Introduction
microRNAs (miRs) are a class of non-coding small
RNAs that serve important roles in carcinogenesis. They regulate
gene expression by binding to the 3′untranslated regions (3′UTR) of
target genes (1,2), which leads to gene transcription,
regulation or mRNA degradation (3,4). A number
of the regulated genes are oncogenes or tumor suppressors (5–7). miRs
regulation may inhibit cancer cell proliferation and induce cell
apoptosis (8,9). A previous study indicated that miR
modulation therapy may affect multiple target genes, which may
potentially improve clinical treatments (10). In previous decades, studies have
identified that in human hepatocellular carcinoma (HCC), there are
a number of important aberrantly expressed miRs, and that these
abnormal miRs were associated with HCC development (11–16).
However, the majority of the biological roles of miRs in HCC remain
incompletely understood.
HCC is a type of cancer that originates in the
hepatocytes, and there are >500,000 people diagnosed with liver
cancer each year globally (17). It
occurs most commonly in countries where viral Hepatitis B and C
infections are common (16,18) and with no perfected targeted
therapies, the <5-year survival rate of HCC is 5% (19). Previous studies have indicated that
miRs regulate essential signal pathways in liver cancer: miR-21 is
highly overexpressed in liver cancer and the downregulation of
miR-21 inhibits HCC cell proliferation, migration and invasion by
targeting the PTEN tumor suppressor (5,20,21). Previous studies have also indicated
that the expression of miR-34a is downregulated in human HCC
(22,23), and that miR-34a regulates the
biological function of HCC cells by targeting the tumor suppressor
p53 (24). These studies demonstrated
that miRs may serve important roles in human HCC tumorigenesis by
regulating the expression of genes. miR-375 was identified to be
abnormally expressed in numerous types of cancer (25–27);
however, the biological role of miR-375 in human HCC remains
incompletely understood. The present study aimed to investigate
whether the miR-375 is involved in the human HCC tumorigenesis, and
to identify the mechanism of action.
Materials and methods
HCC cell lines and patient
samples
Human liver cancer cell lines (Huh7, SK-HEP-1,
MHCC97-H, MHCC97-L and Hep3B2.1–7) were purchased from the Cell
Bank of Type Culture Collection of the Chinese Academy of Sciences
(Shanghai, China). Primary human hepatocyte (PHH) cells (cat. no.
M00995) were purchased from the Research Institute for Liver
Diseases Shanghai, Co., Ltd. (Shanghai, China). Liver cancer cell
lines were cultured in Dulbecco's modified Eagle medium (DMEM;
Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.), 100 IU/ml penicillin (Gibco; Thermo Fisher
Scientific, Inc.) and 100 mg/ml streptomycin (Gibco; Thermo Fisher
Scientific, Inc.). The PHH incubation media (cat. no. PY-HMD-01)
was also purchased from the Research Institute for Liver Diseases
Shanghai, Co., Ltd. Patients with HCC were collected from Daqing
Longnan Hospital (Daqing, China) and 43 pairs (23 male; 20 female;
age range, 31–63; median age, 47) of HCC and noncancerous normal
tissue samples (>30 mm away from the tumor) were obtained from
these patients via surgical resection. Samples were stored in
RNAlater™ (Ambion; Thermo Fisher Scientific, Inc.) at
−80°C until use. The collection of patient tissues was performed
following the Ethical and Institutional Guidelines (Daqing LongNan
Hospital, Daqing, China) and subsequent to provision of written
informed consent from all patients. The present study was approved
by the Medical Ethics Committee of Daqing Longnan Hospital.
Transfection assay
Huh7 cells were cultured in RPMI-1640 medium (Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% FBS at 37°C
overnight, then the miR-375 mimics were transfected into the cells
(10 nM final concentration) using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) as previously
described (28); the control mimic
was used as the control. The miR-375 and control mimics were
purchased from Thermo Fisher Scientific, Inc. Small interfering
(si)RNA (cat. no. sc-400138-KO-2) was purchased from Santa Cruz
Biotechnology, Inc., (Dallas, TX, USA). The protocol used for
receptor tyrosine-protein kinase erbB-2 (ErbB2) knockdown was
performed as previously described (50 nM final concentration)
(29).
miR-375 quantification using reverse
transcription quantitative polymerase chain reaction (RT-qPCR)
Total RNA for miR-375 quantification was extracted
from human HCC tissue samples and cells using TRIzol®
reagent (Thermo Fisher Scientific, Inc.) and reverse transcribed
(42°C, 60 min; 85°C, 5 min) into cDNA with the TaqMan miRNA Reverse
Transcription kit (Applied Biosystems; Thermo Fisher Scientific,
Inc.) with a miRNA-specific looped RT primer (Applied Biosystems;
Thermo Fisher Scientific, Inc.). Then, the expression of miR-375
was evaluated using TaqMan Universal PCR Master Mix with
miRNA-specific TaqMan minor groove binder probes (Thermo Fisher
Scientific, Inc.). The qPCR primers used were commercially
available (cat. no. Hs04231554_s1; Applied Biosystems; Thermo
Fisher Scientific, Inc.). qPCR was performed using the followed
program: 95°C for 10 min; 95°C for 10 sec, 57°C for 20 sec and 72°C
for 10 sec for 40 cycles, using the LightCycler480 Real-Time PCR
System (Roche Diagnostic, Basel, Switzerland). RNA U6 (cat. no.
Hs00984809_m1; Applied Biosystems; Thermo Fisher Scientific, Inc.)
was used as an internal control. Expression of miR-375 relative to
U6 was determined using the 2−ΔΔCq method (30).
Cell viability assay
Human HCC Huh7 cells were cultured in 96-well plate
(3,000 cell/well); the Cell Counting Kit-8 (CCK8) assay was
performed to test the cell viability every 24 h (0, 24, 48, 72 and
96 h). Firstly, CCK8 reagent (Beyotime Institute of Biotechnology,
Haimen, China) was added in to the well at 1:10 dilution, and then
incubated with the cells for an additional 2 h at 37°C. Finally,
the absorbance (OD450) of the 96-well plate was measured. The
absorbance was expressed as the cell viability.
Colony formation assay
A total of 24 h following transfection, Huh7 cells
were seeded into 6-well plates at ~550 cells/well. The culture
medium was changed every other day, and the 6-well plates were
cultured for 2 weeks. Colonies were fixed with 100% methanol at
room temperature for 15 min and stained using crystal violet (0.5%)
at 4°C for 30 min. The colony formation ability was evaluated by
counting the number of colonies formed with a light microscope
(magnification ×4).
Cell apoptosis assay
A total of 24 h following transfection, Huh7 cells
were seeded onto 6-well plates (1,200,000/well) and cultured at
37°C for an additional 48 h. Then, the cells were harvested and
stained with Annexin V (1 µg/ml) and propidium iodide (2 µg/ml) at
4°C for 15 min (Beyotime Institute of Biotechnology). Cell
apoptosis was evaluated using flow cytometry and CellQuest Pro
software (version 5.1; BD FACSCalibur; BD Biosciences, CA,
USA).
MiRNA target predictions
To additionally investigate the potential target of
miR-375, potential genes identified by computer-aided algorithms
were obtained from targetscan (http://www.targetscan.org) and mirbase targets
(http://microrna.sanger.ac.uk/cgi-bin/targets/v5/search.pl).
Dual-luciferase assay
The wild-type (WT) or mutant (Mut) ErbB2 3′-UTRs
reporter vector (Qcbio S&T Co., Ltd, Shanghai, China) were
co-transfected with the miR-375 mimic or control mimic (10 nM final
concentration) into Huh7 cells using Lipofectamine® 2000
in 96-well plates (10,000 cells/well). The duration between
transfection and activity measurement was 24 h. The transfected
cells were cultured at 37°C for an additional 24 h and harvested;
Cells were then lysed as the followed protocol: Removal of the
growth medium; washing the cells with PBS 3 times; adding 20 µl PLB
buffer (Promega Corporation, Madison, WI, USA) into each well;
shaking the solutions via gentle rocking for 15 min; performing
reporter assays directly in the wells of the culture plate (Promega
Corporation). The firefly luciferase activity was examined by the
dual-luciferase reporter assay (Promega Corporation). Relative
luciferase activity was normalized with the Renilla luciferase
activity. The kit used to measure activity was the
Dual-Luciferase® Reporter Assay System (Promega
Corporation).
RT-qPCR for ErbB2
Total RNA was extracted from Huh7 cells using
TRIzol® reagent (Life Technologies; Thermo Fisher
Scientific, Inc.) and reverse transcribed into cDNA with the
Reverse Transcriptase MMLV (Takara Biomedical Technology Co., Ltd.
Dalian, China). Program for reverse transcription: 10 min at 30°C,
60 min at 42°C and 5 min at 95°C. qPCR (SYBR® Green;
FastStart Universal SYBR Green Master; Roche Diagnostics) was
performed using the LightCycler480 Real-Time PCR System (Roche
Diagnostics) and the following thermocycling parameters: 95°C for
10 min, followed by 40 cycles of 15 sec at 95°C, 30 sec at 60°C and
20 sec at 72°C. The primers used were as follows: ErbB2: Forward,
5′-CCAGCCTTCGACAACCTCTATT-3′, and reverse,
5′-TGCCGTAGGTGTCCCTTTG-3′. β-actin: Forward,
5′-ATCTGGCACCACACCTTCTACAAT-3′, and reverse
5′-CCGTCACCGGAGTCCATCA-3′. Expression of ErbB2 relative to β-actin
was determined using the 2−ΔΔCq method (30).
Western blotting assay
A total of 24 h following transfection, Huh7 were
seeded into 6-well plates and cultured at 37°C for an additional 48
h. Cells were harvested and protein was extracted by using the
commercial kit (Cell Lysis Buffer; Applygen Technologies Inc.,
Beijing, China). Subsequently, the proteins (50 µg) were separated
using a 10% gel and SDS-PAGE and the separated proteins were
transferred to a polyvinylidene fluoride (PVDF) membrane. The
transferred PVDF membrane was blocked with blocking buffer (5%
dried milk) at room temperature for 1 h, and then the membrane was
incubated with primary antibodies against ErbB2 (dilution, 1:1,000;
cat. no. 2242; Cell Signaling Technology, Inc., Danvers, MA, USA)
and β-actin (dilution, 1:5,000; cat. no. 4967; Cell Signaling
Technology, Inc.) at room temperature for 2 h. The PVDF membrane
was washed with TBST 3 times and incubated with the secondary
antibody (goat anti-rabbit; dilution, 1:1,000; cat. no. A0208;
Beyotime Institute of Biotechnology) at room temperature for 1 h.
The membrane was then washed again with TBST 3 times, and the
proteins were examined using an electrochemiluminescence kit (cat.
no. P0018; Beyotime Institute of Biotechnology) and exposed to
x-ray film.
Statistical analysis
A one-way analysis of variance and
Student-Newman-Keuls test (post hoc test) were performed to analyze
the statistical difference by using SPSS v13.0 software (SPSS,
Inc., Chicago, IL, USA). Data were expressed as mean ± standard
deviation. Each experiment was performed in triplicate. P<0.05
was considered to indicate a statistically significant
difference.
Results
miR-375 is downregulated in human
HCC
Previous studies have demonstrated that ErbB2 gene
upregulation is an important contributing factor to hepatocellular
growth (31), and that ErbB2
upregulation was associated with miR-375 regulation (32). The present study aimed to examine the
miR-375 level in human liver cancer tissues and cell lines. A total
of 43 pairs of HCC and matched adjacent non-tumor tissues were
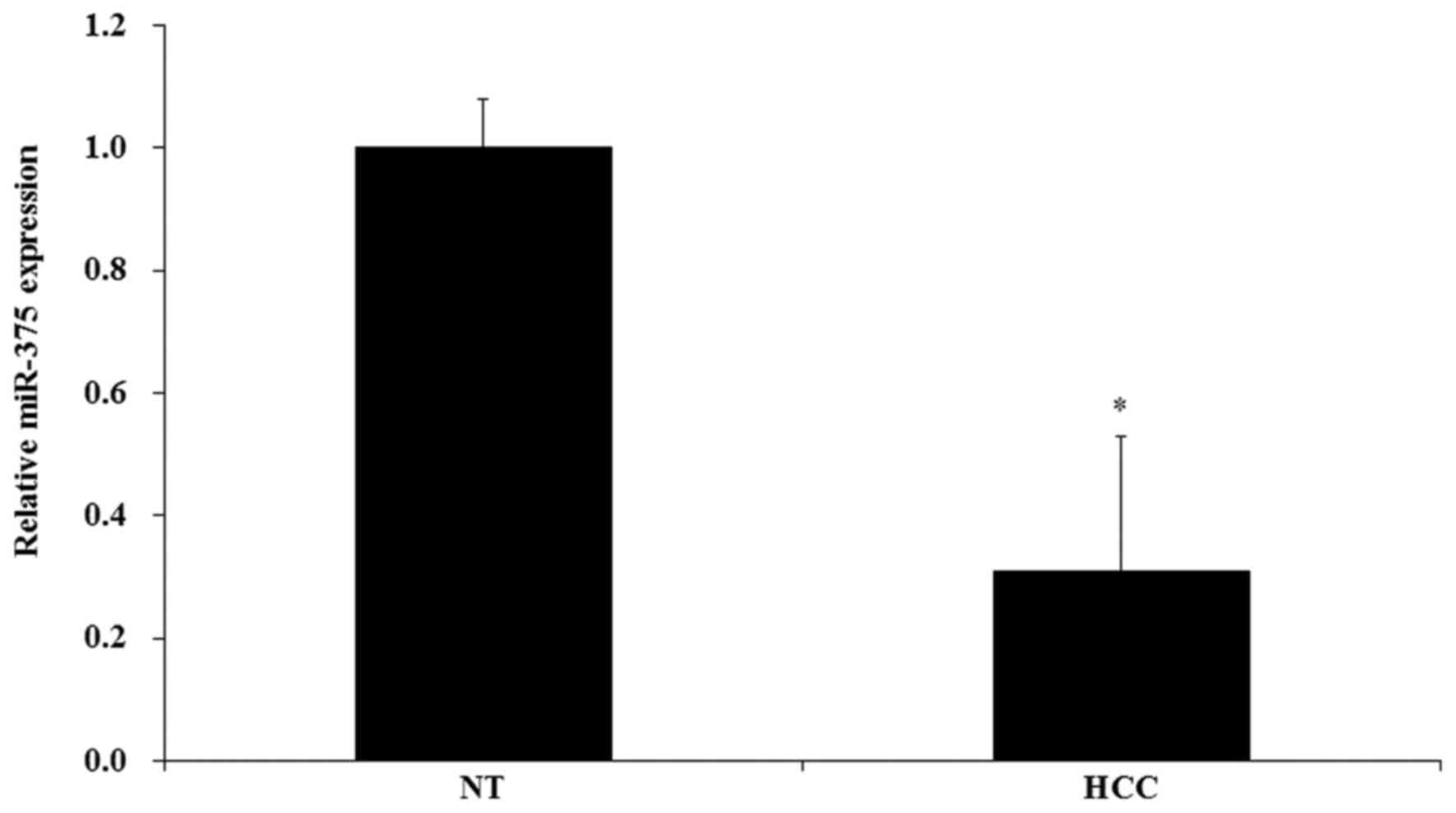
analyzed, and the RT-qPCR results indicated that the level of
miR-375 was significantly decreased in HCC tissues compared with
the non-tumor tissues (P<0.05; Fig.
1).
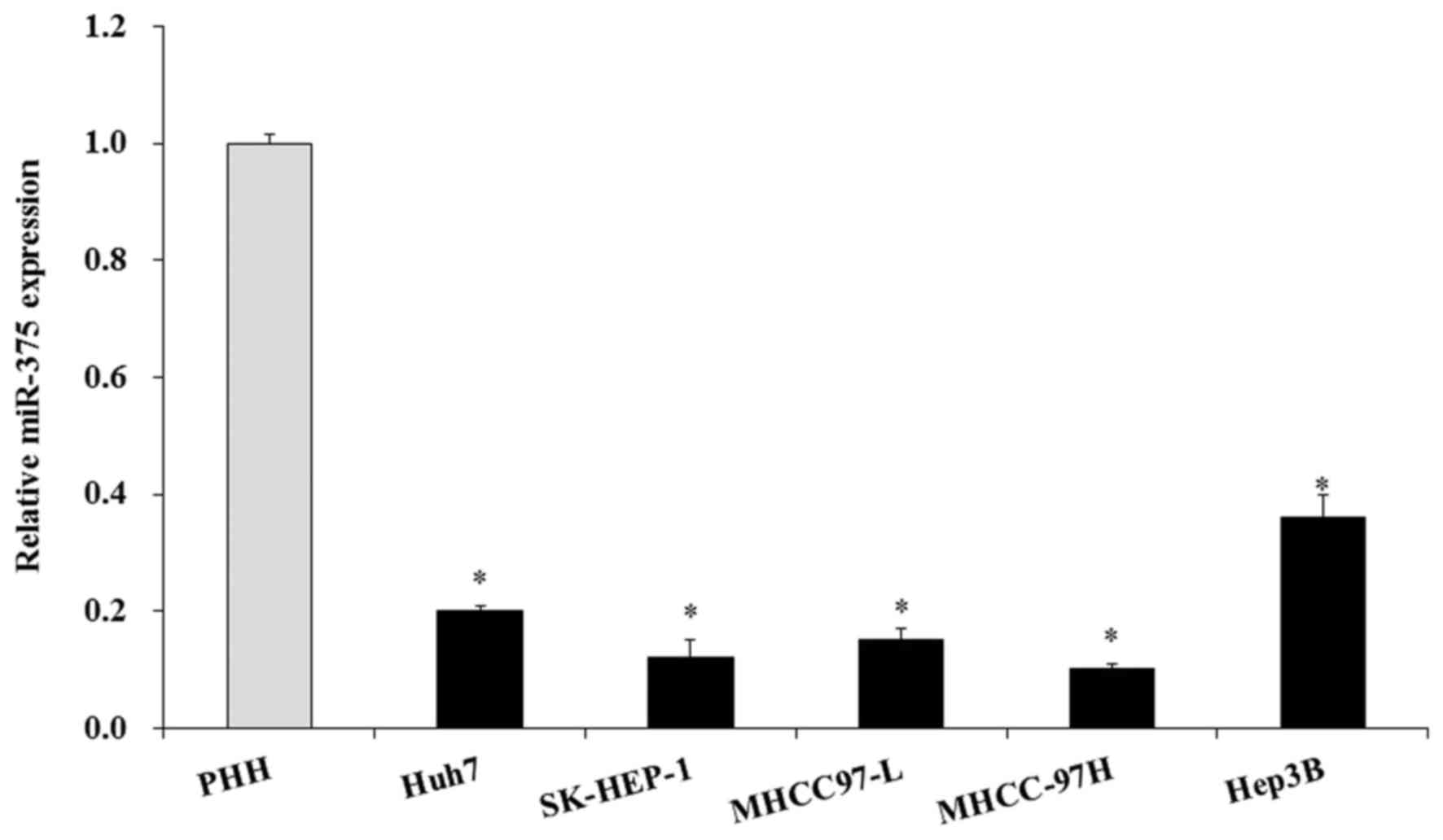
Subsequently, the miR-375 level was examined in
liver cancer cell lines; 5 liver cell lines were used, and PHH was
used as the normal control. The RT-qPCR results indicated that the
miR-375 level was also significantly decreased in the 5 liver
cancer cell lines compared with the normal PHH cell line
(P<0.05; Fig. 2). Therefore,
miR-375 was downregulated in HCC tissues and HCC cell lines.
Manipulation of miR-375 levels in HCC
cells
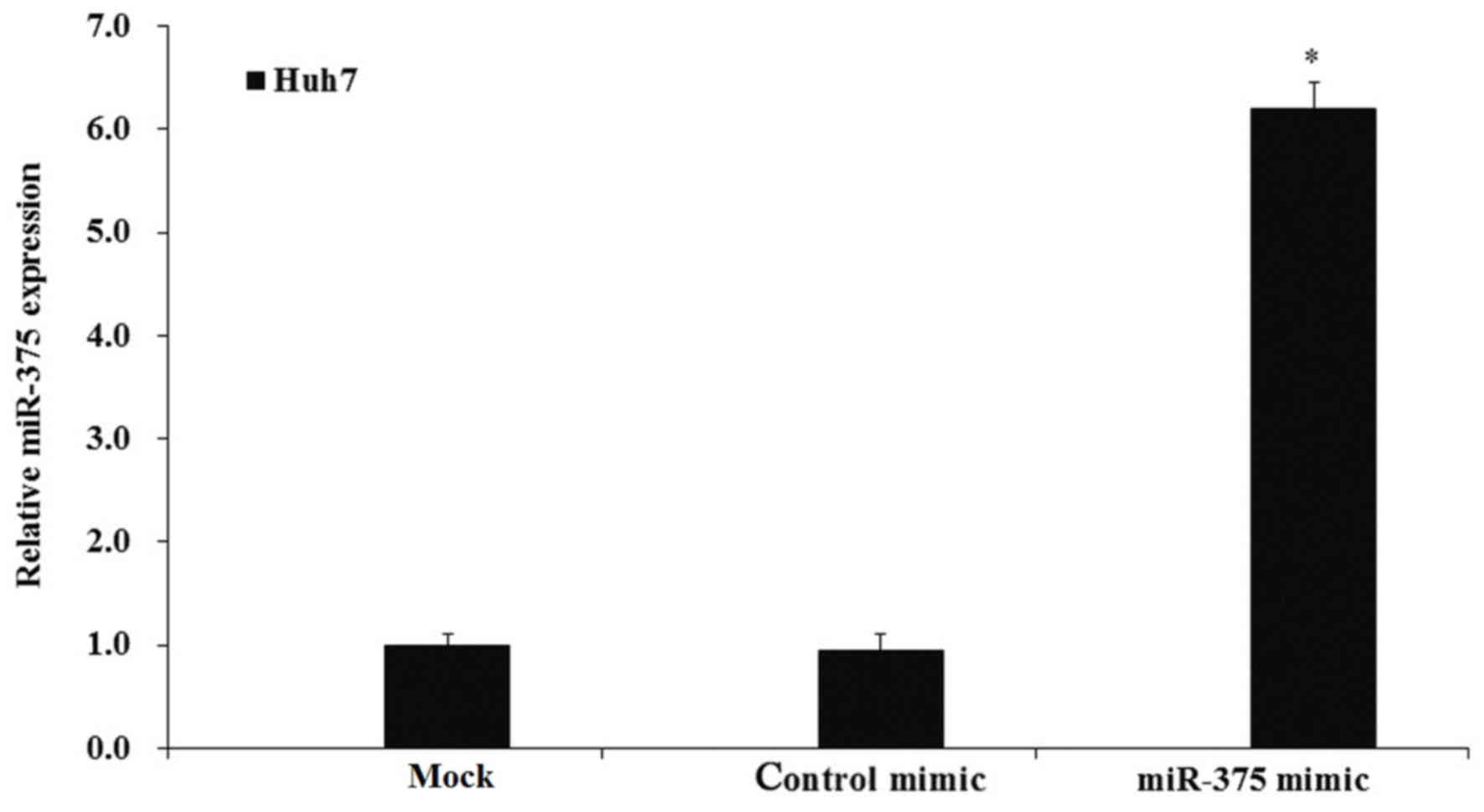
To additionally investigate the biological activity
of miR-375 in HCC cells, the expression of miR-375 was manipulated
by transfection of miR-375 mimics. Huh7 cells were cultured, then
transfected with miR-375 or control mimics and cultured for an
additional 48 h. Then, cells were harvested and an RT-qPCR assay
was performed to examine the expression of miR-375. The RT-qPCR
results demonstrated that transfection of the miR-375 mimic
significantly increased the expression of miR-375 compared with the
control mimic-transfected and mock cells (P<0.05; Fig. 3).
Upregulation of miR-375 inhibits HCC
cell proliferation
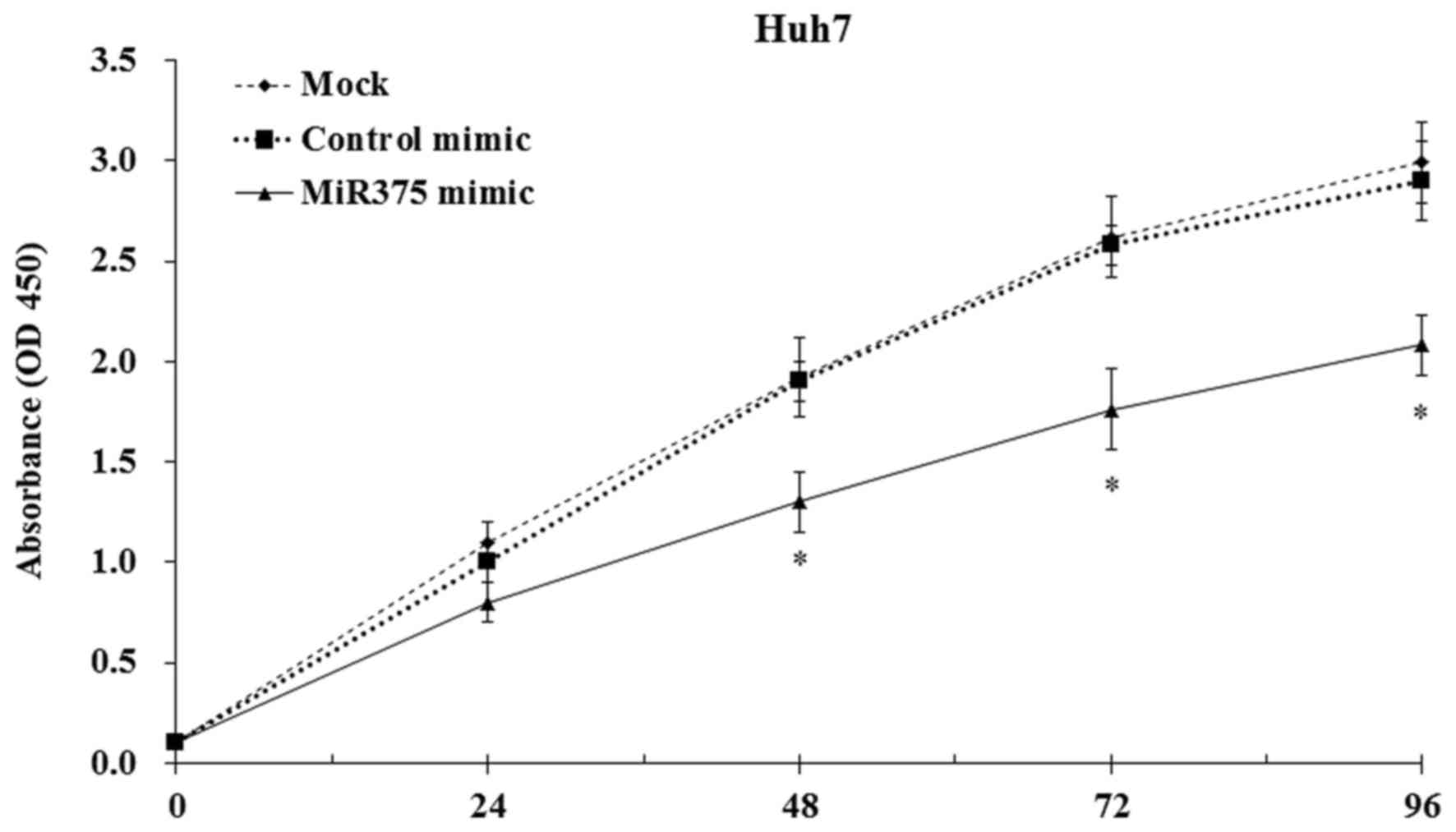
To study the biological activity of miR-375 in HCC
cell proliferation, mock and transfected Huh7 cells were cultured,
and the cell proliferation was evaluated by CCK8 assay. The results
indicated that the induction of miR-375 expression significantly
decreased the HCC cell proliferation compared with the mock and
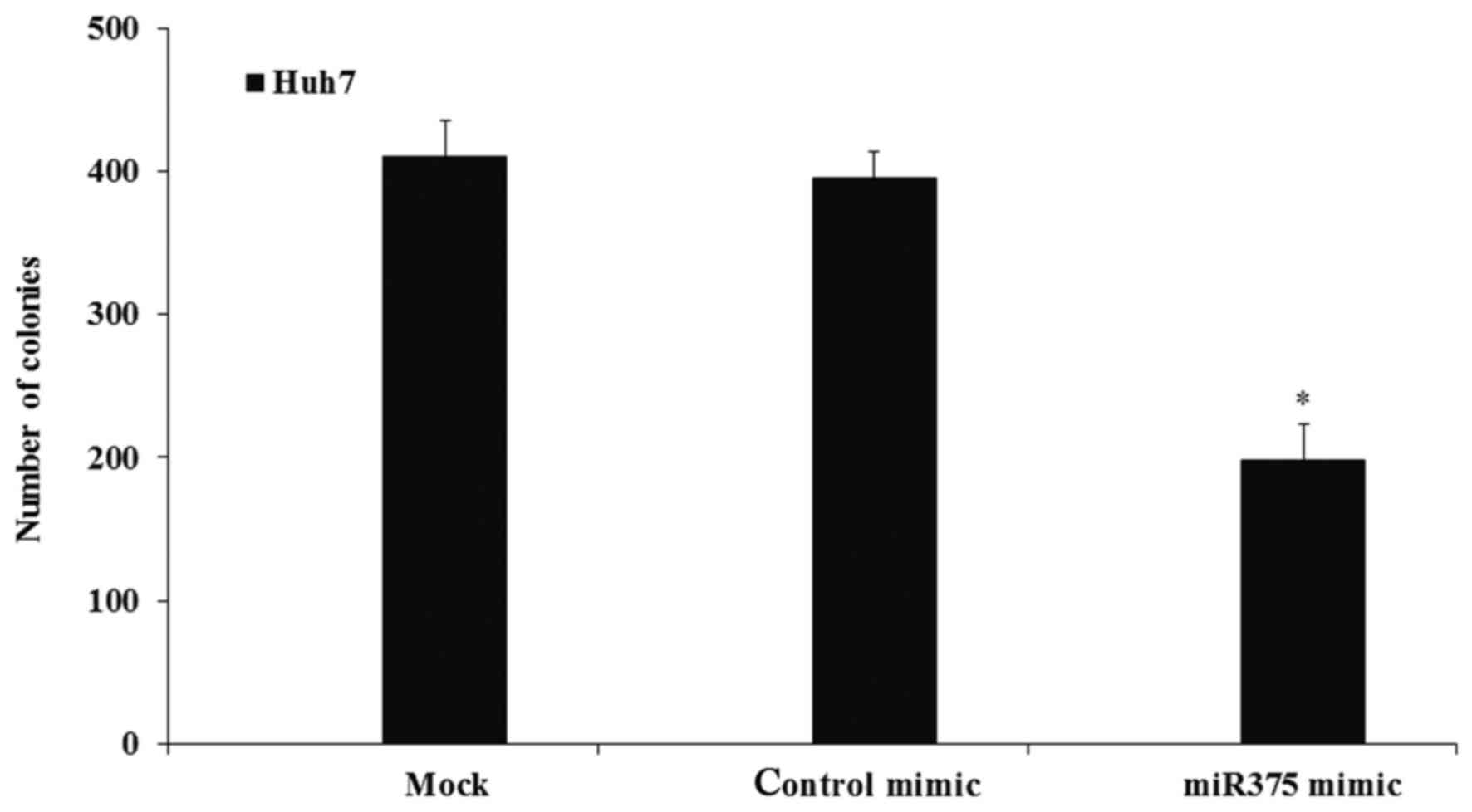
control mimic-transfected cells (P<0.05; Fig. 4). Furthermore, the colony-formation
assay results indicated that the induction of miR-375 expression
significantly decreased the colony numbers compared with the
numbers in the mock cell group (P<0.05), while transfection with
the control mimic did not affect the colony numbers (Fig. 5).
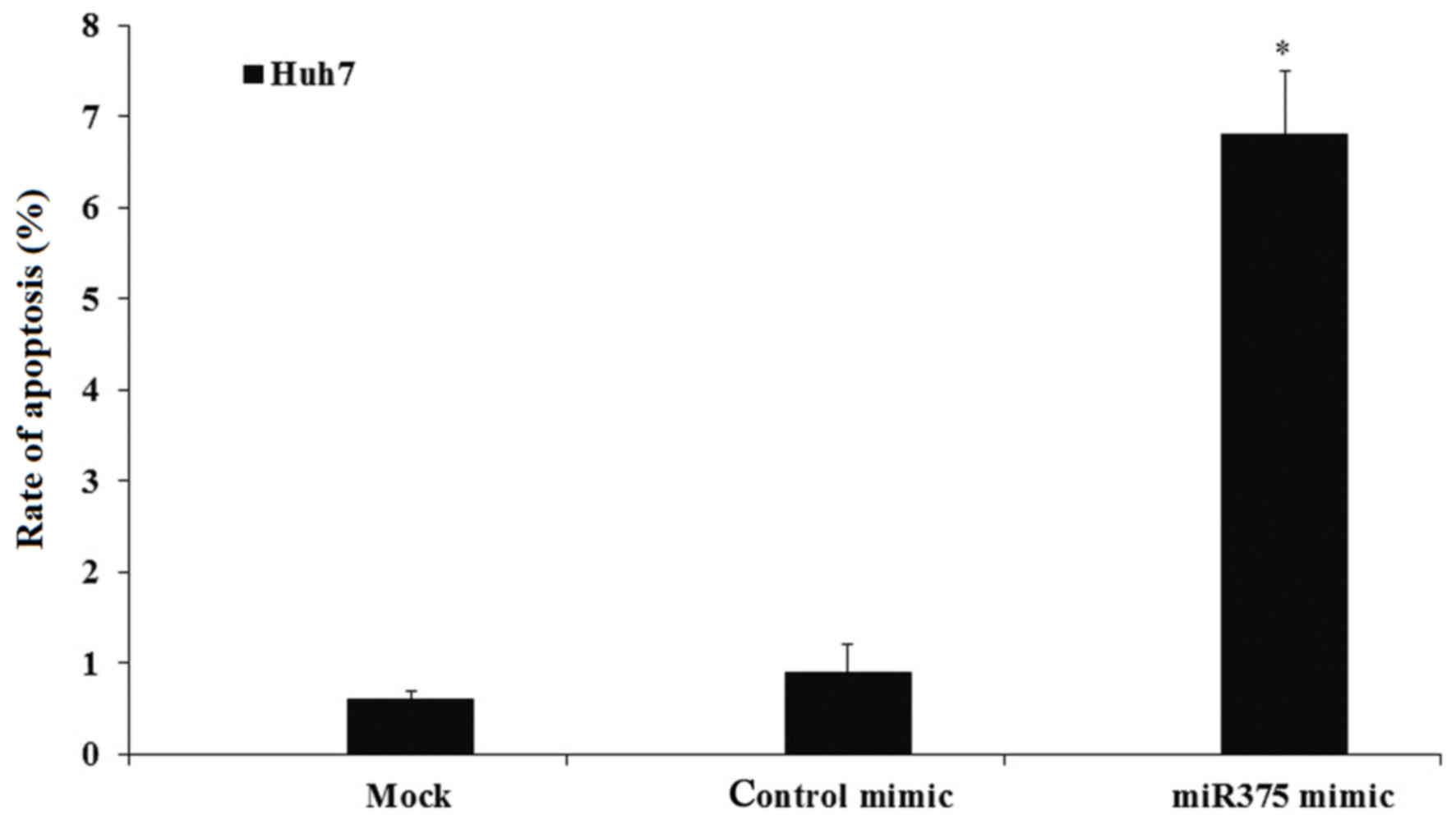
Upregulation of miR-375 induces HCC
cell apoptosis
To confirm whether the proliferation inhibition was
associated with cell apoptosis, the effect of miR-375 induction on
the cell apoptosis was investigated. Mock and transfected Huh7
cells were cultured and the cell apoptosis was evaluated by flow
cytometry. The results indicated that miR-375 mimic transfection
significantly induced the HCC cell apoptosis to ~7%, compared with
0.8% in the mock and 0.9% in control groups (P<0.05; Fig. 6).
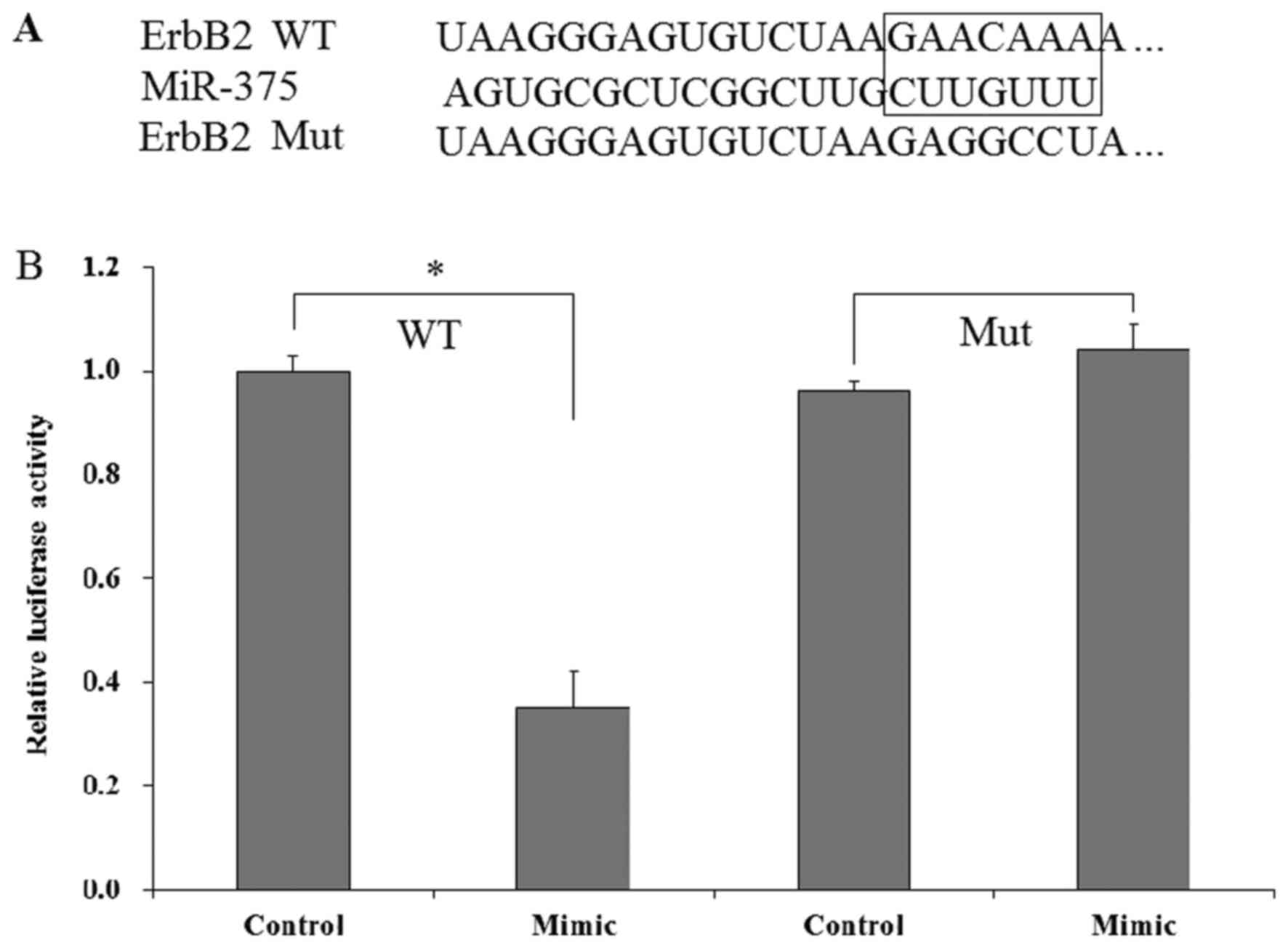
ErbB2 is a direct target of
miR-375
It was identified that ErbB2 is a potential gene of
miR-375 (Fig. 7A). To confirm whether
ErbB2 is a direct target of miR375, a dual luciferase assay was
performed. The miR-375 mimic and pGL2-ErbB2 (WT and Mut) were
co-transfected into Huh7 cells; control mimics were used as the
control. The results indicated that transfection of miR-375 mimic
significantly decreased the luciferase in Huh7 cells compared with
control mimic-transfected Huh7 cells, while the decrease was not
observed in the Mut pGL2-ErbB2 group (Fig. 7B). Therefore, these results
demonstrated that ErbB2 is a direct target gene of miR-375.
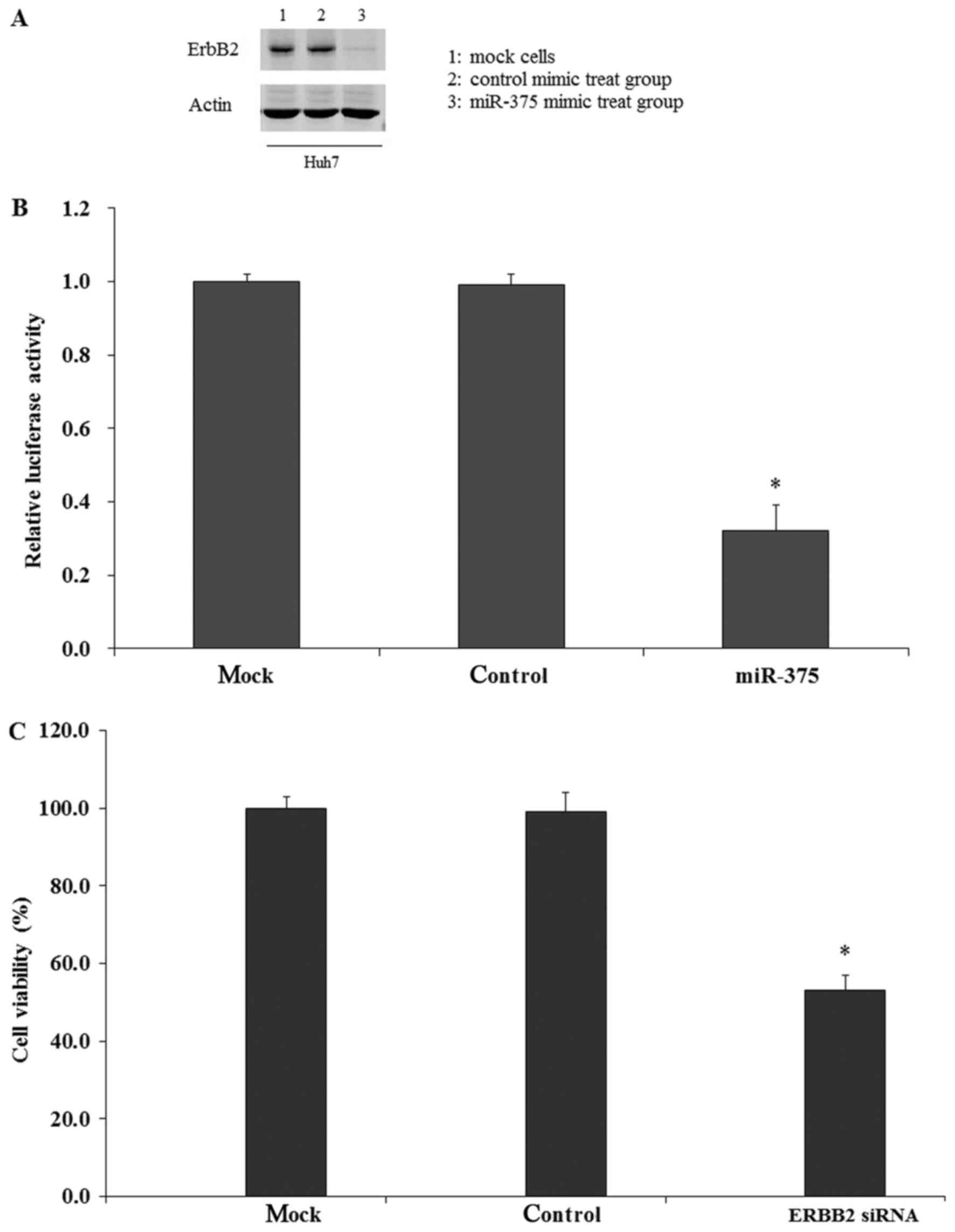
miR-375 modulates HCC cell growth by
repressing ErbB2
To study whether miR-375 regulated HCC cell growth
through targeting the ErbB2 gene, the expression of ErbB2 in HCC
cells was examined by RT-qPCR assay. The results indicated that
ErbB2 expression in Huh7 cells was increased compared with normal
PHH (data not shown). Then, miR-375 mimics were transfected into
Huh7 cells, and it was identified that the expression of ErbB2 was
significantly inhibited at the protein and mRNA levels compared
with the control mimic and mock groups (Fig. 8A and B). To analyze whether the
decrease in ErbB2 expression was associated with HCC cell growth,
HCC cells were transfected with ErbB2 siRNA and the cell viability
was measured by CCK-8 assay. The results indicated that
transfection of ErbB2 siRNA significantly inhibited the cell
viability compared with the control and mock groups (Fig. 8C). Therefore, it was concluded that
the expression of ErbB2 was associated with HCC cell growth, and
that miR-375 modulated the cell growth by repressing the expression
of ErbB2.
Discussion
miRs are a class of non-coding small RNAs measuring
~22 nucleotides in length. They serve important roles in the
translation or degradation of mRNAs (32). Previous studies have demonstrated that
miRs may regulate oncogene expression in tumorigenesis (33–35).
miR-375 was primary identified in the human pancreas in 2004
(36), and Avnit-Sagi et al
(37) suggested that miR-375 was
expressed at a very high level in human pancreatic islets and brain
tissue. Previous studies also identified that miR-375 was involved
in numerous types of cancer (38–40): The
expression of miR-375 was downregulated in human gastric cancer
cells, and the induction of miR-375 expression may affect the
biological function of cells (41);
Zhang et al (42) also
identified that miR-375 expression was abnormally regulated in
pancreatic progenitor cells, and that the regulation of miR-375
expression may inhibit cell proliferation through targeting Yes
associated protein 1. However, the biological effect of miR-375 in
human liver cancer has not been fully studied.
In the present study, the expression level of
miR-375 in human HCC tissues and cell lines was evaluated; it was
demonstrated that miR-375 was significantly downregulated in human
HCC tissues and cell lines compared with normal liver tissues and
cells, therefore we hypothesized that miR-375 served important
biological roles in human liver cancer. Additional analysis
indicated that the induction of miR-375 may inhibit human HCC cell
proliferation and induce apoptosis. Therefore, the regulation of
miR-375 served important roles in human HCC tumorigenesis in
vitro. The in vivo effect is also important: The present
study did not analyze the in vivo effect of miR-375 in HCC
cells; this will be performed as part of future studies.
Furthermore, the present study identified and confirmed that ErbB2
is a direct target of miR-375. ErbB2 belongs to the epidermal
growth factor receptor family; it has been identified to serve
important roles in the development and progression of different
types of human cancer (41,43). Previous studies have indicated that
ErbB2 gene upregulation is an important contributor to
hepatocellular growth, and that ErbB2 upregulation was associated
with miR-375 regulation (29,31). In the present study, it was identified
that downregulation of ErbB2 inhibited HCC cell growth using a
siRNA transfection assay. It was also demonstrated that the
induction of miR-375 significantly decreased the expression of
ErbB2 at mRNA and protein levels. However, the association between
HCC cell apoptosis and decreased ErbB2 remains unknown, and
additional studies are required.
Taken together, the present study demonstrated that
miR-375 was downregulated in human HCC, and the induction of
miR-375 may inhibit cell growth and induce cell apoptosis. The
present study also indicated that miR-375 regulated the biological
functions of HCC cells by targeting the ErbB2 gene, suggesting that
miR-375 may be a potential diagnostic and therapeutic target for
human HCC in the future.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LL, LJ and YD designed the study; LL and LJ
performed the experiments; LL, LJ and YD analyzed the data and
prepared the manuscript. YD reviewed the manuscript. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The Medical Ethics Committee of Daqing Longnan
Hospital approved the present study with written informed consent
from all patients.
Patient consent for publication
The present study was performed following the
Ethical and Institutional Guidelines with written informed consent
from all patients, and in the current manuscript no information of
these patients was disclosed.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chi SW, Zang JB, Mele A and Darnell RB:
Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature.
460:479–486. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hale BJ, Yang CX and Ross JW: Small RNA
regulation of reproductive function. Mol Reprod Dev. 81:148–159.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Farazi TA, Spitzer JI, Morozov P and
Tuschl T: MiRNAs in human cancer. J Pathol. 223:102–115. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Johnson SM, Grosshans H, Shingara J, Byrom
M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D and Slack
FJ: RAS is regulated by the let-7 microRNA family. Cell.
120:635–647. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee YS and Dutta A: The tumor suppressor
microRNA let-7 represses the HMGA2 oncogene. Genes Dev.
21:1025–1030. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ding J, Huang S, Wu S, Zhao Y, Liang L,
Yan M, Ge C, Yao J, Chen T, Wan D, et al: Gain of miR-151 on
chromosome 8q24.3 facilitates tumour cell migration and spreading
through downregulating RhoGDIA. Nat Cell Biol. 12:390–399. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kota J, Chivukula RR, O'Donnell KA,
Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P,
Torbenson M, Clark KR, et al: Therapeutic microRNA delivery
suppresses tumorigenesis in a murine liver cancer model. Cell.
137:1005–1017. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jiang J, Gusev Y, Aderca I, Mettler TA,
Nagorney DM, Brackett DJ, Roberts LR and Schmittgen TD: Association
of MicroRNA expression in hepatocellular carcinomas with hepatitis
infection, cirrhosis, and patient survival. Clin Cancer Res.
14:419–427. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gramantieri L, Ferracin M, Fornari F,
Veronese A, Sabbioni S, Liu CG, Calin GA, Giovannini C, Ferrazzi E,
Grazi GL, et al: Cyclin G1 is a target of miR-122a, a microRNA
frequently down-regulated in human hepatocellular carcinoma. Cancer
Res. 67:6092–6099. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang YS, Dai Y, Yu XF, Bao SY, Yin YB,
Tang M and Hu CX: Microarray analysis of microRNA expression in
hepatocellular carcinoma and non-tumorous tissues without viral
hepatitis. J Gastroenterol Hepatol. 23:87–94. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Budhu A, Jia HL, Forgues M, Liu CG,
Goldstein D, Lam A, Zanetti KA, Ye QH, Qin LX, Croce CM, et al:
Identification of metastasis-related microRNAs in hepatocellular
carcinoma. Hepatology. 47:897–907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Varnholt H, Drebber U, Schulze F,
Wedemeyer I, Schirmacher P, Dienes HP and Odenthal M: MicroRNA gene
expression profile of hepatitis C virus-associated hepatocellular
carcinoma. Hepatology. 47:1223–1232. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kumar V, Fausto N and Abbas A: Robbins
& Cotran Pathologic Basis of Disease. 9th edition. Saunders;
Philadelphia, PA: pp. 870–873. 2015
|
|
17
|
El-Serag HB: Hepatocellular carcinoma. N
Engl J Med. 365:1118–1127. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hao K, Luk JM, Lee NP, Mao M, Zhang C,
Ferguson MD, Lamb J, Dai H, Ng IO, Sham PC and Poon RT: Predicting
prognosis in hepatocellular carcinoma after curative surgery with
common clinicopathologic parameters. BMC Cancer. 9:3892009.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Connolly E, Melegari M, Landgraf P,
Tchaikovskaya T, Tennant BC, Slagle BL, Rogler LE, Zavolan M,
Tuschl T and Rogler CE: Elevated expression of the miR-17-92
polycistron and miR-21 in hepadnavirus-associated hepatocellular
carcinoma contributes to the malignant phenotype. Am J Pathol.
173:856–864. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang F, Li QJ, Gong ZB, Zhou L, You N,
Wang S, Li XL, Li JJ, An JZ, Wang DS, et al: MicroRNA-34a targets
Bcl-2 and sensitizes human hepatocellular carcinoma cells to
sorafenib treatment. Technol Cancer Res Treat. 13:77–86. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dang Y, Luo D, Rong M and Chen G:
Underexpression of miR-34a in hepatocellular carcinoma and its
contribution towards enhancement of proliferating inhibitory
effects of agents targeting c-MET. PLoS One. 8:e610542013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xiao Z, Li CH, Chan SL, Xu F, Feng L, Wang
Y, Jiang JD, Sung JJ, Cheng CH and Chen Y: A small-molecule
modulator of the tumor-suppressor miR34a inhibits the growth of
hepatocellular carcinoma. Cancer Res. 74:6236–6247. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tsukamoto Y, Nakada C, Noguchi T, Tanigawa
M, Nguyen LT, Uchida T, Hijiya N, Matsuura K, Fujioka T, Seto M and
Moriyama M: MicroRNA-375 is downregulated in gastric carcinomas and
regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer
Res. 70:2339–2349. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shen Y, Wang P, Li Y, Ye F, Wang F, Wan X,
Cheng X, Lu W and Xie X: MiR-375 is upregulated in acquired
paclitaxel resistance in cervical cancer. Br J Cancer. 109:92–99.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yan JW, Lin JS and He XX: The emerging
role of miR-375 in cancer. Int J Cancer. 135:1011–1018. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou N, Qu Y, Xu C and Tang Y:
Upregulation of microRNA-375 increases the cisplatin-sensitivity of
human gastric cancer cells by regulating ERBB2. Exp Ther Med.
11:625–630. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Du Y, Zhu M, Zhou X, Huang Z, Zhu J, Xu J,
Cheng G, Shu Y, Liu P, Zhu W and Wang T: MiR-20a enhances cisplatin
resistance of human gastric cancer cell line by targeting NFKBIB.
Tumour Biol. 37:1261–1269. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu J, Ahiekpor A, Li L, Li X, Arbuthnot
P, Kew M and Feitelson MA: Increased expression of ErbB-2 in liver
is associated with hepatitis B × antigen and shorter survival in
patients with liver cancer. Int J Cancer. 125:1894–1901. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sun K and Lai EC: Adult-specific functions
of animal microRNAs. Nat Rev Genet. 14:535–548. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kasinski AL and Slack FJ: Epigenetics and
genetics. MicroRNAs en route to the clinic: Progress in validating
and targeting microRNAs for cancer therapy. Nat Rev Cancer.
11:849–864. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Iorio MV and Croce CM: MicroRNA
dysregulation in cancer: Diagnostics, monitoring and therapeutics.
A comprehensive review. EMBO Mol Med. 4:143–159. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Valencia-Sanchez MA, Liu J, Hannon GJ and
Parker R: Control of translation and mRNA degradation by miRNAs and
siRNAs. Genes Dev. 20:515–524. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Poy MN, Eliasson L, Krutzfeldt J, Kuwajima
S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P
and Stoffel M: A pancreatic islet-specific microRNA regulates
insulin secretion. Nature. 432:226–230. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Avnit-Sagi T, Kantorovich L, Kredo-Russo
S, Hornstein E and Walker MD: The promoter of the pri-miR-375 gene
directs expression selectively to the endocrine pancreas. PLoS One.
4:e50332009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu AM, Poon RT and Luk JM: MicroRNA-375
targets Hippo-signaling effector YAP in liver cancer and inhibits
tumor properties. Biochem Biophys Res Commun. 394:623–627. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Poy MN, Hausser J, Trajkovski M, Braun M,
Collins S, Rorsman P, Zavolan M and Stoffel M: MiR-375 maintains
normal pancreatic alpha- and beta-cell mass. Proc Natl Acad Sci
USA. 106:5813–5818. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kloosterman WP, Lagendijk AK, Ketting RF,
Moulton JD and Plasterk RH: Targeted inhibition of miRNA maturation
with morpholinos reveals a role for miR-375 in pancreatic islet
development. PLoS Biol. 5:e2032007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shen ZY, Zhang ZZ, Liu H, Zhao EH and Cao
H: MiR-375 inhibits the proliferation of gastric cancer cells by
repressing ERBB2 expression. Exp Ther Med. 7:1757–1761. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang ZW, Men T, Feng RC, Li YC, Zhou D
and Teng CB: MiR-375 inhibits proliferation of mouse pancreatic
progenitor cells by targeting YAP1. Cell Physiol Biochem.
32:1808–1817. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Mitri Z, Constantine T and O'Regan R: The
HER2 receptor in breast cancer: Pathophysiology, clinical use, and
new advances in therapy. Chemother Res Pract.
2012:7431932012.PubMed/NCBI
|