 15 min and dehydration in 95 and 80% graded ethanol
at room temperature for 5 min. Activity of endogenous peroxidase
was blocked by 3% H2O2 at room temperature
for 10 min. The sections were then heated to 100°C in 0.1 M citrate
buffer (pH 6.0) for half an hour to retrieve the antigens. These
tissues were incubated with anti-Ki-67 for overnight at 4°C.
Subsequent to washing with PBS three times, tissues were incubated
with secondary antibodies at room temperature for 1 h, which were
horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG
used according to the manufacturer's protocol. Finally, sections
were stained with hematoxylin at room temperature for 10 min. The
stained tissues were photographed using a light microscope at −200
magnification from Carl Zeiss (Axio Observer A1; Carl Zeiss AG,
Oberkochen, Germany). Ki67 antibody was obtained from Bioworld (cat
no. BS9931M; 1:100). The secondary antibody was also obtained from
Bioworld (cat no. BS13278; 1:1,000). In addition, diaminobenzidine
(DAB; Beyotime Institute of Biotechnology) solution was applied for
color development.
Mouse xenograft model
In the present study, five-week old female BALB/c
nude (Nanjing Biomedical Research Institute of Nanjing University,
Nanjing, China) were used and the mean weight of these mice were
17.03 g. Mice were maintained at 25°C with a humidity of ~50% and a
12-h light/12-h dark cycle with free access to food and water.
MDA-MB-231 cells grown at logarithm phase were harvested, washed
and resuspended in PBS at 3×107 cells/ml. A total of 200
µl cell suspension (5×106 cells) was implanted into the
breast (mammary fad pat) of each mouse every other day for 6 days.
For tumor growth analysis, tumor volume was calculated according to
the formula: V=1/2ab2, where a and b stand for the
length and the width of tumor measured with sliding caliper,
respectively. Adenovirus was not injected until tumor volume grew
up to 50 mm3 (25).
Tumor-bearing mice were randomly grouped into three groups
(n=6/group) for Ad-Vector, Ad-ROD1 and Ad-shROD1. All processes
were carried out in a biosafety cabinet (Thermo Fisher Scientific,
Inc.). After 15 days, the mice were sacrificed using cervical
dislocation and the tumors from tumor-bearing mice were
harvested.
Additionally, female TA2 mice with spontaneous
breast cancer were obtained from Tianjin Medical University
(Tianjin, China) and the mean weight of these mice were 18.11 g.
TA2 mice are considered as animal model of breast cancer and in the
present study, 10 TA2 mice were prepared/group. The incidence of
spontaneous breast cancer in TA2 mice is greater than 80% in the
absence of any external chemical stimuli (26,27). The
tumors appeared at the age of 360 days. These mice were sacrificed
using cervical dislocation. Breast cancer tissues and normal
mammary tissues were fixed in 10% formalin buffer at 4°C for 24 h.
Tumors without necrosis of 6 mice were employed for further
analysis.
The present study was approved by the Animal Care
and Protection Committee of the Laboratory Animal Center of Soochow
University (SYXK 2014-0030; Jiangsu, China).
Statistical analysis
All data are expressed as the mean ± standard
deviation. Comparisons between two groups were performed using
Student's t-test. Comparisons among multiple groups were performed
with one-way analysis of variance followed by Tukey's honestly
significant difference test. Statistical analyses were conducted
using SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA). P<0.05
was considered to indicate a statistically significant
difference.
Results
ROD1 expression is downregulated in
breast cancer cells
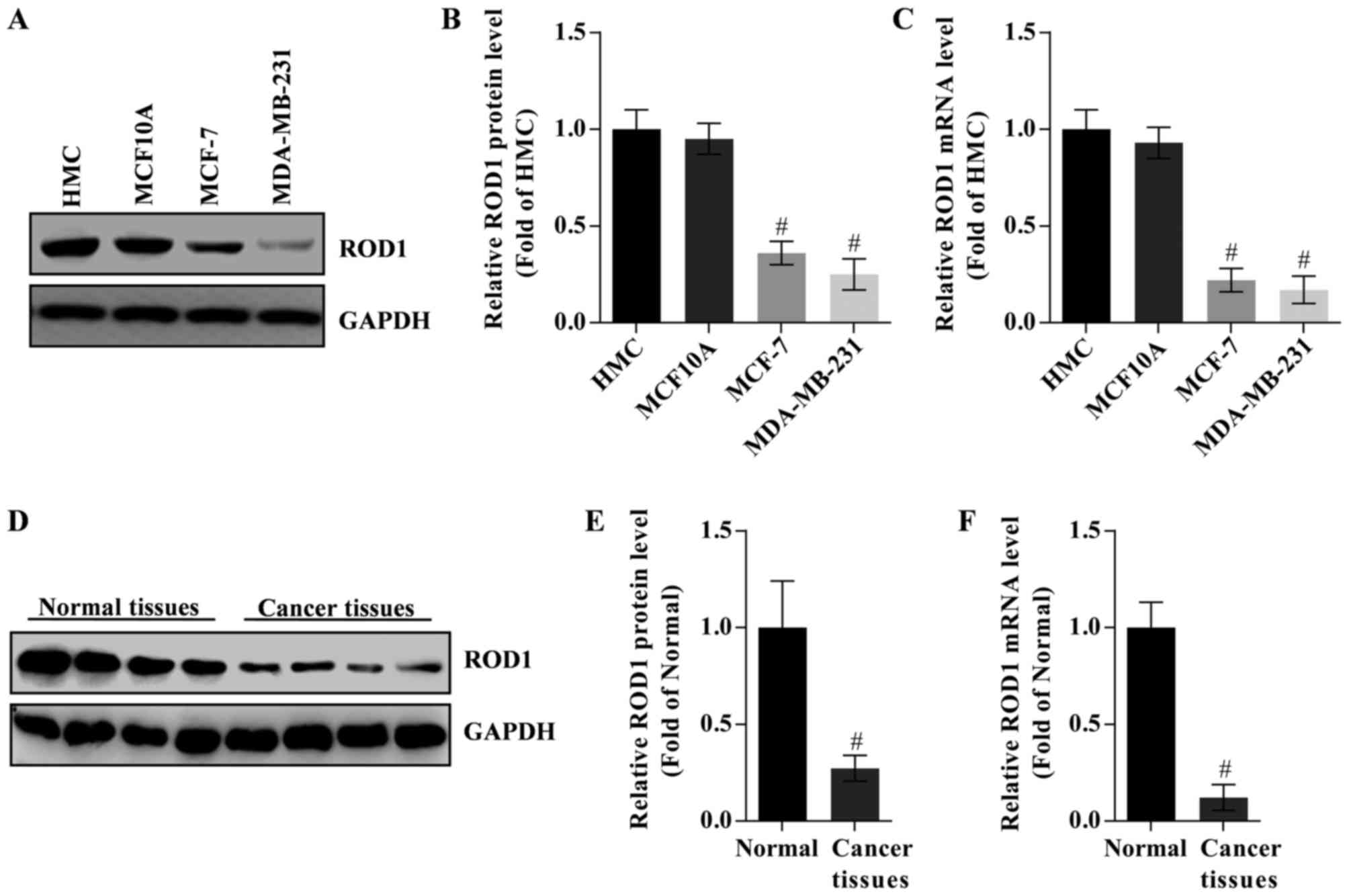
To study the potential role of ROD1 in the invasion
of breast cancer cells, its expression in HMC, MCF10A, MCF-7 and
MDA-MB-231 cells was analyzed by western blot analysis and RT-qPCR.
HMC cells were used as control. The data demonstrated that ROD1
protein and mRNA expression levels were lower in cancer cells
compared with that in the HMC cells. MDA-MB-231 cells exhibited the
lowest expression of ROD1 (Fig.
1A-C). However, there was no difference in the expression of
ROD1 between HMC and MCF10A cells (P>0.5). Notably, ROD1 was
significantly reduced in tumor tissues as evaluated by western blot
analysis (Fig. 1D and E) and RT-qPCR
(Fig. 1F).
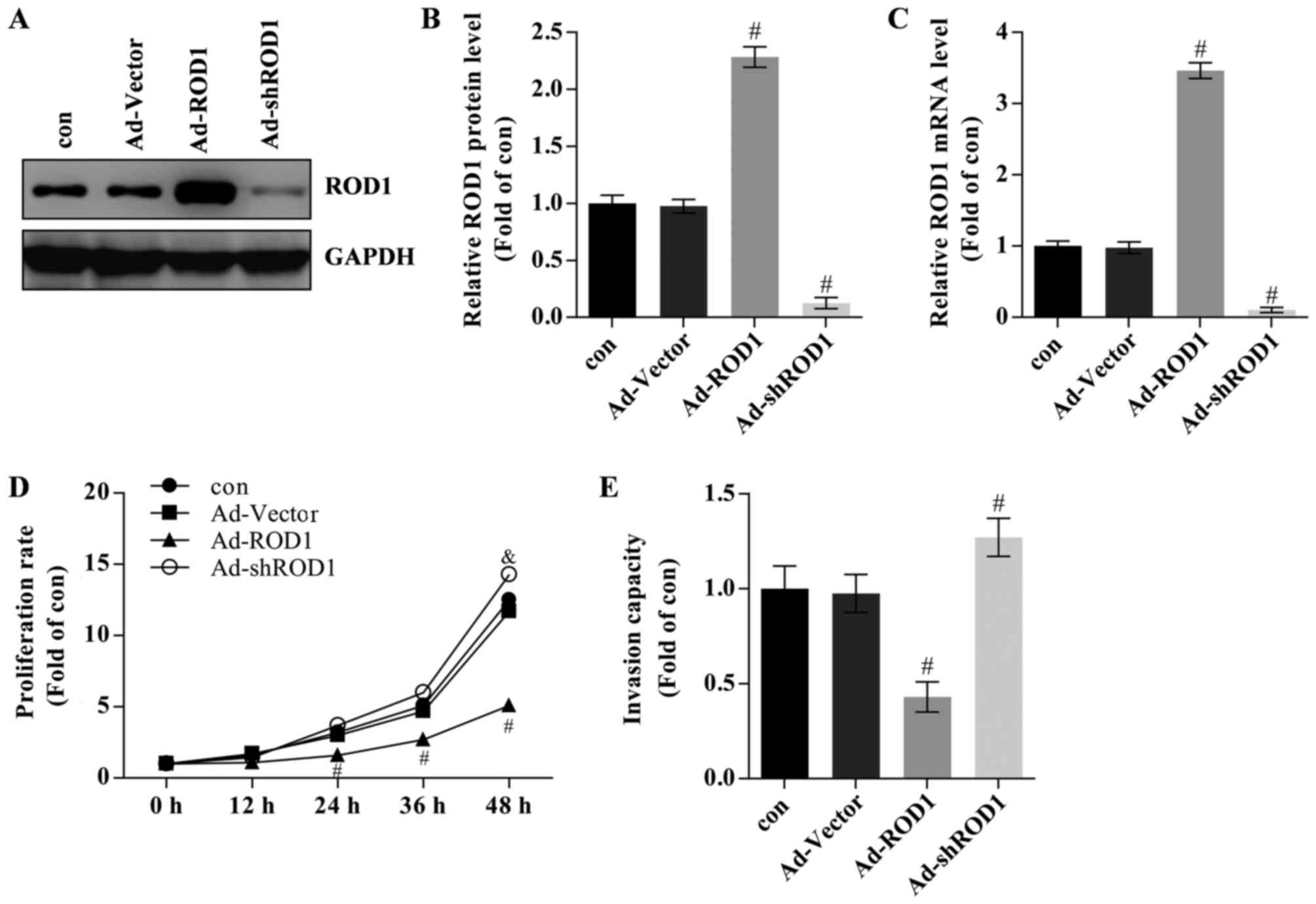
ROD1 inhibits proliferation and
invasion in MDA-MB-231 cells
The effects of ROD1 in breast cancer cells were also
investigated. Ad-ROD1 was used to overexpress ROD1 and Ad-shROD1
was used to knockdown ROD1 in MDA-MB-231 cells. In the present
study, ROD1 was successfully overexpressed compared with Ad-Vector
group) as evaluated by western blotting (Fig. 2A and B) and RT-qPCR (Fig. 2C). The ‘con’ group contained
MDA-MB-231 cells that were not treated with Ad-Vector or other
adenoviral vectors and the Ad-Vector group contained MDA-MB-231
cells that were transfected with Ad-Vector adenoviruses. Breast
cancer cells transfected with Ad-ROD1 exhibited a significantly
lower proliferation rate than the control as assessed by CCK-8
assay following infection for 24, 36 and 48 h (Fig. 2D). The data are presented as folds of
the starting time in each group relative to the control.
Overexpression of ROD1 resulted in a significant reduction in the
invasion rate of MBA-MB-231 cells as determined by a Transwell
assay (Fig. 2E). Cells infected with
Ad-shROD1 exhibited increased cell proliferation and invasion
capacity in vitro (Fig. 2D and
E).
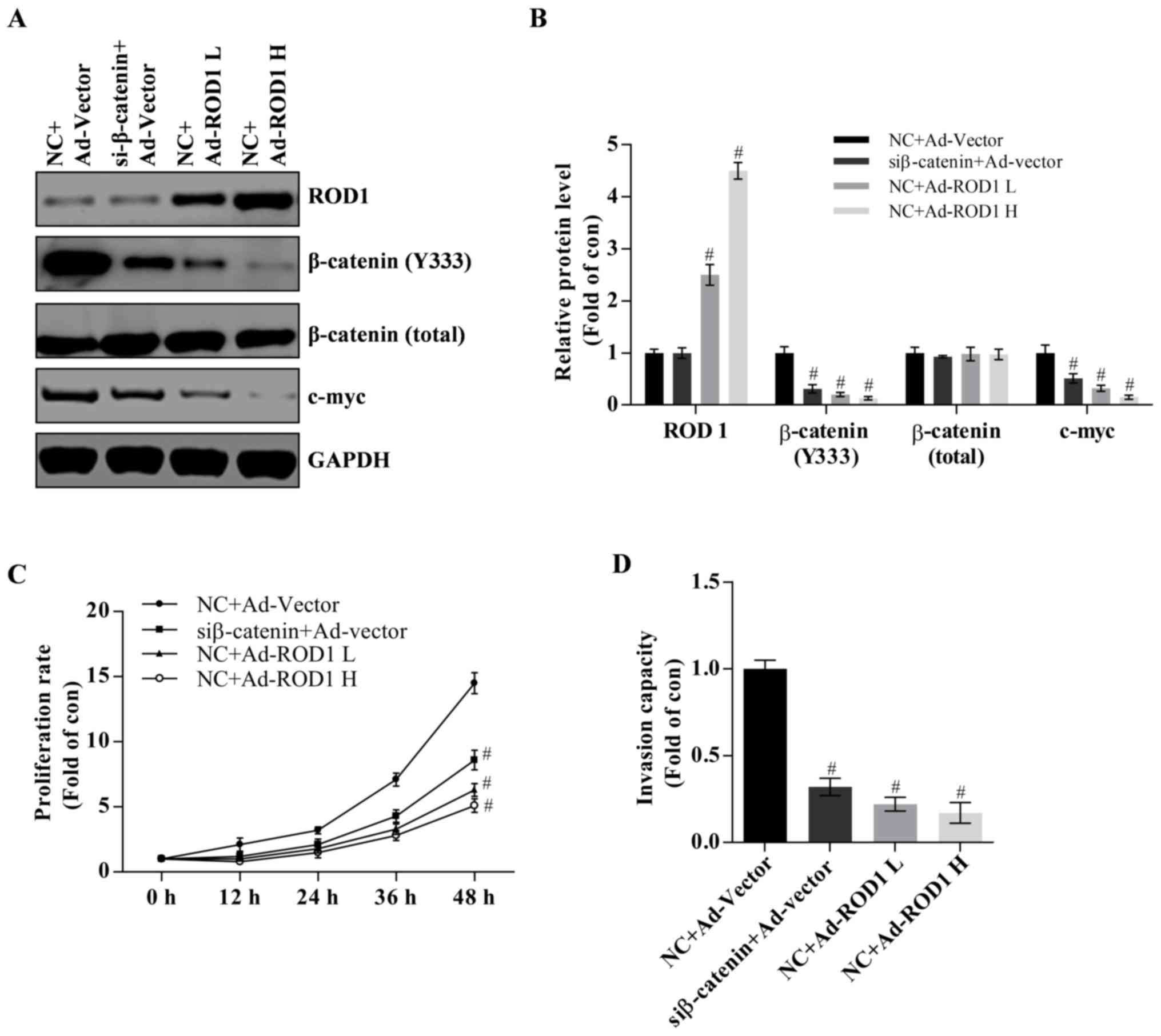
Involvement of Wnt/β-catenin pathway
in proliferation and invasion of MDA-MB-231 cells
It has been reported that activation of the
Wnt/β-catenin signaling pathway serves an essential role in breast
cancer (2–4). In the present study, it was demonstrated
that ROD1 may reduce β-catenin (Y333) and c-Myc protein expression
levels in a dose-dependent manner (Fig.
3A and B). C-Myc is a target of Wnt/β-catenin and often
promotes cancer occurrence (21).
Notably, it was demonstrated that β-catenin was successfully
significantly knocked down as demonstrated by western blot analysis
(P<0.05; Fig. 3A and B). Knockdown
of β-catenin suppressed proliferation and invasion of MDA-MB-231
cells (Fig. 3C and D). Therefore,
these results indicate that wnt/β-catenin pathway is involved in
development of breast cancer.
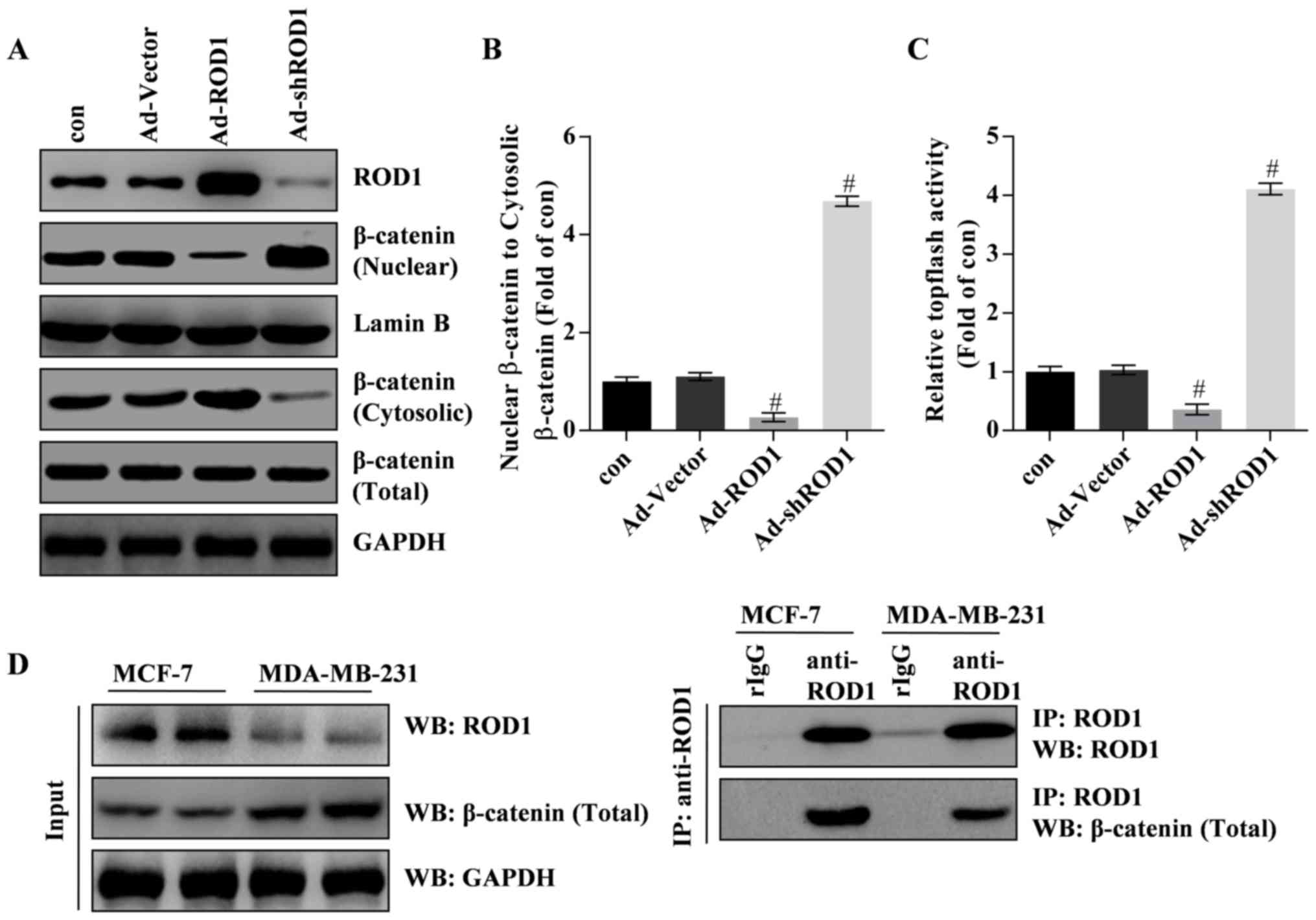
ROD1 inhibits migration of β-catenin
from cytosol to nuclei through interaction with β-catenin
In the present study, the results revealed that ROD1
significantly inactivated Wnt/β-catenin signaling pathway. Western
blotting and TOPflash luciferase assay demonstrated that β-catenin
was inactivated and translocated less into the nucleus in response
to Ad-ROD1, whereas Ad-shROD1 promoted β-catenin translocation into
the nucleus of MDA-MB-231 cells (Fig.
4A-C). Notably, the data indicated that ROD1 interacted with
β-catenin in MDA-MB-231 and MCF-7 cells. Furthermore, ROD1 may
interact with β-catenin in MCF-7 and MDA-MB-231 cells (Fig. 4D). These data indicated that ROD1
interacted with β-catenin leading to a reduction of nuclear
β-catenin protein levels and suppression of Wnt/β-catenin signaling
pathway in cancer cells.
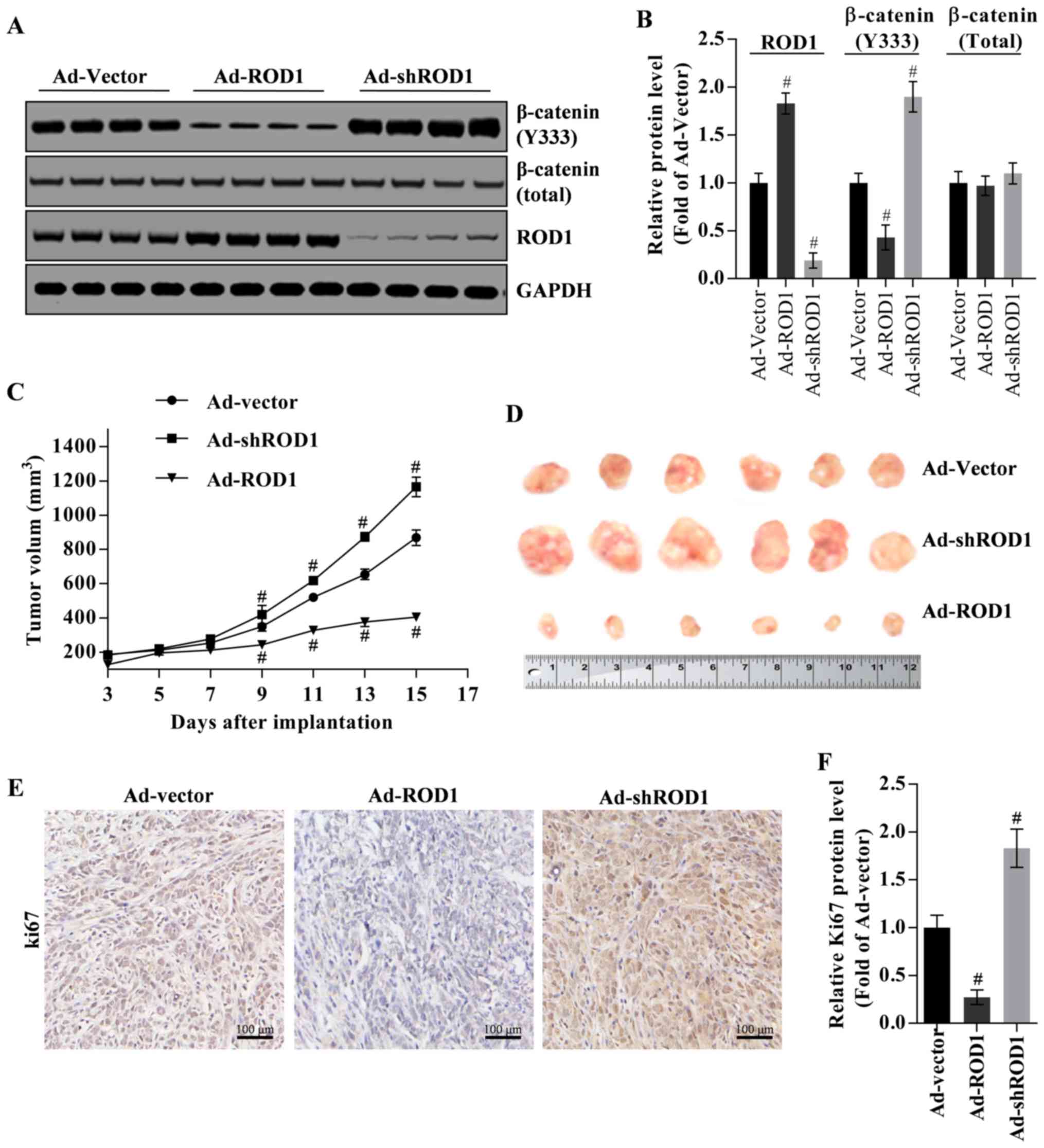
ROD1 suppresses tumor growth in nude
mouse xenograft model
The inhibition of ROD1 on tumor growth was
investigated in a nude mouse xenograft model. β-catenin (Y333) was
commonly located in nucleus. Firstly, β-catenin (Y333) in tumor
tissues was downregulated by overexpression of ROD1 (Ad-ROD1) but
was upregulated by knockdown of ROD1 (Ad-shROD1) as indicated in
Fig. 5A and B. In the present study,
total β-catenin protein was used as an internal control for nuclear
β-catenin (Y333), and GAPDH was used as an internal control for
total β-catenin and ROD1 (Fig. 5A and
B). Notably, in the control group (Ad-Vector group), the volume
of harvested tumors on the third day was ~4.64-fold larger,
compared with that on starting day with injection of Ad-Vector. In
the Ad-ROD1 and Ad-shROD1 groups, the volume of harvested tumors on
the third day was 3.16-fold and 6.33-fold larger, compared with
that on starting day with injection of the adenoviruses. Compared
with the Ad-vector group, Ad-ROD1 demonstrated an inhibitory effect
(P<0.05) on tumor growth (Fig. 5C and
D). Notably, Ki67 was significantly downregulated by Ad-ROD1
(P<0.05) and was significantly upregulated by Ad-shROD1 in
cancer tissues (P<0.05) (Fig. 5E and
F).
Discussion
The Wnt/β-catenin signaling pathway serves a vital
role in stem cell self-renewal and differentiation (28,29), as
well as tumorigenesis (22–24). Wnt/β-catenin signaling pathway is
activated in various types of cancer resulting in abnormal
accumulation of β-catenin in the nucleus, where β-catenin interacts
with the transcription factor TCF/LEF (22–24).
Finally, this interaction promotes the expression of target genes
such as c-Myc and cyclin (22–24).
Additionally, increased β-catenin activity was demonstrated to be
associated with the poor prognosis in patients with breast cancer
(4,5).
Thus, inhibition of Wnt/β-catenin signaling pathway may be a
promising treatment of breast cancer.
ROD1 was initially regarded as an inhibitor of K562
cell differentiation (11). ROD1 also
may promote proliferation but inhibit the differentiation of human
gastric cancer MKN45 cells (16).
However, the role of ROD1 in breast cancer remains unknown. In the
present study, the molecular mechanism of inhibition of breast
cancer cell growth mediated by ROD1 was investigated. The results
revealed that ROD1 inhibited proliferation and migration of
MBA-MB-231 cells. The results demonstrated that the expression of
ROD1 was reduced in breast cancer cells and tumor tissues.
Additionally, ROD1 may inhibit the invasion of MBA-MB-231 cells.
Previous studies (11,16) demonstrated that ROD1 may be a
promising suppressor in breast cancer, however further studies are
needed to confirm the results of the present study.
The molecular mechanism underlying ROD1-mediated
inhibition of the invasion of breast cancer cells was also
investigated. It has been reported that the transcriptional
activity of β-catenin is determined by its protein level and its
cellular localization (30). In the
present study, it was demonstrated that ROD1 may inhibit
translocation of β-catenin into the nucleus leading to decreased
expression of its downstream targets, including c-Myc, in a
dose-dependent manner in MDA-MB-231 cells. Additionally, it was
demonstrated that ROD1 suppressed the migration of β-catenin from
cytosol into the nucleus as assessed using a TOPflash luciferase
assay. Together, these data indicated that ROD1 may suppress the
invasive ability of breast cancer through its interaction with
β-catenin by inhibiting β-catenin migration into the nucleus.
In order to explore the inhibitory effect of ROD1 on
the growth of MDA-MB-231 cells in vivo, these cells were
implanted into nude mice. It was demonstrated that Ad-ROD1
inhibited β-catenin migration into nuclei by reducing
phosphorylated β-catenin (Y333) in tumor tissues, thus leading to
tumor suppression. Additionally, it was demonstrated that Ad-shROD1
may induce tumor growth and β-catenin phosphorylation (Y333).
Furthermore, Ki67 was downregulated by Ad-ROD1 and was upregulated
by Ad-shROD1 in tumor tissues.
Taken together, ROD1 may illustrate its anticancer
effect on breast cancer cells invasion through interacting with
β-catenin by inhibiting its activity and suppressing its migration
to the nucleus. The present study demonstrated that ROD1 may be a
tumor suppressor of breast cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YZ and XZ designed the experiment. YZ, HZ, EW, LH,
RY and YM performed the experiment. YZ and HZ processed the data
and XZ wrote the manuscript. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Animal Care
and Protection Committee of the Laboratory Animal Center of Soochow
University (approval no. SYXK 2014-0030; Jiangsu, China).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
DeSantis C, Ma JM, Bryan L and Jemal A:
Breast Cancer Statistics, 2013. CA Cancer J Clin. 64:52–62. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Polakis P: The oncogenic activation of
beta-catenin. Curr Opin Genet Dev. 9:15–21. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Howe LR and Brown AM: Wnt signaling and
breast cancer. Cancer Biol Ther. 3:36–41. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Incassati A, Chandramouli A, Eelkema R and
Cowin P: Key signaling nodes in mammary gland development and
cancer: β-catenin. Breast Cancer Res. 12:122010
|
|
5
|
Lin SY, Xia WY, Wang JC, Kwong KY, Spohn
B, Wen Y, Pestell RG and Hung MC: beta-catenin, a novel prognostic
marker for breast cancer: Its roles in cyclin D1 expression and
cancer progression. Proc Natl Acad Sci USA. 97:4262–4266. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Behrens J: Control of beta-catenin
signaling in tumor development. Ann Ny Acad Sci. 910:21–35. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Holland JD, Klaus A, Garratt AN and
Birchmeier W: Wnt signaling in stem and cancer stem cells. Curr
Opin Cell Biol. 25:254–264. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Turashvili G, Bouchal J, Burkadze G and
Kolar Z: Wnt signaling pathway in mammary gland development and
carcinogenesis. Pathobiology. 73:213–223. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Verras M and Sun ZJ: Roles and regulation
of Wnt signaling and beta-catenin in prostate cancer. Cancer Lett.
237:22–32. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Amado NG, Fonseca BF, Cerqueira DM, Neto
VM and Abreu JG: Flavonoids: Potential Wnt/beta-catenin signaling
modulators in cancer. Life Sci. 89:545–554. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yamamoto H, Tsukahara K, Kanaoka Y, Jinno
S and Okayama H: Isolation of a mammalian homologue of a fission
yeast differentiation regulator. Mol Cell Biol. 19:3829–3841. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Spellman R, Llorian M and Smith CW:
Crossregulation and functional redundancy between the splicing
regulator PTB and its paralogs nPTB and ROD1. Mol Cell. 27:420–434.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Maniatis T and Tasic B: Alternative
pre-mRNA splicing and proteome expansion in metazoans. Nature.
418:236–243. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hagen RM and Ladomery MR: Role of splice
variants in the metastatic progression of prostate cancer. Biochem
Soc Trans. 40:870–874. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Brazao TF, Demmers J, van IJcken W,
Strouboulis J, Fornerod M, Romão L and Grosveld FG: A new function
of ROD1 in nonsense-mediated mRNA decay. FEBS Lett. 586:1101–1110.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen B, Zhao AG, Shao J, Mu XY, Jiang L
and Liu JW: The effects of PTBP3 silencing on the proliferation and
differentiation of MKN45 human gastric cancer cells. Life Sci.
114:29–35. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Luo J, Deng ZL, Luo X, Tang N, Song WX,
Chen J, Sharff KA, Luu HH, Haydon RC, Kinzler KW, et al: A protocol
for rapid generation of recombinant adenoviruses using the Ad Easy
system. Nat Protoc. 2:1236–1247. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lv K, Guo YJ, Zhang YL, Wang K, Li K, Zhu
Y and Sun S: Transient inhibition of foot-and-mouth disease virus
replication by siRNAs silencing VP1 protein coding region. Res Vet
Sci. 86:443–452. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Anderson RD, Haskell RE, Xia H, Roessler
BJ and Davidson BL: A simple method for the rapid generation of
recombinant adenovirus vectors. Gene Ther. 7:1034–1038. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lu FI, Sun YH, Wei CY, Thisse C and Thisse
B: Tissue-specific derepression of TCF/LEF controls the activity of
the Wnt/β-catenin pathway. Nat Commun. 5:53682014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gaspar C and Fodde R: APC dosage effects
in tumorigenesis and stem cell differentiation. Int J Dev Biol.
48:377–386. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen GP, Shukeir N, Potti A, Sircar K,
Aprikian A, Goltzman D and Rabbani SA: Up-regulation of Wnt-1 and
β-catenin production in patients with advanced metastatic prostate
carcinoma-Potential pathogenetic and prognostic implications.
Cancer. 101:1345–1356. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang F, Zeng Q, Yu G, Li S and Wang CY:
Wnt/beta-catenin signaling inhibits death receptor-mediated
apoptosis and promotes invasive growth of HNSCC. Cell Signal.
18:679–687. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fiebig HH, Berger DP, Winterhalter BR and
Plowman J: In vitro and In vivo evaluation of Us-Nci compounds in
human tumor xenografts. Cancer Treat Rev. 17:109–117. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang D, Fei F, Li S, Zhao Y, Yang Z, Qu
J, Zhang X, Yin Y and Zhang S: The role of β-catenin in the
initiation and metastasis of TA2 mice spontaneous breast cancer. J
Cancer. 8:2114–2123. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sun B, Zhang S, Zhang D, Liu Y, Li Y, Rong
Z, Zhu Y and Jia X: Clusterin is associated with spontaneous breast
cancer in TA2 mice. FEBS Lett. 581:3277–3282. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lindvall C, Evans NC, Zylstra CR, Li Y,
Alexander CM and Williams BO: The Wnt signaling receptor Lrp5 is
required for mammary ductal stem cell activity and Wnt1-induced
tumorigenesis. J Biol Chem. 281:35081–35087. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Reya T and Clevers H: Wnt signalling in
stem cells and cancer. Nature. 434:843–850. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Rosano L, Cianfrocca R, Spinella F, Di
Castro V, Nicotra MR, Lucidi A, Ferrandina G, Natali PG and Bagnato
A: Acquisition of chemoresistance and EMT phenotype is linked with
activation of the endothelin A receptor pathway in ovarian
carcinoma cells. Clin Cancer Res. 17:2350–2360. 2011. View Article : Google Scholar : PubMed/NCBI
|