Introduction
The morbidity and mortality rates of lung cancer
rank first in malignant tumors, among which non-small cell lung
cancer (NSCLC) occupies a major position in lung cancer, so finding
treatment for NSCLC is extremely urgent (1). At present, the treatment of NSCLC has
been developed from traditional comprehensive treatment, such as
surgery and chemotherapy, to individualized treatment. However, the
cure rate is not high and NSCLC cannot be radically cured, so
better treatment means and methods are still needed (2,3). A large
number of studies have shown that the poor prognosis of patients
with NSCLC may be related to a low response rate to chemotherapy or
primary/secondary drug resistance produced during the process of
combined chemotherapy (4). With the
deepening of research on related genes to the prediction of
curative effect of chemotherapeutic and targeted drugs, the
development and application of individualized chemotherapy and
individualized targeted therapy for NSCLC have been greatly
promoted (5). Genes, such as
epithelial growth factor receptor (EGFR), breast cancer 1
(BRCA1) and receptor-associated protein 80 (RAP80),
are all effective genes in the treatment of NSCLC. The combined
detection of several genes that can predict the curative effect can
provide an important reference basis for the development of NSCLC
treatment programs (6). In the
present study, expression of BRCA1 and RAP80 and
EGFR gene mutations in 51 NSCLC patients were detected, and
their association were analyzed, so as to understand the
association of EGFR gene mutations with expression of BRCA1
and RAP80 in NSCLC patients, and provide a basis for further
exploring more effective individualized treatment programs for
NSCLC patients.
Materials and methods
Patients
General data
In the present study, 51 NSCLC patients admitted and
treated in the Thoracic Surgery Department of The Affiliated
Jiangyin Hospital of Southeast University Medical College (WuXi,
China) and who underwent biopsy or surgery from September 2014 to
September 2016 were selected, and general data and smoking status
were recorded. The study was approved by the Ethics Committee of
The Affiliated Jiangyin Hospital of Southeast University Medical
College and informed consents were signed by the patients or
guardians.
Main reagents
EGFR primer was provided by Guangzhou RiboBio Co.,
Ltd. (Guangzhou, China). Polymerase chain reaction (PCR) primers
for BRCA1 and RAP80 were synthesized by Sangon Biotech Co., Ltd.
(Shanghai, China). Tissue protein extraction kits were purchased
from Nanjing KeyGen Biotech Development Co., Ltd. (Nanjing, China).
RNAiso Plus, PrimeScript® RT reagent kit with gDNA
Eraser and SYBR®Premix Ex Taq™ II (Tli RNaseH Plus) were
purchased from Takara Biotechnology Co., Ltd. (Dalian, China).
TRIzol total RNA extraction kits were from Tiangen Biotech Co.,
Ltd. (Beijing, China). Reverse transcription-polymerase chain
reaction (RT-PCR) kits were from Tiangen Biotech Co., Ltd.
Bicinchoninic acid (BCA) protein quantification kits and BeyoECL
Plus kits were from Beyotime Institute of Biotechnology (Haimen,
China). Rabbit anti-human glyceraldehyde-3-phosphate dehydrogenase
(cat. no. 2118; 1:800), BRCA1 (cat. no. 14823; 1:800) and RAP80
(cat. no. 14466; 1:800) monoclonalantibodies, goat anti-rabbit HRP
(cat. no. 7074; 1:1,000) and fluorescence secondary polyclonal
antibodies (cat. no. 4412; 1:100), all were purchaced from Cell
Signaling Technology Europe (B.V., Leiden, The Netherlands).
Experimental methods
Hematoxylin and eosin (H&E)
histopathological staining
Tissues in control and NSCLC group were dehydrated,
embedded in paraffin, and cut into 5 µm slices for section making
and staining. After H&E staining and sealing, sections in
control and NSCLC group were observed under an upright microscope
(Olympus, Tokyo, Japan) for pathological differences of tissue
sections in the two groups, followed by photography and
analysis.
Immunofluorescence staining
The paraffin sections of lung tissues in control and
NSCLC group were dewaxed via xylene and dehydrated with gradient
alcohol, followed by antigen retrieval. Then sections were washed
with 0.01 M phosphate buffered saline (PBS) (pH 7.4) 3 times (5 min
each time), sealed in a wet box containing 10% bull serum albumin
at 37°C for 30 min. Sections were added dropwise with the
fluorescence-labeled antibody appropriately diluted at 1:100,
placed in the wet box and incubated at 4°C overnight. After being
washed with PBS (pH 7.4) 3 times (5 min each time), sections were
added dropwise with the fluorescence-labeled secondary antibody
(diluted at 1:100) in the dark, and incubated in a wet box at 37°C
for another 2 h. Finally, sections were sealed with buffered
glycerol, observed and photographed under an upright fluorescence
microscope (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
RT-PCR
An appropriate number of lung tissues in control and
NSCLC group were rapidly transferred into 1 ml TRIzol reagent,
fully ground and homogenized, let stand at room temperature for 5
min and lysed completely, and centrifuged at 12,000 × g and 4°C for
5 min. Then the supernatant was carefully taken and added with
chloroform. The mixture was mixed evenly, let stand at room
temperature for 5 min, and centrifuged at 12,000 × g at 4°C for 15
min. The supernatant was carefully taken, added with the same
volume of isopropanol, let stand at room temperature for 10 min,
and centrifuged at 12,000 × g at 4°C for 10 min. The sediment was
retained, added with 75% ethanol and mixed evenly. Finally,
RNase-free water was added to completely dissolve the sediment.
Then optical density (OD)260/OD280 ratio and the RNA concentration
were measured. Stepwise amplification was performed according to
the instructions and primer sequence templates shown in Table I, and the reaction products were
subjected to RT-PCR analysis.
 | Table I.RT-PCR primer sequences of BRCA1,
RAP80 and β-actin mRNA. |
Table I.
RT-PCR primer sequences of BRCA1,
RAP80 and β-actin mRNA.
| Gene name | Primer sequence |
|---|
| BRCA1 | F:
5′-ACAGCTGTGTGGTGCTTCT-GTG-3′ |
|
| R:
3′-CATTGTCCTCTGTCCAGGCATC-5′ |
| RAP80 | F:
5′-ACATCAAGTCTTCAGAAACAGGAGC-3′ |
|
| R:
3′-TGCAGCCTGCCTCTTFCCAT-5′ |
| β-actin | F:
5′-GAGCCGGGAAATCGTGCGT-3′ |
|
| R:
3′-GGAAGGAAGGCTGGAAGATG-5′ |
Western blot analysis
Lung tissues from the control and NSCLC group were
washed twice with ice normal saline, respectively. According to
instructions of the total protein extraction kit, tissues were
added with lysis buffer homogenized using a tissue homogenizer for
1 min and centrifuged, and the supernatant was collected. The
concentration of protein was measured using the BCA protein
concentration assay kit. Total protein extracting solution and 2X
loading buffer were mixed evenly at a volume ratio of 1:1, treated
with boiling water bath for 5 min and naturally cooled. Sodium
dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE)
separation gel in an appropriate proportion was prepared according
to the molecular weight of target protein, and frozen for about 1
h. Then 5% SDS-PAGE concentration gel was prepared and frozen for
about 30 min. Electrophoretic buffer solution and denatured protein
samples were added into the loading wells for loading based on the
protein concentration, and the total protein content in each well
was kept the same. The electrophoresis was performed under constant
pressure of 220 V until the bromophenol blue reached the bottom of
the gel. According to the molecular weight of target protein, the
gel was cut and placed into transfer buffer. A layer of
polyvinylidene fluoride (PVDF) membrane and six layers of filter
paper were cut according to the size of the gel. PVDF membrane was
immersed into the methanol for 10 sec, and PVDF membrane and filter
paper were placed into the transfer buffer. Then the positive pole
- three layers of filter paper - PVDF membrane - gel - three layers
of filter paper - negative pole were placed on the membrane
transfer instrument in this order. Their edges were aligned to
prevent blistering. After the membrane transfer under constant
pressure of 110 V for 2 h, the membrane attached with protein was
sealed using 5% skim milk powder at room temperature for 2 h. The
sealed membrane was washed with Tris-buffered saline with Tween-20
(TTBS) for 5 min, and incubated in the primary antibody in
corresponding proportion at 4°C overnight. After the membrane was
washed with TTBS for 5 min (10 min each time), it was incubated
using the corresponding secondary antibody on a shaking table at
room temperature for 3 h, and it was washed again with TTBS 3 times
(10 min each time). After the gel imager was warmed up for 30 min,
reagent A and B in electrochemiluminescence (ECL) kit were evenly
mixed at a volume ratio of 1:1, added dropwise onto the PVDF
membrane, followed by color development in the dark for 1 min.
Excess liquid around the membrane was sucked dry with the filter
paper and the membrane was placed into the gel imager, followed by
photography under the dynamic integral mode and observation of
results. Image analysis software (V3 Western Workflow™; Bio-Rad
Laboratories, Inc.) was used to analyze the images.
EGFR gene detection
Blood specimens of NSCLC patients were collected to
extract gDNA and detect EGFR gene in peripheral leucocytes.
Peripheral venous blood was extracted from the participants of the
study to extract DNA. EGFR gene was detected via PCR and gel
electrophoresis. The forward primer and reverse primer of EGFR gene
are as follows: 5′-CTTCGGGGAGCAGCGATGCGAC-3′ (forward) and
5′-ACCAATACCTATTCCGTTACAC-3′ (reverse).
Statistical analysis
Experimental data are presented as mean ± standard
deviation (mean ± SD), and SSPSS 17.0 software (SPSS, Inc.,
Chicago, IL, USA) was used for the statistical analysis of
experimental results. Data were analyzed using analysis of variance
or t-test and the post-hoc test was Dunnett's test. P<0.05 was
considered to indicate a statistically significant analysis.
Results
General data of patients
The general conditions of NSCLC patients were
recorded, and statistical results are shown in Table II relating to sex, age, history,
stage and grade of 51 NSCLC patients.
 | Table II.General data of patients. |
Table II.
General data of patients.
| Groups | No. | (%) |
|---|
| Sex |
|
|
| Male | 37 | 72.55 |
|
Female | 14 | 27.45 |
| Age |
|
|
|
<60 | 30 | 58.82 |
| ≥60 | 21 | 41.18 |
| History |
|
|
|
Adeno | 36 | 70.59 |
| SCC | 15 | 29.41 |
| Stage |
|
|
| IIIA | 3 | 5.88 |
| IIIB | 12 | 23.53 |
| IV | 36 | 70.59 |
| Grade |
|
|
| G1 | 2 | 3.92 |
| G1–2 | 3 | 5.88 |
| G2 | 8 | 15.69 |
| G2-3 | 3 | 5.88 |
| G3 | 12 | 23.53 |
|
Unknown | 23 | 45.10 |
Smoking status of patients
The smoking status of NSCLC patients was recorded.
As shown in Table III, NSCLC
patients with a smoking history accounted for 64.71%, those who
smoked for more than 20 years accounted for 81.82%, and those who
had quit smoking accounted for 78.13%, suggesting that NSCLC has a
certain association with smoking.
 | Table III.Smoking status of patients. |
Table III.
Smoking status of patients.
| Groups | No. | (%) |
|---|
| Smoking history |
|
|
| Yes | 33 | 64.71 |
| No | 18 | 35.29 |
| Smoking
pack-year |
|
|
|
<20 | 5 | 15.15 |
| ≥20 | 27 | 81.82 |
|
Unknown | 1 | 3.03 |
| Quit smoking |
|
|
|
Yes | 25 | 78.13 |
| No | 1 | 3.13 |
|
Unknown | 6 | 18.75 |
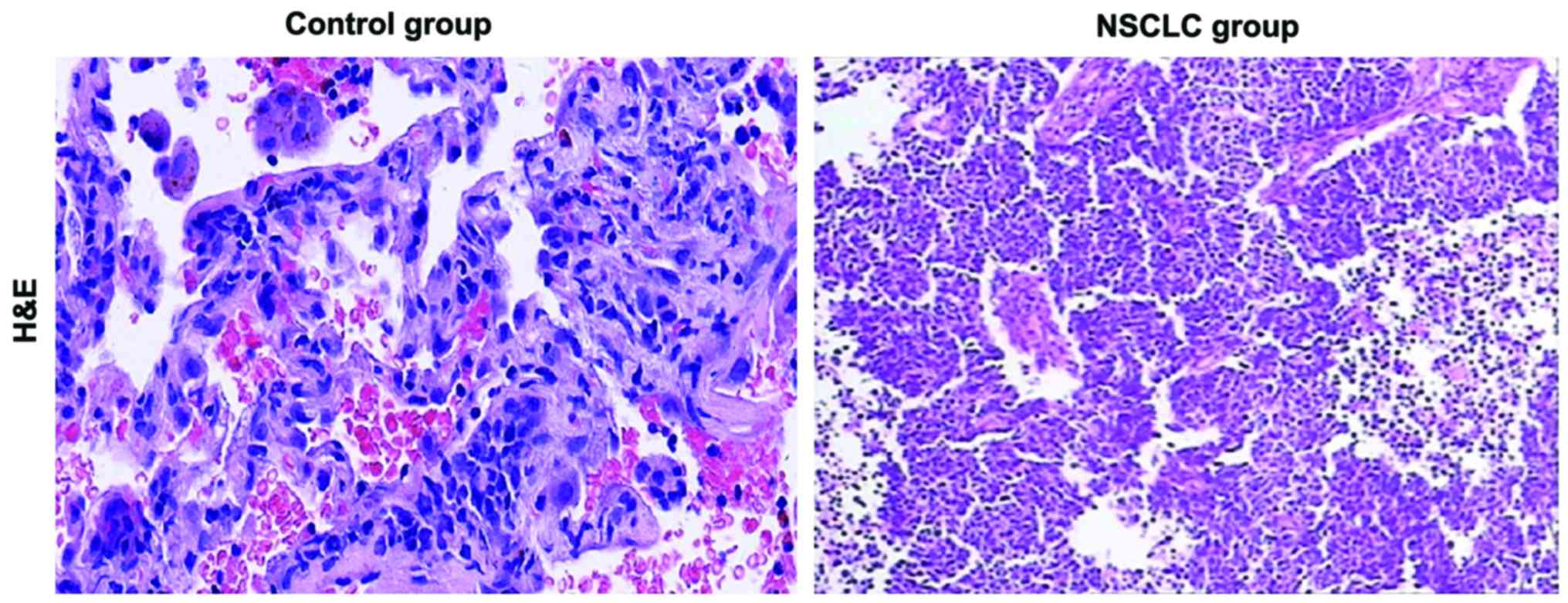
H&E staining results
H&E staining showed that there were significant
pathological differences in lung tissues between control and NSCLC
group. Compared with those in control group, the structure of lung
tissue was destroyed, nuclear chromatin became darker, and a large
number of cancer cells were produced in NSCLC group (Fig. 1).
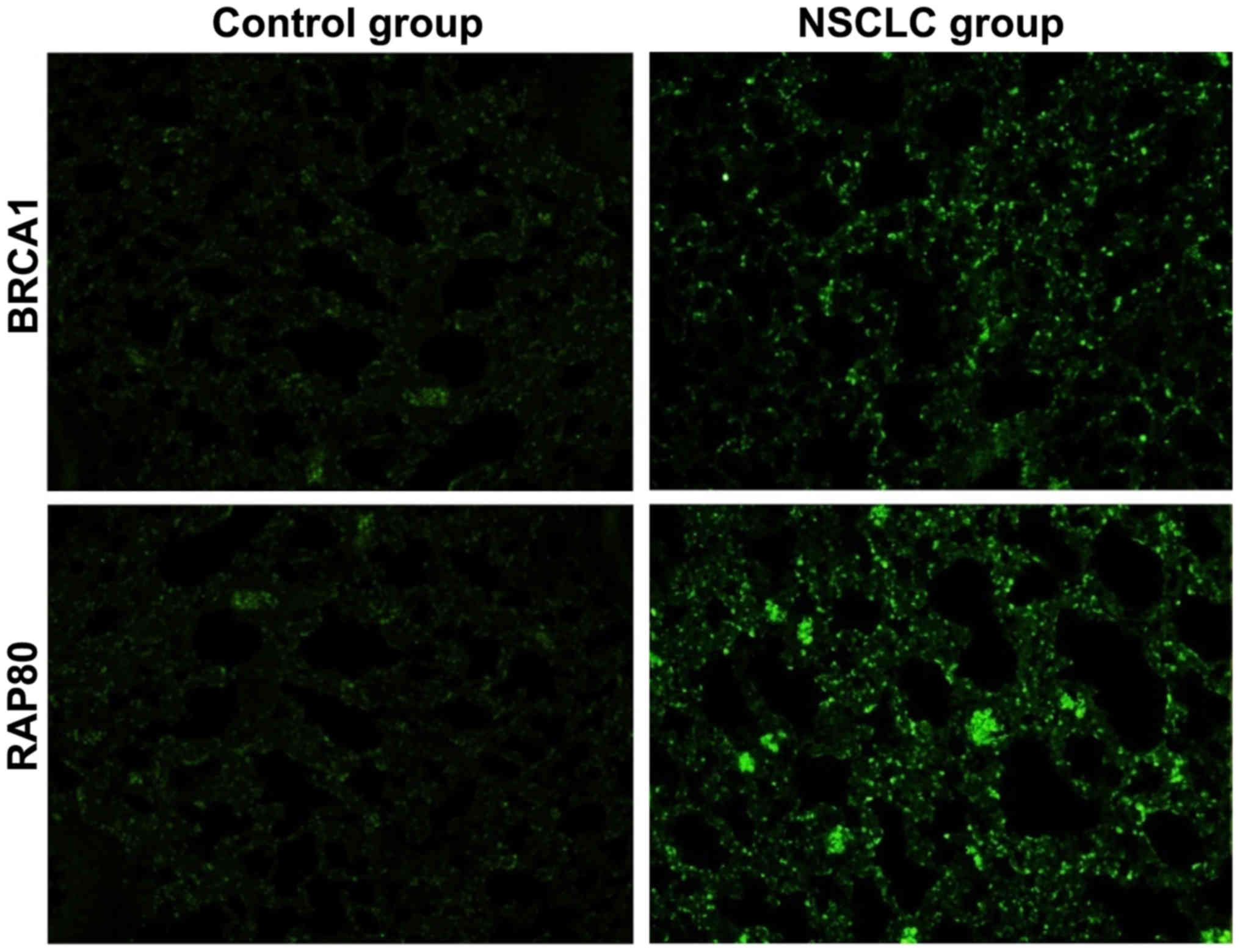
Immunofluorescence staining
results
The expression of BRCA1 and RAP80 in lung tissues in
control and NSCLC group were detected via immunofluorescence
method. Compared with those in control, the expression of BRCA1 and
RAP80 in NSCLC group were significantly decreased, indicating that
BRCA1 and RAP80 are involved in the occurrence and development of
NSCLC (Fig. 2).
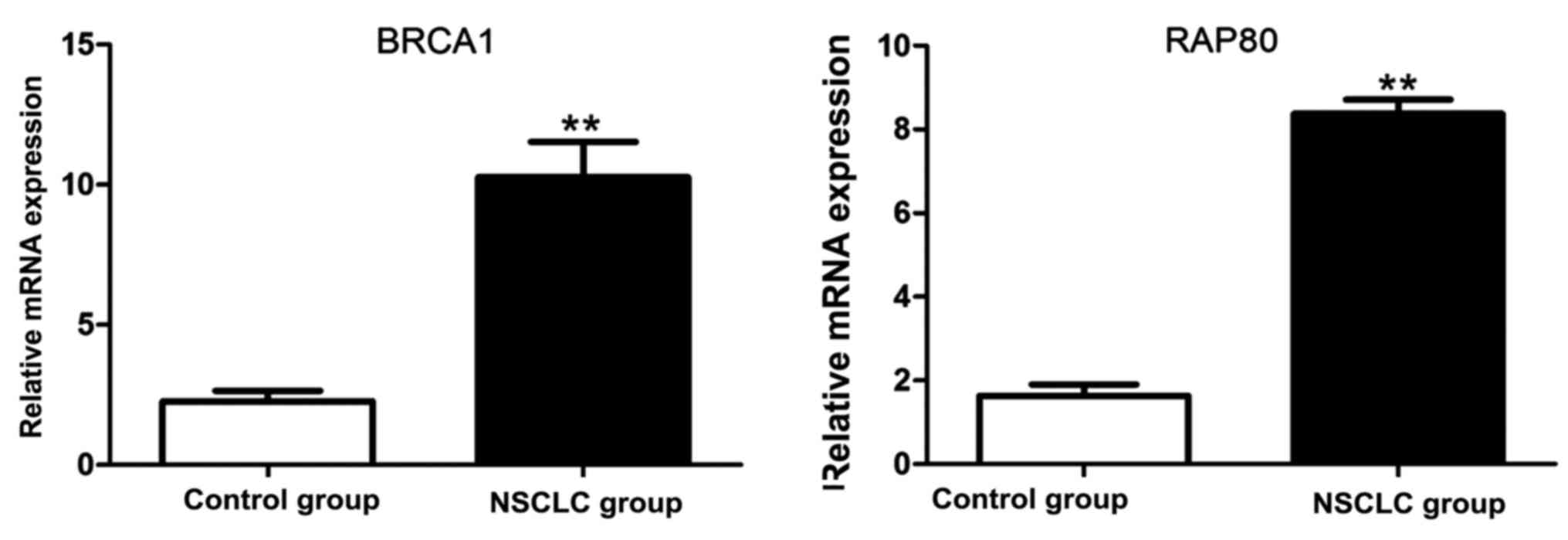
RT-PCR results of BRCA1 and RAP80
mRNA
The total RNA was extracted from lung tissue samples
in control and NSCLC group, respectively. Results of RT-PCR
revealed that BRCA1 and RAP80 mRNA in NSCLC were significantly
increased compared with those in control group (Fig. 3).
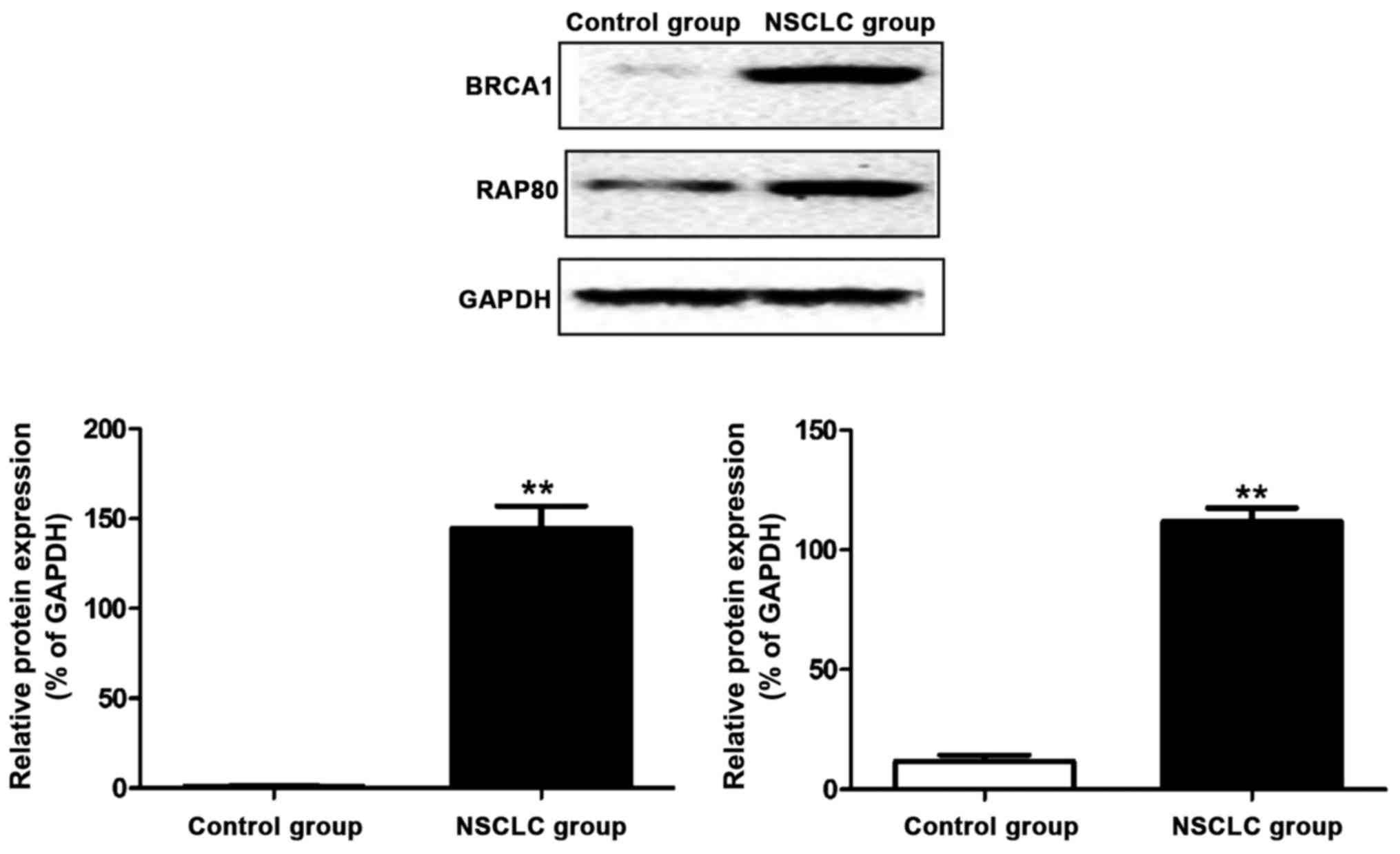
Western blot results of BRCA1 and
RAP80 proteins
The protein was extracted from lung tissue samples
in control and NSCLC group, respectively. Results of western blot
showed that the BRCA1 and RAP80 protein expressions in NSCLC were
obviously increased compared with those in control group (Fig. 4).
EGFR gene mutations
EGFR gene mutations were detected in 14 out
of 51 NSCLC patients, including 1 case of Exon20: s768I, 1 case of
Exon 21: L861Q, 1 case of L858R, 1 case of DEL19, 1 case of L858R,
1 case of EXON 20 Q787Q, 1 case of EXON 21, 4 cases of DEL19, and 5
cases of wild-type (Table IV).
 | Table IV.EGFR gene mutations. |
Table IV.
EGFR gene mutations.
| Type | Νo. | (%) |
|---|
| Wild-type | 5 | 35.72 |
| Exon 20: s768I;
Exon 21: L861Q | 1 | 7.14 |
| L858R | 1 | 7.14 |
| Del19 | 4 | 28.58 |
| Del19, L858R | 1 | 7.14 |
| Exon 20 Q787Q | 1 | 7.14 |
| Exon 21 | 1 | 7.14 |
Discussion
NSCLC, a common malignant tumor, seriously threatens
human health (7). According to large
data in China, the incidence rate of NSCLC in men ranks first in
malignant tumors, while that in women is second only to that of
breast cancer (8). With the
development of economy and science and technology, the treatment of
NSCLC has entered the stage of individualized treatment. In
particular, research and development of drugs targeting EGFR, BRCA1
and RAP80 have milestone significance in the diagnosis and
treatment of NSCLC (9–11).
EGFR is an expression product of proto-oncogene
c-erbB1, a member of the human epidermal receptor family (12). EGFR gene plays an important
role in many physiological processes, including cell growth,
proliferation and differentiation. Abnormalities in the EGFR
gene will lead to a variety of diseases, such as cancer, diabetes
mellitus, immunodeficiency and cardiovascular diseases (13). BRCA1 is a gene discovered in
1990, which is directly related to hereditary breast cancer.
BRCA1 is a gene that inhibits the occurrence of malignant
tumors, which plays an important role in regulating the replication
of human cells, genetic material DNA damage repair and normal
growth of cells (14–17). Moreover, RAP80 also plays a critical
role in DNA damage response, which can promote a series of repair
proteins to be located to the correct DNA damage site (18–20). In
conclusion, EGFR, BRCA1 and RAP80 play key roles in
the occurrence and development of tumors, and they may be target
genes for tumor treatment.
In the present study, 51 NSCLC patients admitted and
treated in the Thoracic Surgery Department of the hospital and who
underwent biopsy or surgery from September 2014 to September 2016
were selected, and general data and smoking status were recorded.
It was found that NSCLC had a association with smoking. H&E
histopathological staining method was used to detect pathological
differences in lung tissues between control and NSCLC group.
H&E histopathological staining showed that the structure of
lung tissues was destroyed, nuclear chromatin became darker and a
large number of cancer cells were produced in NSCLC compared with
those in control group. Immunofluorescent staining method was used
to detect the fluorescence expression of BRCA1 and RAP80 proteins
in both groups, and results revealed that the expression of
fluorescence of BRCA1 and RAP80 proteins in NSCLC group was
significantly increased. RT-PCR and western blot proved that the
BRCA1 and RAP80 mRNA and protein expression in NSCLC group were
significantly increased. Besides, EGFR gene mutations were
detected in 14 out of 51 patients. In summary, NSCLC has a
association with smoking, expression of BRCA1 and RAP80 is closely
related to the occurrence and development processes of NSCLC, and
EGFR mutation is also significantly associated with NSCLC. It is
expected that the key roles of EGFR, BRCA1 and RAP80 in NSCLC in
further research will clarify that they can be the targets of
prevention, diagnosis and treatment of this disease.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors contributions
XS and FC collected and analyzed general data of
patients. XS and HY helped with H&E histopathological staining.
DW performed PCR. NW and MY were responsible for immunofluorescence
staining. YF and QW contributed to western blot analysis. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
The Affiliated Jiangyin Hospital of Southeast University Medical
College (Wuxi, China) and informed consents were signed by the
patients or guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Fossella F, Pereira JR, von Pawel J,
Pluzanska A, Gorbounova V, Kaukel E, Mattson KV, Ramlau R, Szczesna
A, Fidias P, et al: Randomized, multinational, phase III study of
docetaxel plus platinum combinations versus vinorelbine plus
cisplatin for advanced non-small-cell lung cancer: The TAX 326
study group. J Clin Oncol. 21:3016–3024. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Scagliotti GV, Parikh P, von Pawel J,
Biesma B, Vansteenkiste J, Manegold C, Serwatowski P, Gatzemeier U,
Digumarti R, Zukin M, et al: Phase III study comparing cisplatin
plus gemcitabine with cisplatin plus pemetrexed in
chemotherapy-naive patients with advanced-stage non-small-cell lung
cancer. J Clin Oncol. 26:3543–3551. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cobo M, Isla D, Massuti B, Montes A,
Sanchez JM, Provencio M, Viñolas N, Paz-Ares L, Lopez-Vivanco G,
Muñoz MA, et al: Customizing cisplatin based on quantitative
excision repair cross-complementing 1 mRNA expression: A phase III
trial in non-small-cell lung cancer. J Clin Oncol. 25:2747–2754.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Guha U, Chaerkady R, Marimuthu A,
Patterson AS, Kashyap MK, Harsha HC, Sato M, Bader JS, Lash AE,
Minna JD, et al: Comparisons of tyrosine phosphorylated proteins in
cells expressing lung cancer-specific alleles of EGFR and KRAS.
Proc Natl Acad Sci USA. 105:14112–14117. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lynch TJ, Bell DW, Sordella R,
Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat
SM, Supko JG, Haluska FG, et al: Activating mutations in the
epidermal growth factor receptor underlying responsiveness of
non-small-cell lung cancer to gefitinib. N Engl J Med.
350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Paez JG, Jänne PA, Lee JC, Tracy S,
Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et
al: EGFR mutations in lung cancer: Correlation with clinical
response to gefitinib therapy. Science. 304:1497–1500. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pao W, Miller V, Zakowski M, Doherty J,
Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, et al:
EGF receptor gene mutations are common in lung cancers from ‘never
smokers’ and are associated with sensitivity of tumors to gefitinib
and erlotinib. Proc Natl Acad Sci USA. 101:13306–13311. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sequist LV, Martins RG, Spigel D, Grunberg
SM, Spira A, Jänne PA, Joshi VA, McCollum D, Evans TL, Muzikansky
A, et al: First-line gefitinib in patients with advanced
non-small-cell lung cancer harboring somatic EGFR mutations. J Clin
Oncol. 26:2442–2449. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Taron M, Ichinose Y, Rosell R, Mok T,
Massuti B, Zamora L, Mate JL, Manegold C, Ono M, Queralt C, et al:
Activating mutations in the tyrosine kinase domain of the epidermal
growth factor receptor are associated with improved survival in
gefitinib-treated chemorefractory lung adenocarcinomas. Clin Cancer
Res. 11:5878–5885. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Costa DB, Kobayashi S, Tenen DG and
Huberman MS: Pooled analysis of the prospective trials of gefitinib
monotherapy for EGFR-mutant non-small cell lung cancers. Lung
Cancer. 58:95–103. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Marchetti A, Martella C, Felicioni L,
Barassi F, Salvatore S, Chella A, Camplese PP, Iarussi T, Mucilli
F, Mezzetti A, et al: EGFR mutations in non-small-cell lung cancer:
Analysis of a large series of cases and development of a rapid and
sensitive method for diagnostic screening with potential
implications on pharmacologic treatment. J Clin Oncol. 23:857–865.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lafarge S, Sylvain V, Ferrara M and Bignon
YJ: Inhibition of BRCA1 leads to increased chemoresistance to
microtubule-interfering agents, an effect that involves the JNK
pathway. Oncogene. 20:6597–6606. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Husain A, He G, Venkatraman ES and Spriggs
DR: BRCA1 up-regulation is associated with repair-mediated
resistance to cis-diamminedichloroplatinum(II). Cancer Res.
58:1120–1123. 1998.PubMed/NCBI
|
|
14
|
Bhattacharyya A, Ear US, Koller BH,
Weichselbaum RR and Bishop DK: The breast cancer susceptibility
gene BRCA1 is required for subnuclear assembly of Rad51 and
survival following treatment with the DNA cross-linking agent
cisplatin. J Biol Chem. 275:23899–23903. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Abbott DW, Thompson ME, Robinson-Benion C,
Tomlinson G, Jensen RA and Holt JT: BRCA1 expression restores
radiation resistance in BRCA1-defective cancer cells through
enhancement of transcription-coupled DNA repair. J Biol Chem.
274:18808–18812. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mullan PB, Quinn JE, Gilmore PM,
McWilliams S, Andrews H, Gervin C, McCabe N, McKenna S, White P,
Song YH, et al: BRCA1 and GADD45 mediated G2/M cell cycle arrest in
response to antimicrotubule agents. Oncogene. 20:6123–6131. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Quinn JE, Kennedy RD, Mullan PB, Gilmore
PM, Carty M, Johnston PG and Harkin DP: BRCA1 functions as a
differential modulator of chemotherapy-induced apoptosis. Cancer
Res. 63:6221–6228. 2003.PubMed/NCBI
|
|
18
|
Chabalier C, Lamare C, Racca C, Privat M,
Valette A and Larminat F: BRCA1 downregulation leads to premature
inactivation of spindle checkpoint and confers paclitaxel
resistance. Cell Cycle. 5:1001–1007. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sobhian B, Shao G, Lilli DR, Culhane AC,
Moreau LA, Xia B, Livingston DM and Greenberg RA: RAP80 targets
BRCA1 to specific ubiquitin structures at DNA damage sites.
Science. 316:1198–1202. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim H, Chen J and Yu X: Ubiquitin-binding
protein RAP80 mediates BRCA1-dependent DNA damage response.
Science. 316:1202–1205. 2007. View Article : Google Scholar : PubMed/NCBI
|