Introduction
Gastric cancer (GC), as one of the leading causes of
morbidity and mortality worldwide, demonstrates a relative poor
prognosis to other cancers with an overall 5-year survival rate
below 30% (1). The TNM
(tumor/node/metastasis) staging system is routinely used in the
classification of GC. However, even GC patients with the same
stages and treatments tend to demonstrate variations in clinical
outcomes. Therefore, the efficacy of TNM staging may be unable to
distinguish higher and lower risk patients, and new markers
accompanying this system are urgently needed to define more robust
prognostic subtypes of GC (2).
With the development of high-throughput
technologies, including genomics, transcriptomics and proteomics,
an increasing number of prognostic molecules have been identified,
but most of them cannot be clinically applied due to low
effectiveness and poor reproducibility when validated in larger
cohorts (3). It has been proposed
that the construction of gene signature with multiple biomarkers
can greatly enhance the predictive and prognostic ability over
single biomarkers; a panel of gene signatures has been introduced
for clinical application. Currently, several potential prognostic
gene signatures have been developed for the prognosis of GC
(4–8).
However, these signatures always involve scores of genes, which are
not clinically applicable. Furthermore, although these prognostic
signatures can define high-risk GC subtypes, most of them do not
contribute to the development of personalized treatment for these
patients.
The hedgehog (Hh) signaling pathway acts as a
driving factor in the progression of cancer and may be considered
as a potential therapeutic target; considerable efforts have been
performed to develop its inhibitors to treat multiple cancers, such
as GC (9). Several previous studies
have evaluated the prognostic significance of the Hh signaling
pathway members in small scale clinical samples of GC, the
prognostic power of the combination of Hh-associated genes at a
larger scale has never been reported (10–12).
Therefore, we hypothesized that the Hh signaling pathway-based gene
signature may have greater prognostic power than a single
Hh-associated gene, and may also provide more biological and
clinical significance.
In this study, using The Cancer Genome Atlas (TCGA)
gene expression profiling, we developed a 3-Hh-gene signature that
may classify patients with GC into prognostic subgroups with
differing clinical outcomes. We further validated the potential of
this signature in silico using a large cohort of combined GC
transcriptomic data, and experimentally investigated these findings
in clinical GC samples.
Materials and methods
Training and validation data sets
Gene expression profiling data of TCGA GC (STAD) in
RSEM format (RNA-SeqV2) were downloaded from cBioPortal (http://www.cbioportal.org/) (13). TCGA STAD comprising 386 cases of GC
samples with overall survival data served as the training data set.
The REEM values of gene expression levels were log-transformed
before further analysis.
Combined gene expression microarray datasets
(GSE14210, GSE15459, GSE22377, GSE29272, GSE51105 and GSE62254)
from KM Plotter (http://kmplot.com/analysis) was used for in
silico validation (14). This
combined data set comprised 631 GC samples with survival data was
analyzed on Affymetrix Human Genome U133/U133A 2.0/HG-U133_Plus_2
platforms.
One hundred and twenty-six cases of pathologically
proven primary GC specimens at stage I–III were included for
experimental confirmation in our established gene classification.
All the samples were obtained with from patients with GC who
received standard curative surgery at Zhejiang Hospital of
Traditional Chinese Medicine from January 2004 and December 2011
with informed consent. All the patients received postoperative
chemotherapy (FOLFOX regimen), and no adjacent treatment was
administered before surgery. The study was approved by the
institutional review boards of Zhejiang Hospital of Traditional
Chinese Medicine and Zhejiang Police College.
Development of prognostic Hh signature
in the training data set
Nine canonical Hh signaling pathway-associated
genes, including three secreted ligands [SHH, indian hedgehog (IHH)
and DHH], three transmembrane receptors or co-receptors [patched
(PTCH)1, PTCH2 and smoothened frizzled class receptor (SMO)], and
three transcription factors (GLI1, GLI2 and GLI3) were selected for
analysis. The Cutoff Finder online tool (http://molpath.charite.de/cutoff/) was used to
determine the optimized threshold of gene expression to obtain most
the significant prognostic power as described before (15). According the established cutoff value,
patients were divided into high- and low-risk subgroups, and were
subject to univariate Cox proportional hazards analysis; The Cox
regression coefficients for each gene was calculated.
The expression value (G) of significant
Hh-associated genes identified in univariate Cox proportional
hazards analysis and their corresponding Cox regression coefficient
were utilized to reconstruct the prognostic gene signature, as
described in the following equation: Risk score=∑Cox coefficient of
gene Gi × expression value of gene Gi. The patients in the training
data set were divided into two groups (high- and low-risk)
according to the cutoff value for risk using Cutoff Finder.
Survival times were compared between the two groups using the
Kaplan-Meier analysis and log-rank test.
Assessment of the prognostic Hh gene
signature in the combined GEO validation data set
The potential of the Hh-gene signature for
predicting the outcome of patients with GC in the training data set
was externally invetsigated in the combined six-microarray cohort
derived from the GEO database. The workflow analysis was similar to
the training data set. Risk scores were determined in the
validation samples based on the above equation. The coefficient
values derived from the training data set were directly applied for
validation in the cohort. Two subgroups were constituted based on
the median risk score, and their survival times were compared with
the Kaplan-Meier analysis. The analysis was performed using the KM
Plotter online tool (http://kmplot.com/analysis).
Experimental validation of the
prognostic significance in our GC cohort by immunohistochemistry
(IHC)
Immunohistochemical staining for IHH, PTCH1 and SMO
on GC tumors were performed by the avidin-biotin complex method.
Briefly, 4 µm of sections of paraffin-embedded tissue blocks were
deparafnizated and rehydrated. After blocking of endogenous
peroxidases activity and heat-induced antigen-retrieval procedures,
the sections were incubated with rabbit anti-human antibodies
against IHH (1:150 dilution), PTCH1 (1:100 dilution) and SMO (1:100
dilution; all Abcam, Cambridge, UK) overnight at 4°C. Then, a
HRP-conjugated secondary antibody substrate (Dako, Copenhagen,
Denmark) was added, and 3,3′-diaminobenzidine (DAB) was used as the
chromogen.
The widely accepted German scoring system was used
to evaluate the IHC staining semi-quantitatively as previously
described (16). Each tumor was
assigned an IHC score (from 0 to 300), which was determined by
multiplying the intensity score by the score for the expression
extent of stained cells.
Statistical analysis
As for web-based bioinformatic tools, statistical
analysis was automatically obtained and conducted online. The
association between clinical variables and gene expression levels
generated from IHC-based validation were evaluated using the
Chi-square test. Cumulative survival curves were computed by using
the Kaplan-Meier method, and evaluated between groups using the
log-rank test. Multivariate survival analysis was performed using
Cox's regression model. All reported P-values were two-sided, and
P<0.05 was considered to indicate a statistically significant
difference. All the statistical analyses were performed using SPSS
22.0 software (IBM SPSS Statistics, Armonk, NY, USA).
Results
Detection of the Hh signaling
pathway-based genes is associated with overall survival and
development of prognostic gene signature in the training data
set
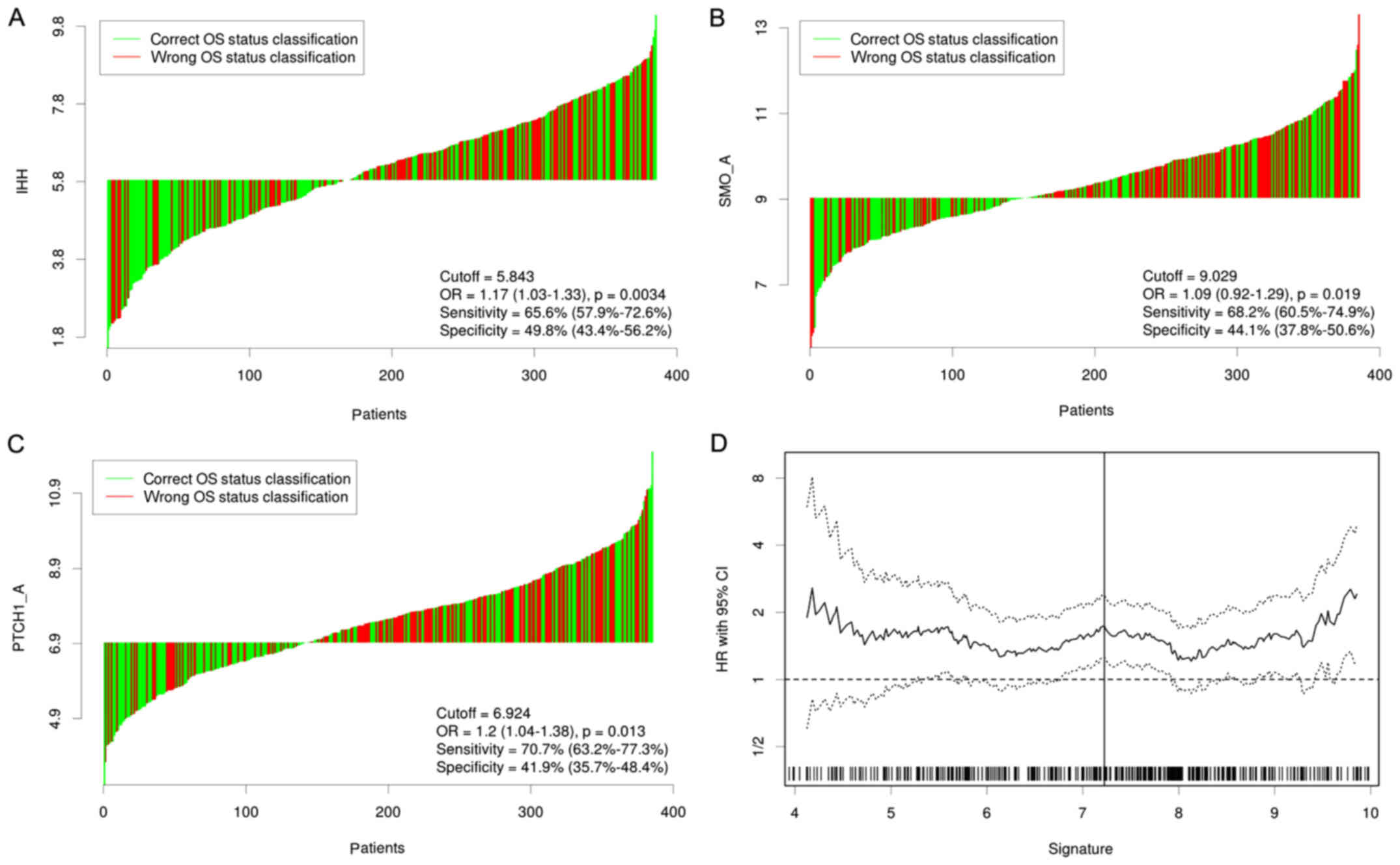
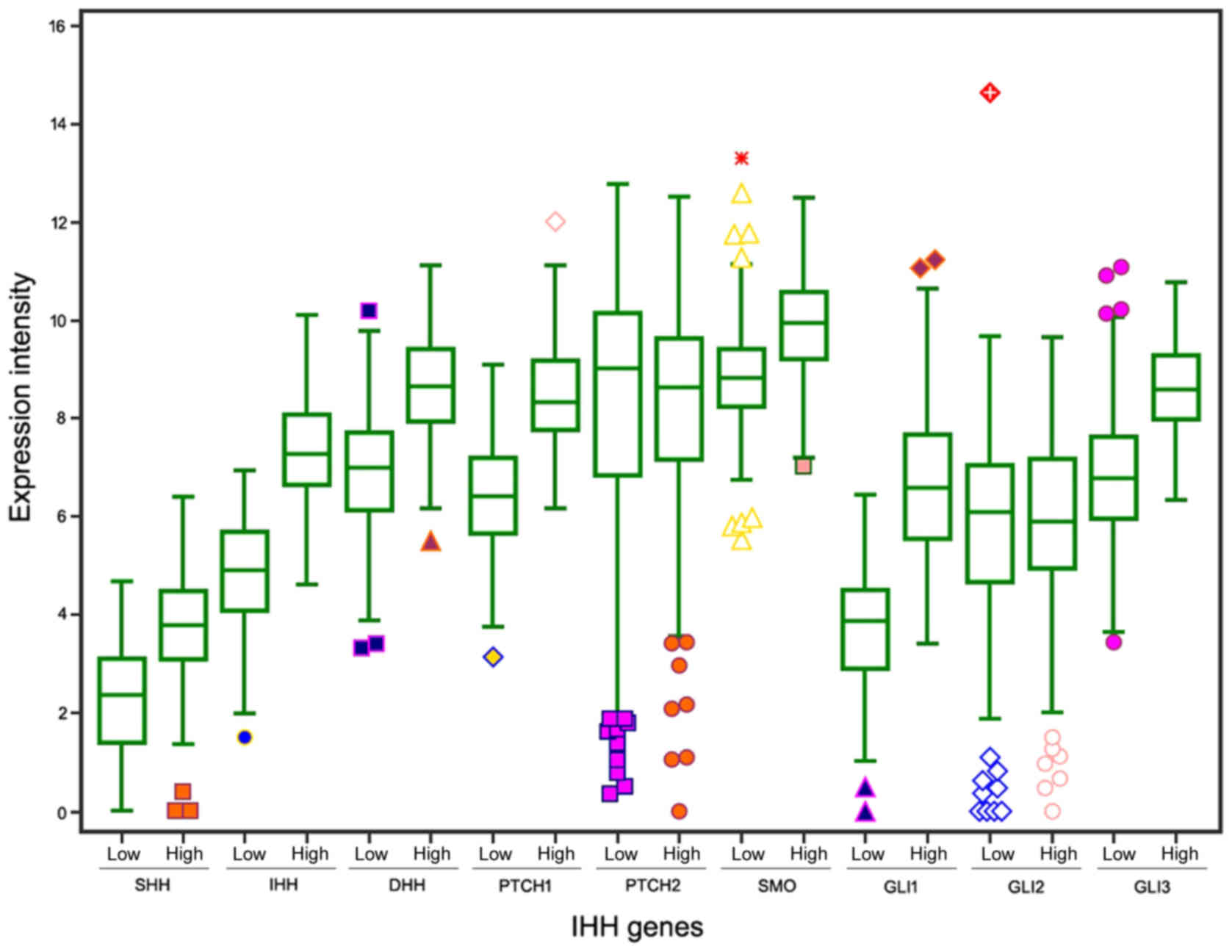
We first evaluated the prognostic values of 9
canonical Hh signaling pathway-associated genes for patients with
GC in TCGA STAD data set. Three members IHH, PTCH1 and SMO were
identified to have significant prognostic value at cutoff values of
5.843, 6.924 and 9,029 (Fig. 1); high
expression of IHH, PTCH1 and SMO was significantly associated with
mortality and shorter overall survival (Fig. 2). Subsequently, a univariate Cox
regression analysis was carried out to calculate the coefficient
for each of the three Hh-associated biomarkers. The prognostic risk
for each case was then scored by summing the coefficient-weighted
expression of the IHH-PTCH1-SMO signature as follows: 3-gene
signature score=(0.553xIHH value) + (0.457xPTCH1 value) +
(0.411xSMO value). We found that this signature had the best
prognostic ability at a cutoff value of 7.225; patients with higher
risk scores (n=201) had a significantly shorter survival than those
in the lower risk group (n=186) [Hazard Ratio (HR)=1.73, 95%
confidence interval (CI)=1.26–2.39, log-rank test, P=0.00069]
(Figs. 1 and 2). The expression levels of the 9
Hh-associated genes in the high- and low-risk subgroups were
demonstrated in Fig. 3.
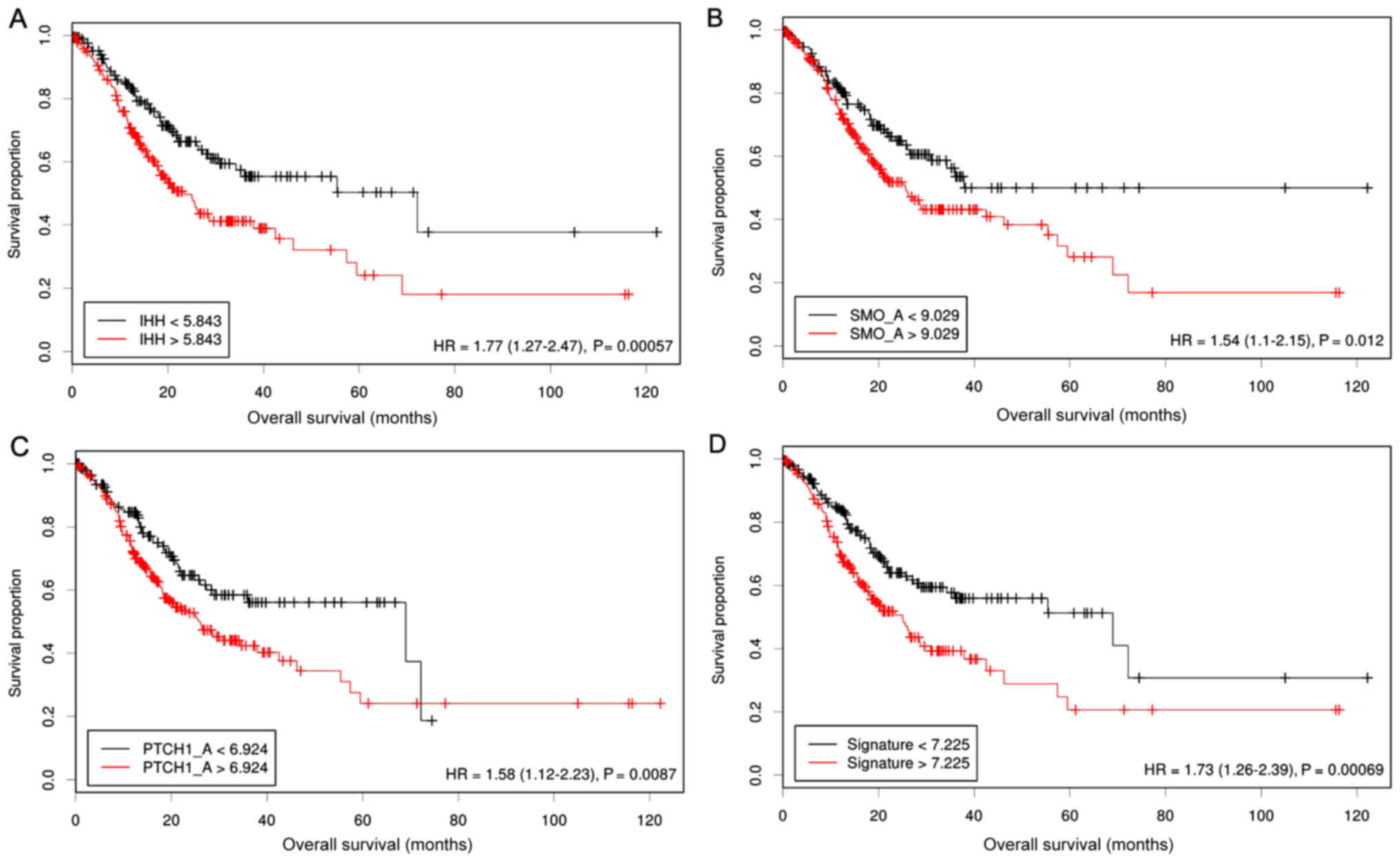
Prediction performance of the
3-Hh-gene signature in the combined GEO dataset
To reliably estimate the prediction potential, we
extracted the expression data of these 3 Hh-associated genes from
another large cohort of the GEO database. When the same coefficient
in the training data set was used, patients with GC of the
validation cohort were dichotomized according to the risk scores;
overall survival rates were significantly elevated in the subgroup
with high risk scores (HR=1.45, 95% CI=1.17–1.81, P=0.00068)
(Fig. 4).
To test whether the 3-gene signature is independent
of the current staging systems and other prognostic predictors, the
validation cohort were further stratified according to stages, HER2
status and treatment therapies. We found that the risk score
successfully separated patients into prognostic subgroups in all
the stages except stage IV (Fig. 4).
In addition, we also observed that this gene signature may also be
effectively used to identify high-risk individuals in the
HER2-negative subgroup (HR=1.65, 95% CI=1.26–2.16, P=0.00024) and
patients with GC receiving surgery alone (HR=1.49, 95%
CI=1.11–1.99, P=0.0075). These findings indicate that the 3-Hh-gene
signature may provide more prognostic information to the current
classification systems.
Experimental validation of the 3-Hh
gene signature in discriminating high-risk GC patients by IHC
Next, we evaluated whether this mRNA-based gene
signature retained its effectiveness in an independent cohort by
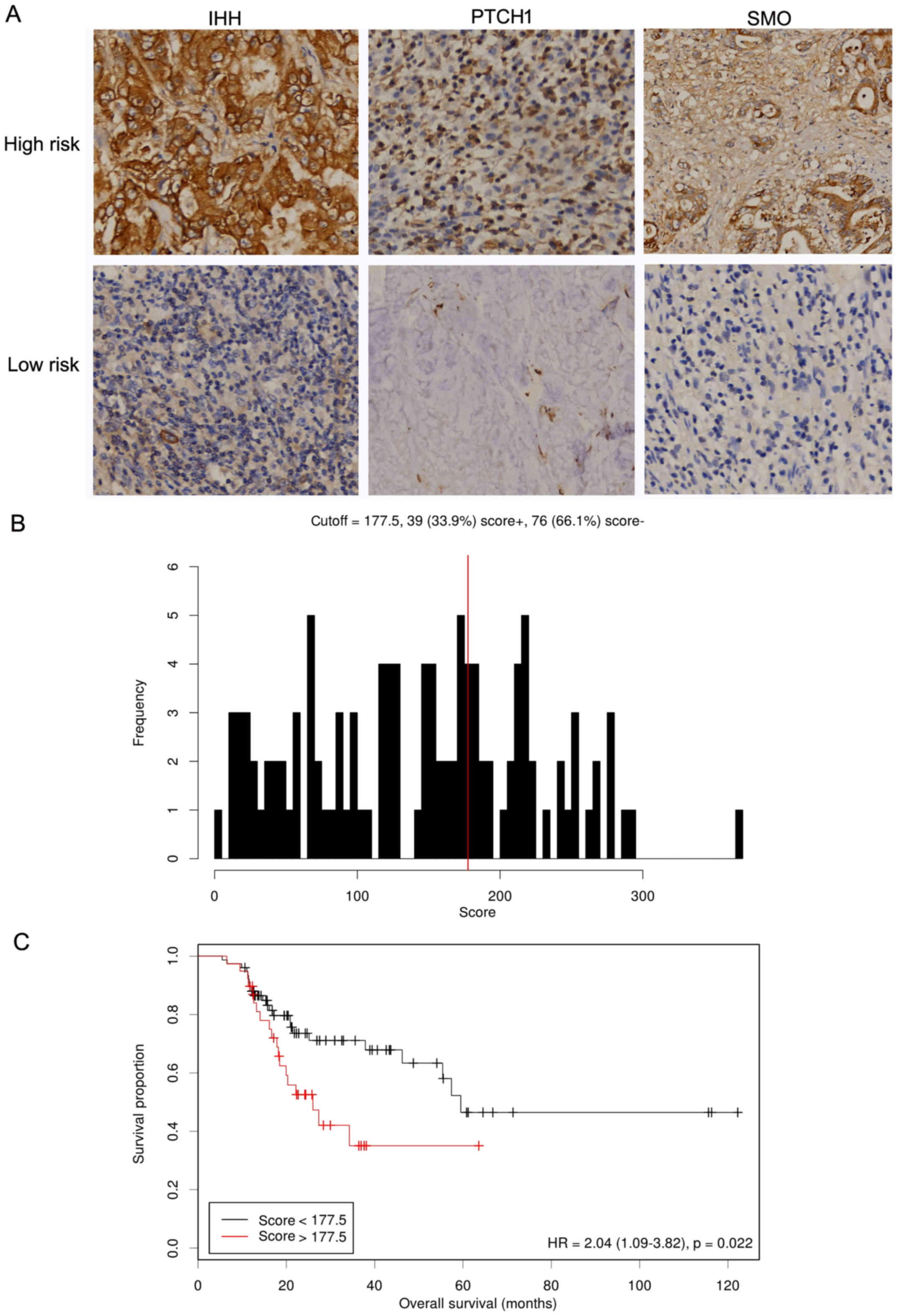
the IHC-detected strategy. Representative staining images of IHH,
PTCH1 and SMO were demonstrated in Fig.
5A. Each case was scored; univariate Cox regression analysis
indicated that high expression levels of these markers were
associated with decreased overall survival, but only reaching
marginal significance. We then derived the same formula in training
dataset to calculate a risk score for each case based on IHC
scores. Using the formula, our cohort was divided into high- (n=39)
and low-risk (n=76) subgroups with an optimal cutoff of 2.04 using
the Cutoff Finder tool (Fig. 5B).
Distribution of the demographic and clinicopathological
characteristics did not significantly vary between the high- and
the low-risk subgroups (Table I).
Furthermore, we found that patients with high-risk scores exhibited
shorter survival compared with in those with low-risk scores
(HR=2.04, 95% CI=1.09–3.82-, P=0.02) (Fig. 5C). In the multivariate Cox regression
analyses, age, tumor size and high-risk scores of gene signature
were independently associated with shorter overall survival
(HR=2.133, 95% CI=1.110–4.099, P=0.02) (Table II).
 | Table I.Clinical characteristics of GC
patients dichotomised by IHC-detected gene signature in
experimental validation cohort. |
Table I.
Clinical characteristics of GC
patients dichotomised by IHC-detected gene signature in
experimental validation cohort.
|
| 3-Hh-gene
signature |
|---|
|
|
|
|---|
| Characteristics | High risk (n=39) | Low risk (n=76) | P-value |
|---|
| Age (years) | 66.82±11.016 | 63.05±10.611 | 0.805 |
| Sex |
|
|
|
| Male | 23 | 52 | 0.314 |
|
Female | 16 | 24 |
|
| Differentiation
grade |
|
|
|
| Well or
moderately | 20 | 34 | 0.506 |
|
Poorly | 19 | 42 |
|
| Tumor size |
|
|
|
| T1-2 | 14 | 18 | 0.483 |
| T3-4 | 25 | 58 |
|
| Lymph node |
|
|
|
|
Negative | 17 | 28 | 0.166 |
|
Positive | 22 | 48 |
|
| TNM Stage |
|
|
|
| I | 11 | 11 |
|
| II | 13 | 26 | 0.178 |
| III | 15 | 39 |
|
 | Table II.Cox regression analysis of IHC-based
gene signature score and clinicopathological covariates with
overall survival. |
Table II.
Cox regression analysis of IHC-based
gene signature score and clinicopathological covariates with
overall survival.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Characteristics | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age | 1.044
(1.011–1.078) | 0.009 | 1.041
(1.110–1.076) | 0.016 |
| Sex | 0.883
(0.458–1.701) | 0.710 | 0.742
(0.382–1.443) | 0.379 |
| Grade | 1.550
(0.836–2.874) | 0.164 | 1.659
(0.883–3.120) | 0.116 |
| Tumor size | 2.471
(1.036–5.890) | 0.041 | 3.182
(1.268–7.986) | 0.014 |
| Lymph node | 1.081
(0.567–2.061) | 0.812 | 0.755
(0.378–1.509) | 0.427 |
| 3-Hh-gene
signature | 1.957
(1.045–3.661) | 0.036 | 2.133
(1.110–4.099) | 0.023 |
Discussion
Aberrant activation of the Hh signaling pathway has
been associated with a variety of human cancers, including GC
(17–19). As potential targets in the treatment
of GC, the prognostic impact of the Hh signaling pathway genes in
GC have been widely explored previously. However, a potent,
reproducible Hh-associated prognostic factor for clinical use is
yet to be confirmed. This is may be due to the fact that most the
previous prognostic studies of Hh-associated factors in GC are
limited to small cohorts and focus on signle not combined Hh
biomarker in combination.
To overcome these limitations, in this study, we
used two large gene expression cohorts (both comprising more than
300 samples with survival data) to identify and validate the most
potent Hh-associated pathway factors for the prognoses of GC. Among
current canonical Hh-associated genes, we identified IHH, PTCH1 and
SMO as the most powerful prognostic factors for GC. Furthermore, we
developed a signature with these three Hh-associated genes, which
demonstrated a stable and reliable potential to identify high-risk
GC patients with poor clinical outcome. These reproducible effects
were confirmed in a variety independent cohorts and different gene
expression platforms (RNA-seq, microarray, and IHC). Furthermore,
we demonstrated that this signature was independent of known
clinical prognostic factors, and in particular, its robustness was
effective in early and advanced stages; HER2-negative or surgery
alone patients exhibited the most heterogeneous clinical outcomes.
HER2-positive GCs represent a subgroup with poor prognosis that can
be treated using anti-HER2 drugs, such as trastuzumab (20). Our signature further identified a
poor-prognostic subgroup from the HER2-negative GCs, but did not
benefit from treatment with anti-HER2 drugs; therefore, alternative
treatments may be considered for these patients. Therefore, our
signature may provide more details for the current risk
classification system.
Compared with previous established prognostic gene
signatures, our signature only has 3 genes, which is simple and
practical for further clinical use. Secondly, although our
3-Hh-gene signature is derived from RNA-seq and microarray-based
platforms, in this study, we also identified a high-risk GC
subgroup with poor prognosis by using simple IHC technology.
Therefore, this signature may have notable biological importance.
PTCH1 has been reported to be closely associated with the
chemoresistance of GC cell lines (21). Recently, Ma et al (22) linked high SMO expression levels with
paclitaxel-resistant GC clinical samples; IPI-926, an inhibitor of
SMO, may sensitize paclitaxel-resistant tumors. SMO has been
confirmed to be a target with the highest therapeutic potential; a
panel of SMO inhibitors have been investigated for the treatment of
cancer (23–24). Therefore, it may be reasonable to
speculate that high-risk patients defined by our 3-Hh gene
signature may benefit most from SMO inhibitors.
The limitation of the present study is that this
signature was investigated in a retrospective manner, therefore,
future study may require the integration of this model with
prospective randomized trials to further validate its clinical
relevance.
In summary, we identified a stable Hh signaling
pathway-based 3 gene set that may provide additional prognostic
information to improve clinical classification, and transfer to
IHC-based assay. We propose that it may be developed into a robust,
practical and inexpensive molecular diagnostic tool for clinical
use in the near future.
Acknowledgements
Not applicable.
Funding
The present study was supported by Key Research
Program of Science and Technology Planning Project of Zhejiang
Province (grant no. 2017C3026) and Zhejiang Provincial Natural
Science Foundation of China (grant no. LQ14H160014).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
WL and JW designed the project and were responsible
for the data analysis. WL and XW performed the IHC experiments. JW
and WL wrote the manuscript. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The study was approved by the institutional review
boards of Zhejiang Hospital of Traditional Chinese Medicine and
Zhejiang Police College. Written informed consent was obtained from
all patients.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
References
|
1
|
Miller KD, Siegel RL, Lin CC, Mariotto AB,
Kramer JL, Rowland JH, Stein KD, Alteri R and Jemal A: Cancer
treatment and survivorship statistics, 2016. CA Cancer J Clin.
66:271–289. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Figueiredo C, Camargo MC, Leite M,
Fuentes-Pananá EM, Rabkin CS and Machado JC: Pathogenesis of
gastric cancer: Genetics and molecular classification. Curr Top
Microbiol Immunol. 400:277–304. 2017.PubMed/NCBI
|
|
3
|
Ghosh D and Poisson LM: ‘Omics’ data and
levels of evidence for biomarker discovery. Genomics. 93:13–16.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cho JY, Lim JY, Cheong JH, Park YY, Yoon
SL, Kim SM, Kim SB, Kim H, Hong SW, Park YN, et al: Gene expression
signature-based prognostic risk score in gastric cancer. Clin
Cancer Res. 17:1850–1857. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang P, Wang Y, Hang B, Zou X and Mao JH:
A novel gene expression-based prognostic scoring system to predict
survival in gastric cancer. Oncotarget. 7:55343–55351.
2016.PubMed/NCBI
|
|
6
|
Hou JY, Wang YG, Ma SJ, Yang BY and Li QP:
Identification of a prognostic 5-Gene expression signature for
gastric cancer. J Cancer Res Clin Oncol. 143:619–629. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee J, Cristescu R, Kim KM, Kim K, Kim ST,
Park SH and Kang WK: Development of mesenchymal subtype gene
signature for clinical application in gastric cancer. Oncotarget.
8:66305–66315. 2017.PubMed/NCBI
|
|
8
|
Wang X, Liu Y, Niu Z, Fu R, Jia Y, Zhang
L, Shao D, Du H, Hu Y, Xing X, et al: Prognostic value of a 25-gene
assay in patients with gastric cancer after curative resection. Sci
Rep. 7:75152017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu F, Zhang Y, Sun B, McMahon AP and Wang
Y: Hedgehog signaling: From basic biology to cancer therapy. Cell
Chem Biol. 24:252–280. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Abdel-Rahman O: Hedgehog pathway
aberrations and gastric cancer; evaluation of prognostic impact and
exploration of therapeutic potentials. Tumour Biol. 36:1367–1374.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee SJ, Do IG, Lee J, Kim KM, Jang J, Sohn
I and Kang WK: Gastric cancer (GC) patients with hedgehog pathway
activation: PTCH1 and GLI2 as independent prognostic factors.
Target Oncol. 8:271–280. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang ZS, Shen Y, Li X, Zhou CZ, Wen YG,
Jin YB and Li JK: Significance and prognostic value of Gli-1 and
Snail/E-cadherin expression in progressive gastric cancer. Tumour
Biol. 35:1357–1363. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Szász AM, Lánczky A, Nagy Á, Förster S,
Hark K, Green JE, Boussioutas A, Busuttil R, Szabó A and Győrffy B:
Cross-validation of survival associated biomarkers in gastric
cancer using transcriptomic data of 1,065 patients. Oncotarget.
7:49322–49333. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Budczies J, Klauschen F, Sinn BV, Győrffy
B, Schmitt WD, Darb-Esfahani S and Denkert C: Cutoff finder: A
comprehensive and straightforward Web application enabling rapid
biomarker cutoff optimization. PLoS One. 7:e518622012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ishibashi H, Suzuki T, Suzuki S, Moriya T,
Kaneko C, Takizawa T, Sunamori M, Handa M, Kondo T and Sasano H:
Sex steroid hormone receptors in human thymoma. J Clin Endocrinol
Metab. 88:2309–2317. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Peukert S and Miller-Moslin K:
Small-molecule inhibitors of the hedgehog signaling pathway as
cancer therapeutics. ChemMedChem. 5:500–512. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wan J, Zhou J, Zhao H, Wang M, Wei Z, Gao
H, Wang Y and Cui H: Sonic hedgehog pathway contributes to gastric
cancer cell growth and proliferation. Biores Open Access. 3:53–59.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lee DH, Lee SY and Oh SC: Hedgehog
signaling pathway as a potential target in the treatment of
advanced gastric cancer. Tumour Biol. 39:10104283176922662017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bang YJ, Van Cutsem E, Feyereislova A,
Chung HC, Shen L, Sawaki A, Lordick F, Ohtsu A, Omuro Y, Satoh T,
et al: Trastuzumab in combination with chemotherapy versus
chemotherapy alone for treatment of HER2-positive advanced gastric
or gastro-oesophageal junction cancer (ToGA): A phase 3,
open-label, randomised controlled trial. Lancet. 376:687–697. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yu B, Gu D, Zhang X, Li J, Liu B and Xie
J: GLI1-mediated regulation of side population is responsible for
drug resistance in gastric cancer. Oncotarget. 8:27412–27427.
2017.PubMed/NCBI
|
|
22
|
Ma H, Tian Y and Yu X: Targeting
smoothened sensitizes gastric cancer to chemotherapy in
experimental models. Med Sci Monit. 23:1493–1500. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Khoo AB, Ali FR and Lear JT: Defining
locally advanced basal cell carcinoma and integrating smoothened
inhibitors into clinical practice. Curr Opin Oncol. 28:180–184.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Doan HQ, Silapunt S and Migden MR:
Sonidegib, a novel smoothened inhibitor for the treatment of
advanced basal cell carcinoma. Onco Targets Ther. 9:5671–5678.
2016. View Article : Google Scholar : PubMed/NCBI
|