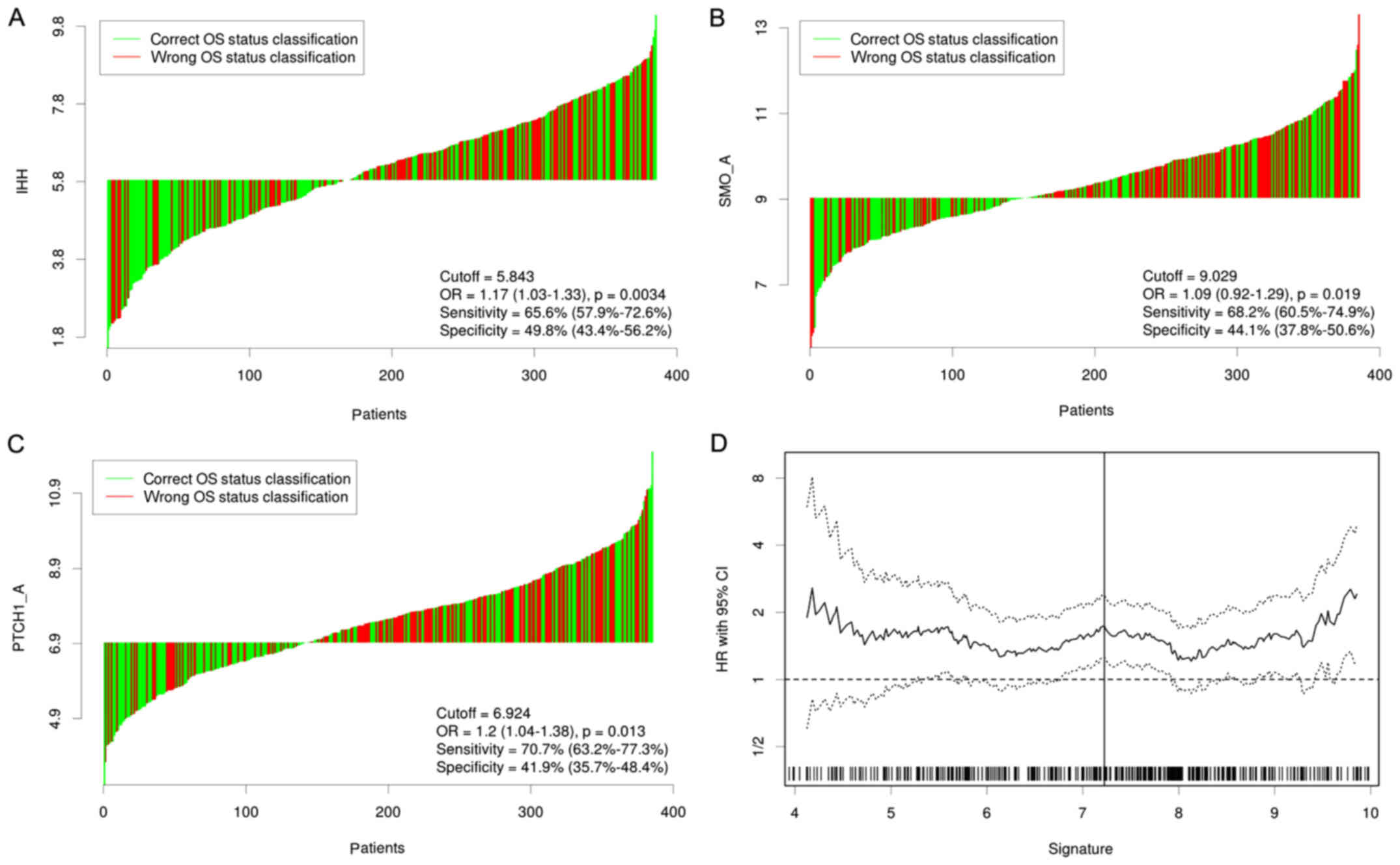
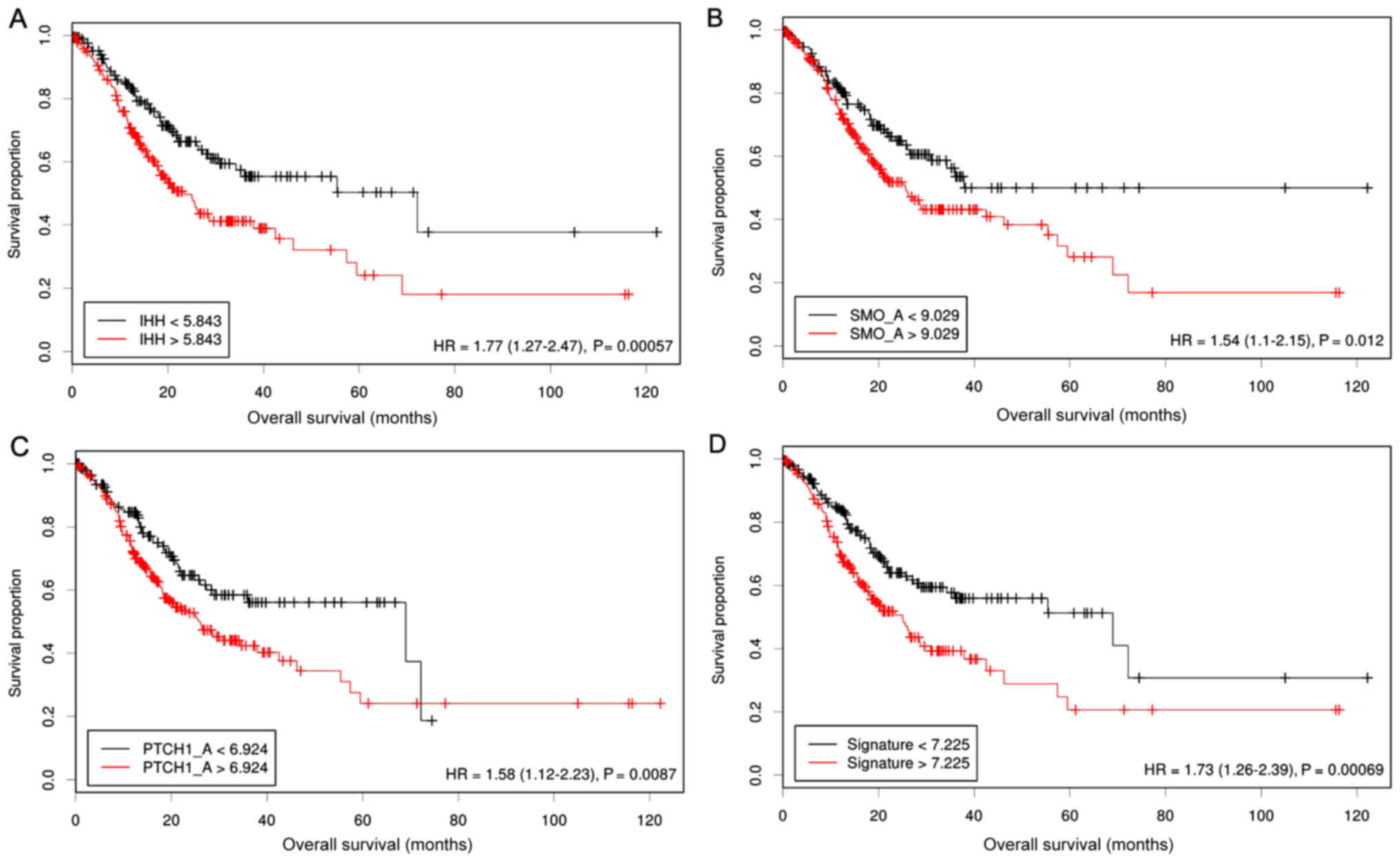
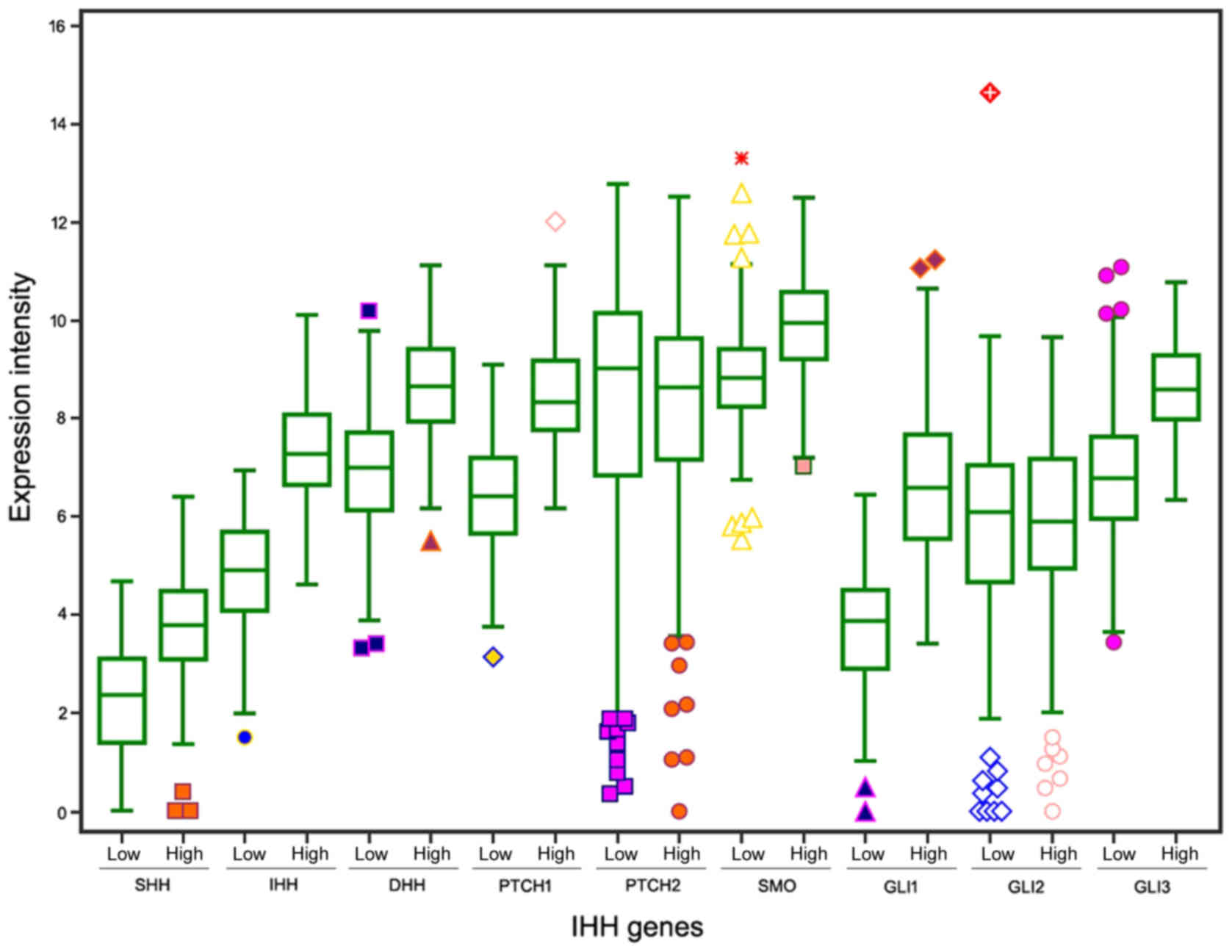
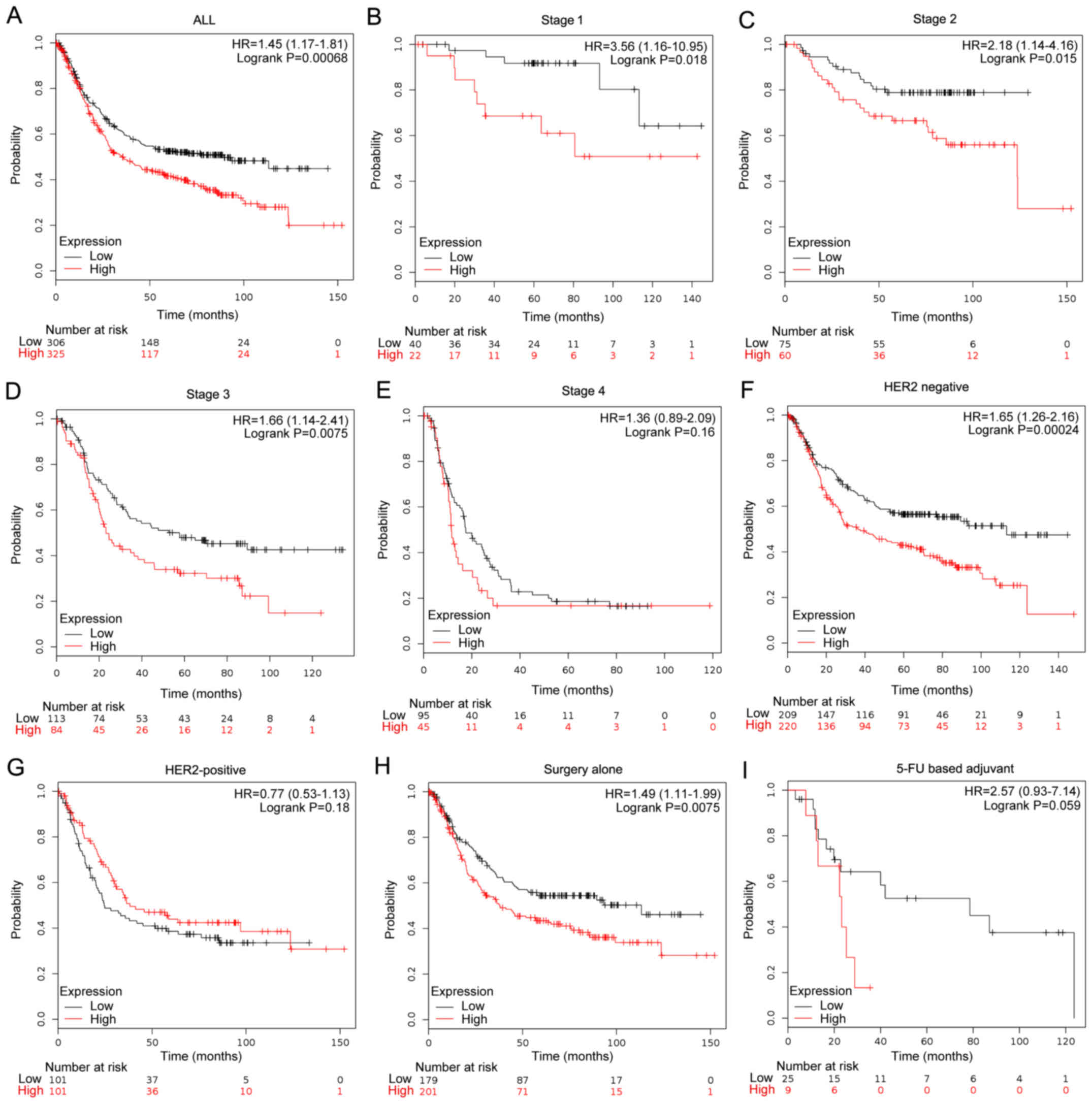
|
1
|
Miller KD, Siegel RL, Lin CC, Mariotto AB,
Kramer JL, Rowland JH, Stein KD, Alteri R and Jemal A: Cancer
treatment and survivorship statistics, 2016. CA Cancer J Clin.
66:271–289. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Figueiredo C, Camargo MC, Leite M,
Fuentes-Pananá EM, Rabkin CS and Machado JC: Pathogenesis of
gastric cancer: Genetics and molecular classification. Curr Top
Microbiol Immunol. 400:277–304. 2017.PubMed/NCBI
|
|
3
|
Ghosh D and Poisson LM: ‘Omics’ data and
levels of evidence for biomarker discovery. Genomics. 93:13–16.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cho JY, Lim JY, Cheong JH, Park YY, Yoon
SL, Kim SM, Kim SB, Kim H, Hong SW, Park YN, et al: Gene expression
signature-based prognostic risk score in gastric cancer. Clin
Cancer Res. 17:1850–1857. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang P, Wang Y, Hang B, Zou X and Mao JH:
A novel gene expression-based prognostic scoring system to predict
survival in gastric cancer. Oncotarget. 7:55343–55351.
2016.PubMed/NCBI
|
|
6
|
Hou JY, Wang YG, Ma SJ, Yang BY and Li QP:
Identification of a prognostic 5-Gene expression signature for
gastric cancer. J Cancer Res Clin Oncol. 143:619–629. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee J, Cristescu R, Kim KM, Kim K, Kim ST,
Park SH and Kang WK: Development of mesenchymal subtype gene
signature for clinical application in gastric cancer. Oncotarget.
8:66305–66315. 2017.PubMed/NCBI
|
|
8
|
Wang X, Liu Y, Niu Z, Fu R, Jia Y, Zhang
L, Shao D, Du H, Hu Y, Xing X, et al: Prognostic value of a 25-gene
assay in patients with gastric cancer after curative resection. Sci
Rep. 7:75152017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu F, Zhang Y, Sun B, McMahon AP and Wang
Y: Hedgehog signaling: From basic biology to cancer therapy. Cell
Chem Biol. 24:252–280. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Abdel-Rahman O: Hedgehog pathway
aberrations and gastric cancer; evaluation of prognostic impact and
exploration of therapeutic potentials. Tumour Biol. 36:1367–1374.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee SJ, Do IG, Lee J, Kim KM, Jang J, Sohn
I and Kang WK: Gastric cancer (GC) patients with hedgehog pathway
activation: PTCH1 and GLI2 as independent prognostic factors.
Target Oncol. 8:271–280. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang ZS, Shen Y, Li X, Zhou CZ, Wen YG,
Jin YB and Li JK: Significance and prognostic value of Gli-1 and
Snail/E-cadherin expression in progressive gastric cancer. Tumour
Biol. 35:1357–1363. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Szász AM, Lánczky A, Nagy Á, Förster S,
Hark K, Green JE, Boussioutas A, Busuttil R, Szabó A and Győrffy B:
Cross-validation of survival associated biomarkers in gastric
cancer using transcriptomic data of 1,065 patients. Oncotarget.
7:49322–49333. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Budczies J, Klauschen F, Sinn BV, Győrffy
B, Schmitt WD, Darb-Esfahani S and Denkert C: Cutoff finder: A
comprehensive and straightforward Web application enabling rapid
biomarker cutoff optimization. PLoS One. 7:e518622012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ishibashi H, Suzuki T, Suzuki S, Moriya T,
Kaneko C, Takizawa T, Sunamori M, Handa M, Kondo T and Sasano H:
Sex steroid hormone receptors in human thymoma. J Clin Endocrinol
Metab. 88:2309–2317. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Peukert S and Miller-Moslin K:
Small-molecule inhibitors of the hedgehog signaling pathway as
cancer therapeutics. ChemMedChem. 5:500–512. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wan J, Zhou J, Zhao H, Wang M, Wei Z, Gao
H, Wang Y and Cui H: Sonic hedgehog pathway contributes to gastric
cancer cell growth and proliferation. Biores Open Access. 3:53–59.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lee DH, Lee SY and Oh SC: Hedgehog
signaling pathway as a potential target in the treatment of
advanced gastric cancer. Tumour Biol. 39:10104283176922662017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bang YJ, Van Cutsem E, Feyereislova A,
Chung HC, Shen L, Sawaki A, Lordick F, Ohtsu A, Omuro Y, Satoh T,
et al: Trastuzumab in combination with chemotherapy versus
chemotherapy alone for treatment of HER2-positive advanced gastric
or gastro-oesophageal junction cancer (ToGA): A phase 3,
open-label, randomised controlled trial. Lancet. 376:687–697. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yu B, Gu D, Zhang X, Li J, Liu B and Xie
J: GLI1-mediated regulation of side population is responsible for
drug resistance in gastric cancer. Oncotarget. 8:27412–27427.
2017.PubMed/NCBI
|
|
22
|
Ma H, Tian Y and Yu X: Targeting
smoothened sensitizes gastric cancer to chemotherapy in
experimental models. Med Sci Monit. 23:1493–1500. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Khoo AB, Ali FR and Lear JT: Defining
locally advanced basal cell carcinoma and integrating smoothened
inhibitors into clinical practice. Curr Opin Oncol. 28:180–184.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Doan HQ, Silapunt S and Migden MR:
Sonidegib, a novel smoothened inhibitor for the treatment of
advanced basal cell carcinoma. Onco Targets Ther. 9:5671–5678.
2016. View Article : Google Scholar : PubMed/NCBI
|