Introduction
Colon cancer is the third most common type of cancer
and the fourth leading cause of cancer-associated mortality
worldwide (1,2). Multiple factors are involved in the
occurrence and development of colon cancer, including uncontrolled
cell proliferation, angiogenesis and suppression of apoptosis
(3–5).
Mutations are present in almost all types of colon cancer, and
point mutations in the KRAS gene can lead to activation of
potentially carcinogenic RAS protein activity (6). Therefore, understanding the pathogenesis
of colon cancer and elucidating the functional role of point
mutations in different genes are imperative to improving treatment
strategies for patients.
Non-coding RNAs (ncRNAs) have no protein-coding
abilities but may act as key regulators to control biological and
pathological processes (7). A large
number of ncRNAs that function as central orchestrators of
cell-specific gene networks have been identified (8). An important subclass of these ncRNAs are
long non-coding RNAs (lncRNAs) that are broadly defined as
regulatory non-coding transcripts of longer than 200 nucleotides.
Thus far, >100,000 lncRNA genes have been described in the human
genome, which is greater than the number of protein-coding and
microRNA genes combined. Although their biological roles and
mechanisms of function remain largely elusive, lncRNAs have
critical roles in the development process, cellular homeostasis,
genomic imprinting and pluripotency of embryonic stem cells
(9). The importance of lncRNA
regulation has been emphasized by their roles in the etiology of
human diseases (10–13). Several lncRNAs are involved in
carcinogenesis, disease progression, metastasis and/or
chemoresistance of human cancer types (14,15). Thus,
lncRNAs are novel therapeutic targets in cancer.
The novel lncRNA differentiation antagonizing
non-protein coding RNA (DANCR) is located on human chromosome 4,
with the closest adjacent annotated genes located 54.8 kb upstream
of USP46 and 28.7 kb downstream from ERVMER34-1 and the ANCR locus
(14). Previous studies have
demonstrated that DANCR is involved in the differentiation of
progenitors and osteoblasts (16,17).
Recently, abundant evidence has suggested that DANCR is an oncogene
in various types of cancer, including breast cancer (15), gastric cancer (16), prostate cancer (17) and hepatocellular carcinoma (18). In colon cancer, DANCR expression was
increased, and high DANCR expression was revealed to be correlated
with a poor overall survival (OS) and disease-free survival (DFS)
in patients (19). However, the
biological functions and significance of DANCR in colon cancer have
not yet been established. In the present study, DANCR expression in
colon cancer cell lines was determined by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) and
Cell Counting kit-8 assay. A colony formation assay, flow
cytometry, Hoechst 33258 staining and western blotting were
employed to investigate the effects and mechanisms of DANCR in
regulating colon cancer growth. Establishment of a xenograft tumor
model was followed by terminal deoxynucleotidyl transferase (TdT)
dUTP nick-end labeling (TUNEL) assay. Immunohistochemical staining
was performed to confirm the findings in vitro.
Materials and methods
Cancer cell lines and cell
culture
Human colon cancer SW620, SW480, HCT116, HT29, HCT15
and Caco-2 cell lines and normal human colon epithelial HCoEpiC
cells were obtained from American Type Culture Collection
(Manassas, VA, USA). Cells were cultured in Dulbecco's modified
Eagle's medium (DMEM) supplemented with 10% fetal bovine serum
(FBS; Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA), 100 U/ml penicillin G and 100 µg/ml streptomycin (Thermo
Fisher Scientific, Inc.) in a humidified atmosphere of 5%
CO2 at 37°C.
Generation of stably infected cell
lines
Lentiviral vector-based short hairpin RNA (shRNA)
targeting non-specific control (NC) or human LncRNA DANCR
(5′-GGAGCTAGAGCAGTGACAATG-3′) were purchased from Shanghai Co.,
Ltd., GenePharma (Shanghai, China). Cells achieved 70–80%
confluence in Lv-shNC and Lv-shDANCR infection. The stably infected
cells were selected by adding puromycin (2 µg/ml), and were
collected for further qPCR analysis to confirm the efficient
knockdown of DANCR in colon cancer cell lines.
RNA isolation and RT-qPCR
Total cellular RNA was isolated from colon cancer
cells using TRIzol (Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocols. The RNA was reverse-transcribed into
cDNA by using a reverse transcription kit (RR047A; Takara Bio,
Inc., Otsu, Japan) according to the manufacturer's protocol.
Real-time reverse transcription-PCR was performed with a SYBR Green
PCR kit (RR902A; Takara Bio, Inc.) and ABI 7300 Real-Time PCR
System (Applied Biosystems; Thermo Fisher Scientific, Inc.).
Amplification was performed as follows: Incubation at 95°C for 30
sec, followed by 40 cycles of denaturation at 95°C for 5 sec and
subsequent annealing and extension at 58°C for 30 sec. Relative
gene expression was calculated using the 2−ΔΔCq method
(20). For fold change analysis,
GAPDH (forward, 5′-TGCGTGACATTAAGGAGAA-3′ and reverse,
5′AAGGAAGGCTGGAAGAGT-3′) expression was analyzed to normalize
target gene expression. Primer sequence of DANCR used in this study
are as follow (forward, 5′-GCCACTATGTAGAGGGTTTC-3′ and reverse,
5′-ACCTGCGCTAAGAACTGAGG-3′).
Cell Counting kit-8 assay
Cells (1,000 cells/well) were plated in 96-well
plates. Cell proliferation was measured at 0, 24, 48 and 72 h post
cell plating. Cell Counting kit-8 assay (Beyotime Institute of
Biotechnology, Beijing, China) was used for cell viability
determination at a wavelength of OD 450 nm.
Western blotting
Cells were washed once with ice-cold PBS and lysed
in radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology) containing 1% protease inhibitors (Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany). Protein concentration was
subsequently measured with a BCA kit (Thermo Fisher Scientific,
Inc.), ~10 µg protein was separated with 8–12% SDS-PAGE gel and
blotted onto polyvinylidene fluoride or polyvinylidene difluoride
membranes (Merck KGaA, Darmstadt, Germany). Membranes were
subsequently blocked with 5% milk at room temperature for 1 h and
incubated with primary antibodies against caspase 3 (1:600; cat.
no. 9665), caspase 8 (1:1,000; cat. no. 4790), caspase 9 (1:800;
cat. no. 9508), p53 (1:1,200; cat. no. 2524), Cyto C (1:1,000; cat.
no. 4280), Bcl-2 (1:1,200; cat. no. 3498), Bcl-xL (1:1,000; cat.
no. 2762) and GAPDH (1:5,000; cat. no. 2118) (Cell Signaling
Technology, Inc., Danvers, MA, USA) at 4°C overnight, followed by 3
washes with tris-buffered saline with tween and a 1 h incubation
with horseradish peroxidase-conjugated secondary antibodies at room
temperature (OriGene Technologies Inc., Beijing, China). Protein
bands were visualized with an enhanced chemiluminescence kit (Merck
KGaA). The density of bands was quantified by ImageJ software
(version 1.48; National Institutes of Health, Bethesda, MD,
USA).
Apoptosis analysis by flow
cytometry
Cells were harvested and stained with Annexin
V-FITC/PI Apoptosis Detection kit (Nanjing KeyGen Biotech Co.,
Ltd., Nanjing, China), according to the manufacturer's protocol.
Apoptotic cells were assessed by flow cytometry (De Novo Software,
Glendale, CA, USA). Each experiment was performed three times.
Hoechst 33258 staining
A total of 1×105 cells were plated in
each well of the 6-well plates. After 24 h, the cells were fixed
with 4% paraformaldehyde for 10 min at room temperature. Following
washing with PBS 3 times, the cells were stained with Hoechst 33258
(Beyotime Institute of Biotechnology) in the dark for 15 min at
room temperature. Images of the cells were captured using a
fluorescence microscope (Olympus Corporation, Tokyo, Japan).
Colony formation assay
A total of 1,000 cells were plated in each well of
the 6-well plates containing 2 ml DMEM medium (10% FBS). Following
7–10 days, the cells were fixed with 4% paraformaldehyde for 10 min
at room temperature. Cells were washed with PBS 3 times, and
subsequently stained with purple crystal (Beyotime Institute of
Biotechnology) for 15 min at room temperature. The number of
colonies were counted and analyzed. The assays were replicated
three times.
Animal study
This study was approved by the Ethics Committee of
Dongtai Municipal People's Hospital of Nantong University and
complied with the animal guidelines. Ten 6-week-old female BALB/c
nude mice (weighing between 18 and 20 g) were purchased from Hfkbio
(Beijing, China; http://www.hfkbio.com/) and housed in a specific
pathogen-free room (at a temperature of 22–25°C) with 12 h
dark/light cycle and ad libitum access to food and water.
SW480-shNC and SW480-shDANCR cells (5×106 cells/mouse)
were subcutaneously injected into the right flank of (n=5 per
experimental group). Tumor length and width were measured every
five days from the tenth day post cell injection. Tumor volumes
were calculated as ellipsoids (length × width2 × 0.52).
At 30 days post cell injection, the mice were sacrificed and the
tumors were removed and measured.
Terminal deoxynucleotidyl transferase
dUTP nick end labeling (TUNEL) assay
Tumor tissue sections were examined for the presence
of apoptotic cells using the DeadEnd™ Fluorometric TUNEL system
(Promega Corporation, Madison, WI, USA) according to the
manufacturer's protocol, in which fragmented DNA from apoptotic
cells is end-labeled with the fluorophore. Following
deparaffinization and rehydration, sections were fixed with 4%
paraformaldehyde at room temperature (22–25°C) for 10 min,
permeabilized with proteinase K for 8–10 min at room temperature
(22–25°C), and repeatedly fixed. The sections were then covered
with 50 ml terminal deoxynucleotidyl transferase mix for 1 h at
37°C in a humidified chamber. The coverslips were removed and the
sections were immersed in 2× saline-sodium citrate buffer for 15
min, washed with PBS and mounted with glycerin (Beyotime Institute
of Biotechnology) that included DAPI (Beyotime Institute of
Biotechnology). Four random fluorescence images were captured using
a fluorescence microscope (Olympus Corporation).
Immunohistochemical staining
Collected tumor tissues from mice injected with
SW480 cells were fixed with 4% paraformaldehyde at room temperature
for 48 h. Then, fixed tumor tissues were embedded in paraffin and
sectioned into 4 µm slices. Following removal of the paraffin and
antigen retrieval in citrate buffer (pH 6.0) at high temperature
and pressure for 3 min, slices were blocked with 5% goat serum at
room temperature (22–25°C) for 15 min and incubated overnight with
anti-caspase 3 antibody (1:200; cat. no. 9662; Cell Signaling
Technology, Inc.) at 4°C. Detection (inclusive of secondary
antibody) was performed in an automated slide staining instrument
(SP9001; OriGene Technologies, Inc.) by using a
3,3′-diaminobenzidine staining kit (Maixin, Fuzhou, China) and the
slices were counterstained with hematoxylin (OriGene Technologies,
Inc.) at room temperature for 3 min. All kits were used according
to the manufacturer's protocol. Images of the slices were captured
with a CX51 light microscope (magnification, ×20; Olympus
Corporation). The caspase 3 positive, and total cell number in each
frame were counted.
Statistical analysis
All data are presented as the mean ± standard
deviations. Student's t-tests were performed to compare the
difference between two groups and one-way analysis of variance
followed by Turkey's multiple comparisons analysis was used to
compare the difference among multi-groups. SPSS software (version
21.0, IBM Corp., Armonk, NY, USA), was used. P<0.05 was
considered to indicate a statistically significant difference.
Results
Silencing DANCR inhibits colon cancer
cell growth in vitro
To determine whether DANCR serves a pivotal role in
colon cancer, we investigated the expression levels of lncRNA DANCR
in several human colon cancer cell lines. Healthy human epithelial
cells (HCoEpiC) were used as a control. It was revealed that DANCR
was overexpressed in all the six colon cancer cell lines compared
with HCoEpiC cells (Fig. 1A). Among
the colon cancer cell lines, SW480 and HCT15 cells revealed a
>10-fold increase in DANCR expression level (Fig. 1A). To investigate the biological
effect of DANCR in colon cancer, lentivirus-based shRNA targeting
DANCR was used to infect SW480 and HCT15 cells. Following puromycin
selection, DANCR expression in SW480 and HCT15 cells that had been
infected with shDANCR was decreased compared with shNC group
(Fig. 1B and C). As a result,
silencing DANCR significantly decreased SW480 and HCT15 cell
proliferation (Fig. 1D and E) and
colony formation (Fig. 1F and G),
respectively. These data suggested that DANCR functions as an
oncogene in colon cancer.
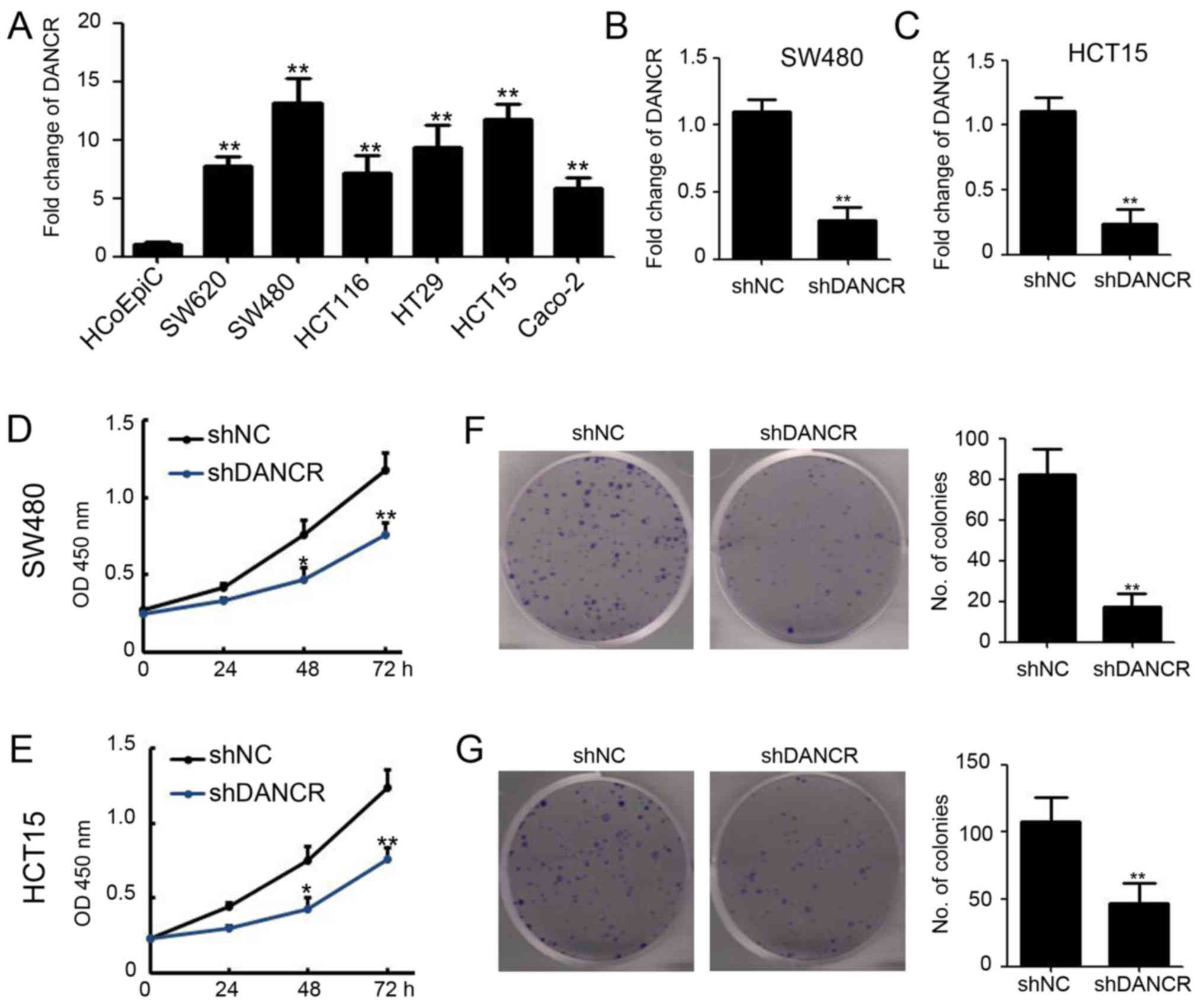 | Figure 1.Silencing DANCR inhibits colon cancer
cell growth in vitro. (A) Fold change of DANCR in human
colon cancer cell lines and normal human epithelial cells
(HCoEpiC). DANCR was examined by RT-qPCR and normalized to GAPDH
expression (P<0.01). (B) SW480 and (C) HCT15 cell lines were
infected with lentivirus-based shRNA targeting DANCR and NC. The
fold change of DANCR were examined by RT-qPCR and normalized to
GAPDH expression (P<0.01). (D) Cell Counting kit-8 assay was
performed to detect the cell viability of SW480-shNC and
SW480-shDANCR cells at 0, 24, 48 and 72 h post cell plating
(P<0.05, P<0.01). (E) Cell Counting kit-8 assay was performed
to detect the cell viability of HCT15-shNC and HCT15-shDANCR cells
at 0, 24, 48 and 72 h post cell plating (P<0.05, P<0.01). (F)
Colony formation assay was performed in SW480-shNC and
SW480-shDANCR cells. The number of colonies was analyzed
(P<0.01). (G) Colony formation assay was performed in HCT15-shNC
and HCT15-shDANCR cells. The number of colonies was analyzed
(P<0.01). *P<0.05 and **P<0.01 vs. shNC. shDANCR, short
hairpin differentiation antagonizing non-protein coding RNA; shRNA,
short hairpin ribonucleic acid; NC, negative control; RT-qPCR,
reverse transcription-quantitative polymerase chain reaction. |
Silencing DANCR promotes cell
apoptosis
To determine the potential mechanism of DANCR
regulation of colon cancer cell proliferation, flow cytometry with
Annexin V and PI staining was performed to detect cell apoptosis
and Annexin V positive and PI negative cells were considered to be
apoptotic cells. As shown in Fig. 2A and
B, more apoptotic cells were observed in SW480-shDANCR
(Fig. 2A; shDANCR vs. shNC:
19.07±1.22 vs. 6.72±0.71%) and HCT15-shDANCR cells (Fig. 2B; shDANCR vs. shNC: 16.17±1.02 vs.
5.63±0.82%), compared with shNC cells. Hoechst 33258 was also used
to detect apoptotic cells and, in concordance with Annexin V/PI
data, indicated that DANCR knockdown promotes cell apoptosis in
SW480 (Fig. 2C) and HCT15 (Fig. 2D) cells. Collectively, these results
suggested that silencing DANCR promotes cell apoptosis in colon
cancer cells.
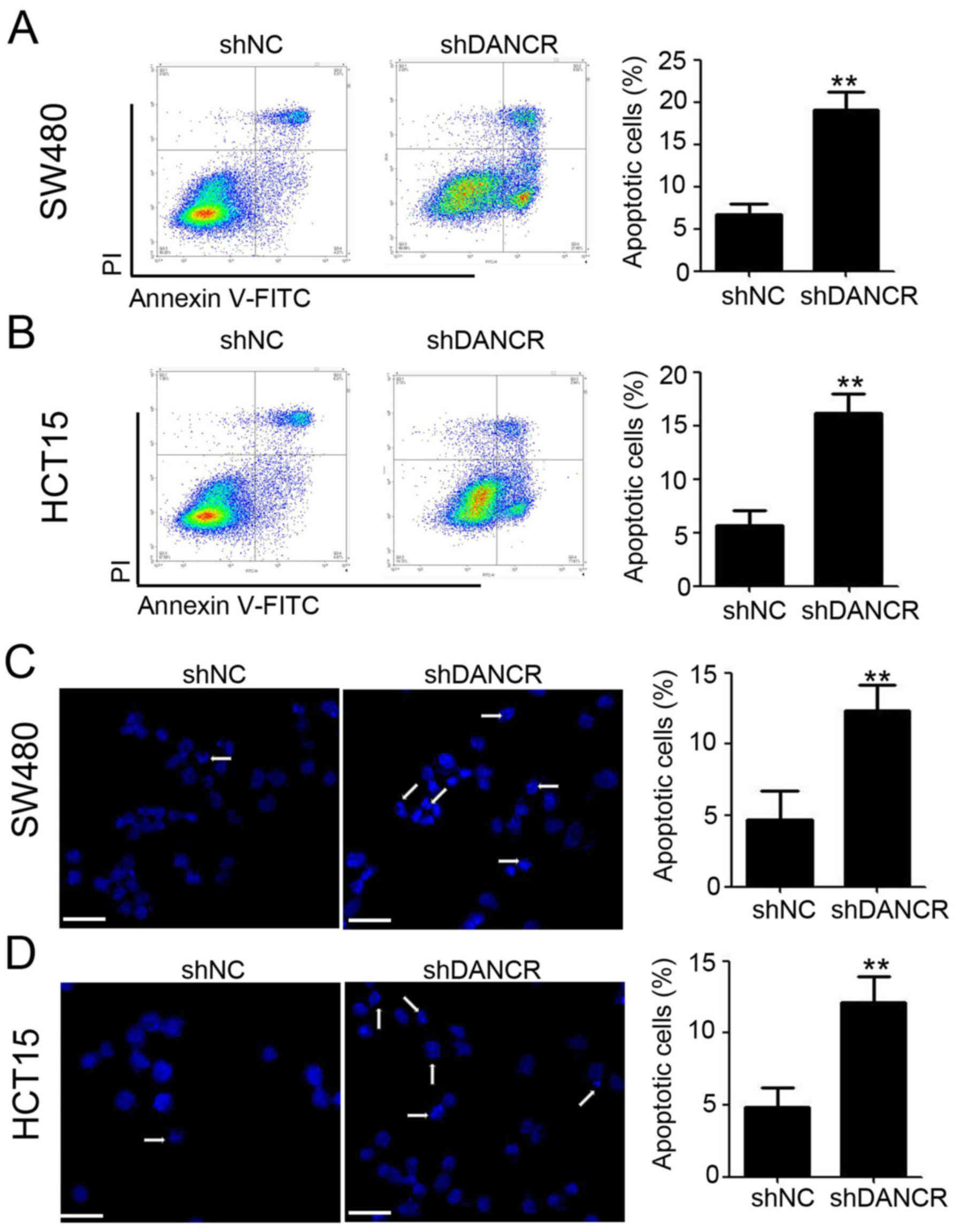 | Figure 2.Silencing DANCR promotes cell
apoptosis in colon cancer cells. (A) Annexin V-FITC and PI staining
were performed to detect apoptotic cells in SW480-shNC and
SW480-shDANCR cells, followed with flow cytometry analysis.
Representative dot plots (left) and cumulative data (right) are
shown (P<0.01). (B) Annexin V-FITC and PI staining were
performed to detect apoptotic cells in HCT15-shNC and HCT15-shDANCR
cells, followed by flow cytometry analysis. Representative dot
plots (left) and cumulative data (right) are shown (P<0.01). (C)
Hoechst 33258 staining was performed to detect apoptotic cells in
SW480-shNC and SW480-shDANCR cells, followed by fluorescence
capturing. Representative images (left) and cumulative data (right)
are shown, scale bar=50 µm (P<0.01). (D) Hoechst 33258 staining
was performed to detect apoptotic cells in HCT15-shNC and
HCT15-shDANCR cells, followed by fluorescence capturing.
Representative images (left) and cumulative data (right) are shown,
scale bar, 50 µm (P<0.01). **P<0.01 vs. shNC. shDANCR, short
hairpin differentiation antagonizing non-protein coding RNA; NC,
negative control; FITC, fluorescein isothiocyanate; PI, propidium
iodide; shRNA, short hairpin ribonucleic acid. |
Silencing DANCR regulates caspase
protein and upstream protein expression in colon cancer cells
To further determine the molecular mechanism behind
the antitumor activity of DANCR knockdown in colon cancer cells,
western blotting was employed to investigate the expression of
caspase proteins and certain upstream proteins. It was revealed
that silencing DANCR significantly induces caspase 3, caspase 8 and
caspase 9 expression, which demonstrated low/no basal expression
levels, in SW480 and HCT15 cells (Fig.
3A). Furthermore, shDANCR was revealed to promote p53 and cyto
C expression and inhibit Bcl-2 and Bcl-xL expression in SW480 and
HCT15 cells (Fig. 3B). These results
suggested that silencing DANCR regulates caspase protein and
upstream protein expression in colon cancer cells.
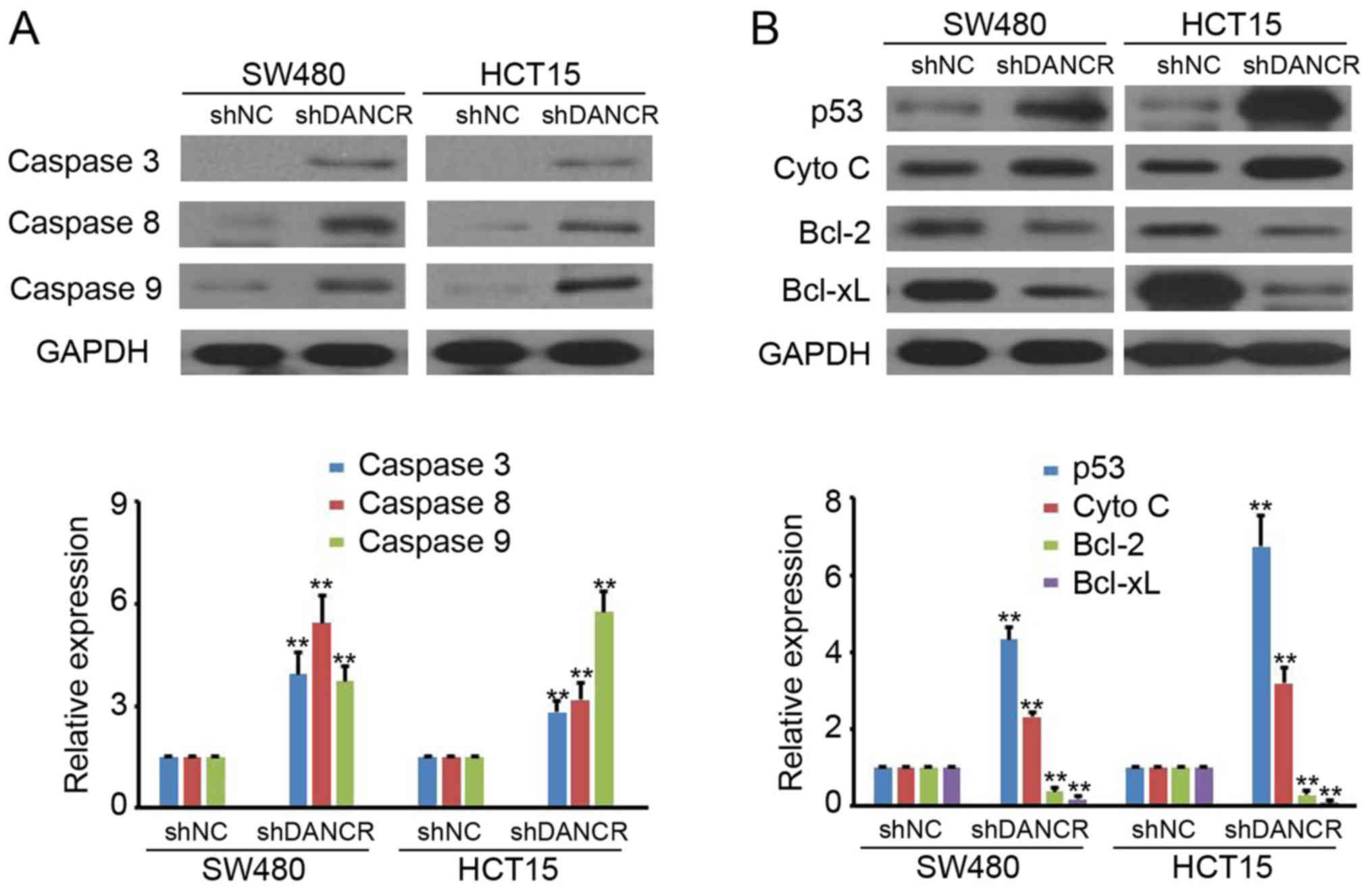 | Figure 3.Silencing DANCR regulates caspase
protein expression. (A) The expression of caspase 3, 8 and 9 in
SW480-shNC, SW480-shDANCR HCT15-shNC and HCT15-shDANCR cells was
detected by western blot analysis. GAPDH was used as a loading
control. The relative expression was analyzed (P<0.01). (B) The
expression of p53, Cyto C, Bcl-2 and Bcl-xL in SW480-shNC,
SW480-shDANCR HCT15-shNC and HCT15-shDANCR cells were detected by
western blot analysis. GAPDH was used as a loading control. The
relative expression was analyzed (P<0.01). **P<0.01 vs. shNC.
shDANCR, short hairpin differentiation antagonizing non-protein
coding RNA; NC, negative control; shRNA, short hairpin ribonucleic
acid; Cyto C, cytochrome c; Bcl-2, B-cell lymphoma 2;
Bcl-xL, B-cell lymphoma-extra-large. |
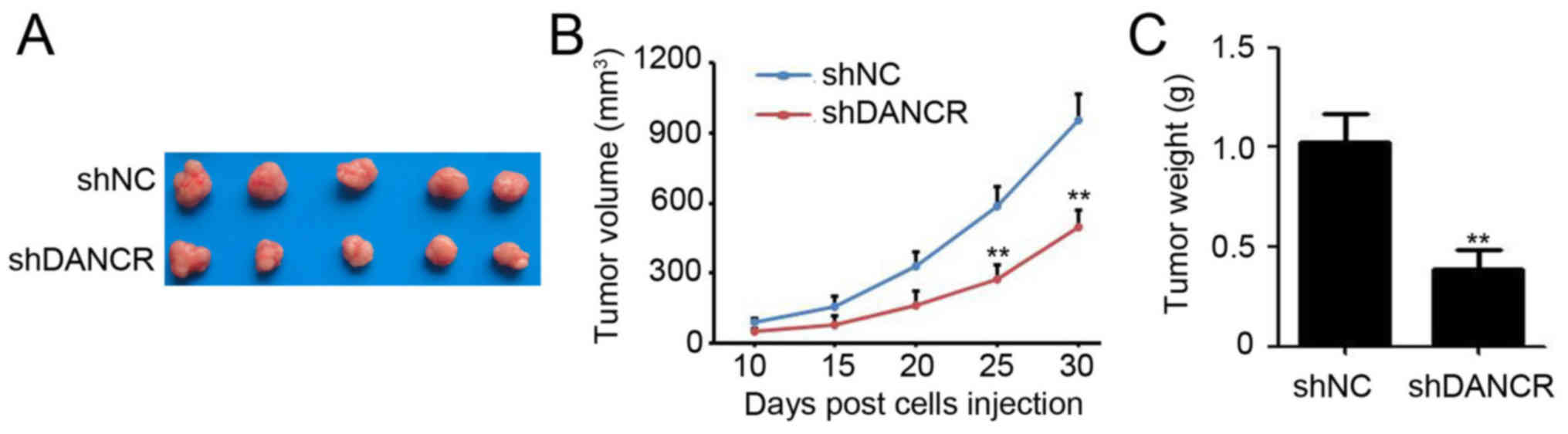
Silencing DANCR impairs colon tumor
growth in vivo
To investigate the effect of DANCR on colon cancer
growth in vivo, SW480 cells that were stably infected with
shDANCR and shNC were subcutaneously injected into the right flank
of 6-week-old female BALB/c nude mice. After 30 days, the mice were
sacrificed and the tumors were visualized (Fig. 4A). The results revealed that tumors
generated from SW480 cells infected with shDANCR grew significantly
slower than the negative control, evidenced by the growth curve
(Fig. 4B; final tumor volume of shNC
vs. shDANCR: 959.2±107.3 vs. 498.5±73.4 mm3).
Furthermore, silencing DANCR also caused a 58.7% reduction in the
weight of SW480 cell-tumor (Fig. 4C;
shNC vs. shDANCR: 1.04±0.13 vs. 0.43±0.06 g). These results
suggested that DANCR was capable of regulating colon tumor growth
generated from SW480 cells.
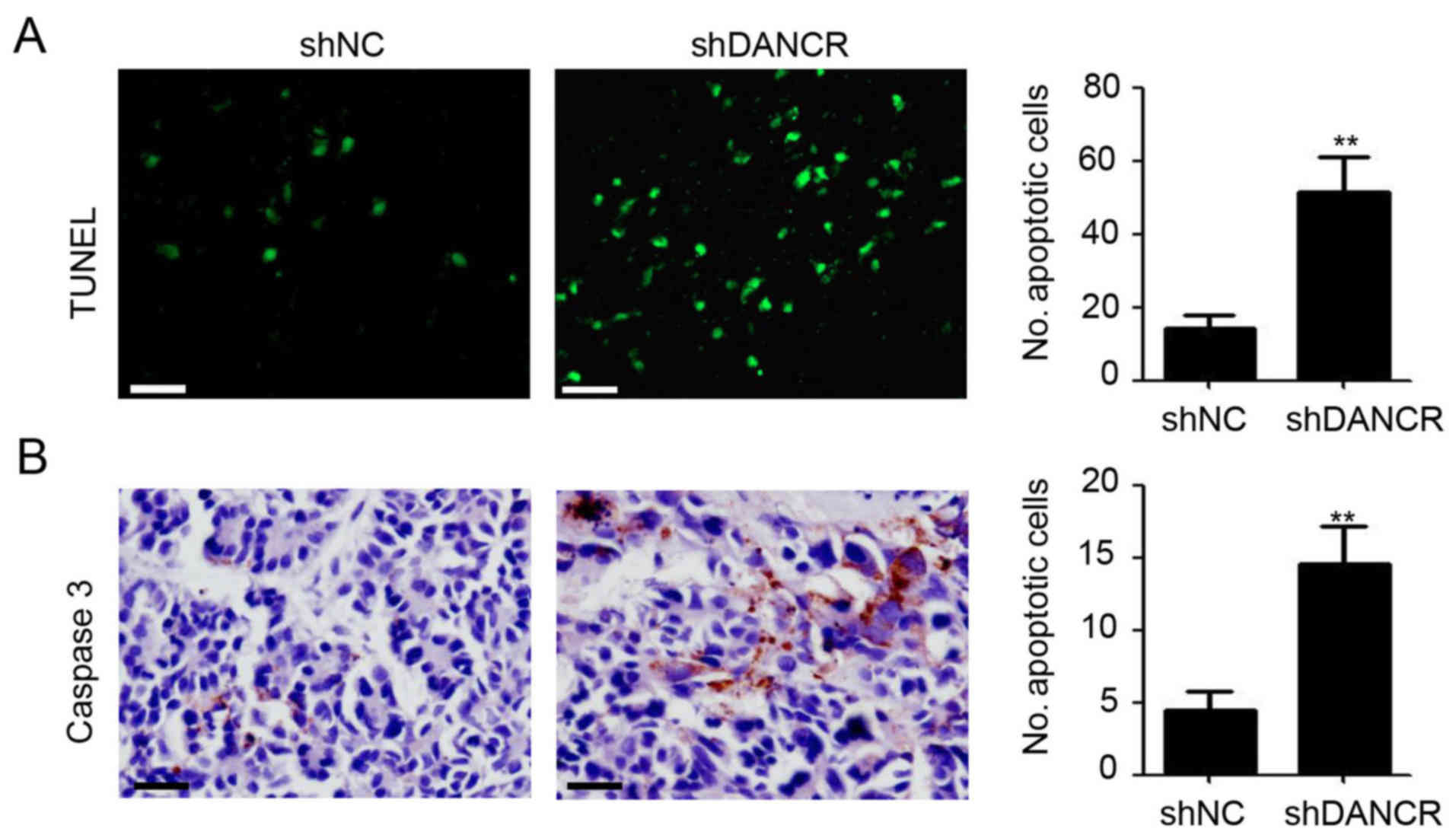
Silencing DANCR promotes tumor cell
apoptosis and caspase 3 expression
As DANCR-knockdown appeared to promote cell
apoptosis through the induction of caspase protein expression in
colon cancer, the apoptotic cell number, and caspase 3 protein
expression in SW480 tumors was further investigated. As shown in
Fig. 5A, more apoptotic cells were
observed in SW480-shDANCR tumors by TUNEL assay, compared with
SW480-shNC tumors. Concurrently, an almost negligible number of
caspase 3-positive tumor cells were observed in SW480-shNC tumor
tissues (Fig. 5B). Of note, DANCR
knockdown significantly promoted caspase 3 expression in SW480
tumors (Fig. 5B). These results
provided further evidence for the role of DANCR knockdown in the
induction of colon cancer cell apoptosis.
Discussion
Due to their critical roles in the development
process, cellular homeostasis, genomic imprinting and pluripotency
of embryonic stem cells (9), lncRNAs
are involved in the carcinogenesis, disease progression, metastasis
or chemoresistance of human cancer types (14,15). In
the present study, it was demonstrated that lncRNA DANCR was
overexpressed in almost all colon cancer cell lines (Fig. 1). Silencing DANCR significantly
inhibited cell proliferation (Fig.
1), colony formation (Fig. 1) and
tumor growth (Fig. 4), and induced
apoptosis (Fig. 2) through regulating
caspase proteins (Figs. 3 and
5) and the expression of upstream
proteins (Fig. 3). The present study
provides evidence of the functional role and mechanism of DANCR in
colon cancer.
In a previous study by Sha et al (15), DANCR expression was revealed to be
increased in triple-negative breast cancer (TNBC) tissues compared
with adjacent normal tissues in 63 TNBC specimens. Patients with
higher DANCR expression correlated with worse tumor-node-metastasis
stages as well as a shorter overall survival (OS) using
Kaplan-Meier analysis (15). DANCR
was upregulated in the tumor tissues and plasma of patients with
HCC, and its expression was highly correlated with microvascular
and liver capsule invasion in hepatocellular carcinoma (HCC)
(18). Further receiver operating
characteristic analysis demonstrated that plasma DANCR exhibited
significantly increased discriminatory power for differentiating
patients with HCC from HVs and non-HCC patients compared with
a-fetoprotein, which has been used as a biomarker for HCC diagnosis
(18). In osteosarcoma tissues, DANCR
was consistently significantly increased and its expression was an
independent poor prognostic factor (21). DANCR was demonstrated to be increased
in colorectal cancer (CRC) tissues compared with that in adjacent
normal tissues and patients with high DANCR expression had a
shorter OS and disease-free survival (DFS) compared with the low
DANCR expression group (19). Of
note, in a multivariate Cox model, DANCR was demonstrated to be an
independent poor prognostic factor for OS and DFS in CRC (19). The present study, also demonstrated an
increased expression of DANCR in the majority of colon cancer cell
lines that were investigated (Fig.
1). These results are consistent with the findings by Liu et
al (19) in patients with colon
cancer.
DANCR has been demonstrated to have multiple
functional roles in regulating cell fate. In synovium-derived MSCs,
overexpression of DANCR can promote the proliferation and
chondrogenesis of MSCs (22).
Furthermore, DANCR functions as an oncogene in various types of
cancer. In HCC, DANCR markedly increased stemness features of
cancer cells to promote tumorigenesis and intra/extra-hepatic tumor
colonization (23). In vivo
xenograft experiments demonstrated that knockdown of DANCR in
MDA-MB-231 cells reduced tumor progression significantly through
inhibiting cell proliferation and invasion, and fewer cancer stem
cells were observed in DANCR-knocked down MDAMB-231 cells (15). DANCR also promotes prostate cancer
invasion and metastasis through repressing the expression of
TIMP2/3 (17). Of note, DANCR could
suppress 786O and ACHN proliferation, migration, and invasion, and
induce apoptosis in renal cell carcinoma (24). In the present study, we demonstrated
that DANCR-knockdown significantly inhibited cell proliferation,
colony formation, and tumor growth in vitro and in
vivo (Figs. 1 and 4). To the best of our knowledge, the present
study is the first to provide evidence for understanding how
knockdown of DANCR promotes apoptosis in colon cancer cells, which
contradicts previous findings in renal cell carcinoma (24). The diverse roles of DANCR in apoptosis
differed between the cells investigated in the present study.
In a previous study, the role of DANCR in regulating
the stemness of HCC relied largely on an association with, and
regulation of, CTNNB1 (23). In TNBC,
knockdown of DANCR was associated with increased binding of EZH2 to
the promoters of CD44 and ABCG2 (15). MiR-1305 serves as a downstream target
of DANCR during the induction of cell proliferation and
chondrogenic differentiation of synovium-derived MSCs (25). Components of the Wnt/b-catenin
signaling pathway were also downstream targets of DANCR during
odontoblast-like differentiation of human dental pulp cells. This
was demonstrated by a decrease in the expression levels of p-GSK-3b
and b-catenin upon DANCR overexpression (26). The present study also demonstrated
that DANCR-knockdown promoted apoptosis in colon cancer through
caspases (Fig. 3). However, further
investigations are needed to determine the direct target of DANCR
during the regulation of apoptosis in colon cancer.
In summary, the present study highlights the
potential role of DANCR in regulating colon cancer growth in
vitro and in vivo. We demonstrated that caspase proteins
mediated apoptosis. These findings suggested that DANCR may be a
novel therapeutic target in colon cancer. However, further
investigations are required to determine the direct target of DANCR
in the regulation of apoptosis in colon cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated and/or analyzed during this study
are included in this published article.
Authors' contributions
XJY and JJZ were involved in the acquisition of the
data. WJC, GGZ and WW were involved in the analysis and
interpretation of the data. HCT was involved in the conception and
design of the present study.
Ethics approval and consent to
participate
The present animal study was approved by the Ethics
Committee of Dongtai Municipal People's Hospital of Nantong
University and complied with the animal guidelines.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD, Fedewa SA, Ahnen DJ,
Meester RGS, Barzi A and Jemal A: Colorectal cancer statistics,
2017. CA Cancer J Clin. 67:177–193. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dai L, Cui X, Zhang X, Cheng L, Liu Y,
Yang Y, Fan P, Wang Q, Lin Y, Zhang J, et al: SARI inhibits
angiogenesis and tumour growth of human colon cancer through
directly targeting ceruloplasmin. Nat Commun. 7:119962016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang Y, Lin C, Liao G, Liu S, Ding J,
Tang F, Wang Z, Liang X, Li B, Wei Y, et al: MicroRNA-506
suppresses tumor proliferation and metastasis in colon cancer by
directly targeting the oncogene EZH2. Oncotarget. 6:32586–32601.
2015.PubMed/NCBI
|
|
5
|
Zeng M, Zhu L, Li L and Kang C: miR-378
suppresses the proliferation, migration and invasion of colon
cancer cells by inhibiting SDAD1. Cell Mol Biol Lett. 22:122017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Frouws MA, Reimers MS, Swets M,
Bastiaannet E, Prinse B, van Eijk R, Lemmens VE, van Herk-Sukel MP,
van Wezel T, Kuppen PJ, et al: The Influence of BRAF and KRAS
mutation status on the association between aspirin use and survival
after colon cancer diagnosis. PLoS One. 12:e01707752017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wilusz JE: Long noncoding RNAs: Re-writing
dogmas of RNA processing and stability. Biochim Biophys Acta.
1859:128–138. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lam MT, Li W, Rosenfeld MG and Glass CK:
Enhancer RNAs and regulated transcriptional programs. Trends
Biochem Sci. 39:170–182. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Boon RA, Jaé N, Holdt L and Dimmeler S:
Long noncoding RNAs: From clinical genetics to therapeutic targets?
J Am Coll Cardiol. 67:1214–1226. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang J, Wang P, Wan L, Xu S and Pang D:
The emergence of noncoding RNAs as Heracles in autophagy.
Autophagy. 13:1004–1024. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dangwal S, Schimmel K, Foinquinos A, Xiao
K and Thum T: Noncoding RNAs in heart failure. Handb Exp Pharmacol.
243:423–445. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bär C, Chatterjee S and Thum T: Long
noncoding RNAs in cardiovascular pathology, diagnosis, and therapy.
Circulation. 134:1484–1499. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Adams BD, Parsons C, Walker L, Zhang WC
and Slack FJ: Targeting noncoding RNAs in disease. J Clin Invest.
127:761–771. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tong X, Gu PC, Xu SZ and Lin XJ: Long
non-coding RNA-DANCR in human circulating monocytes: A potential
biomarker associated with postmenopausal osteoporosis. Biosci
Biotechnol Biochem. 79:732–737. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sha S, Yuan D, Liu Y, Han B and Zhong N:
Targeting long non-coding RNA DANCR inhibits triple negative breast
cancer progression. Biol Open. 6:1310–1316. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mao Z, Li H, Du B, Cui K, Xing Y, Zhao X
and Zai S: LncRNA DANCR promotes migration and invasion through
suppression of lncRNA-LET in gastric cancer cells. Biosci Rep.
37:pii: BSR20171070. 2017. View Article : Google Scholar
|
|
17
|
Jia J, Li F, Tang XS, Xu S, Gao Y, Shi Q,
Guo W, Wang X, He D and Guo P: Long noncoding RNA DANCR promotes
invasion of prostate cancer through epigenetically silencing
expression of TIMP2/3. Oncotarget. 7:37868–37881. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dumars C, Ngyuen JM, Gaultier A, Lanel R,
Corradini N, Gouin F, Heymann D and Heymann MF: Dysregulation of
macrophage polarization is associated with the metastatic process
in osteosarcoma. Oncotarget. 7:78343–78354. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu Y, Zhang M, Liang L, Li J and Chen YX:
Over-expression of lncRNA DANCR is associated with advanced tumor
progression and poor prognosis in patients with colorectal cancer.
Int J Clin Exp Pathol. 8:11480–11484. 2015.PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jiang N, Wang X, Xie X, Liao Y, Liu N, Liu
J, Miao N, Shen J and Peng T: lncRNA DANCR promotes tumor
progression and cancer stemness features in osteosarcoma by
upregulating AXL via miR-33a-5p inhibition. Cancer Lett. 405:46–55.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang L, Yang C, Chen S, Wang G, Shi B,
Tao X, Zhou L and Zhao J: Long noncoding RNA DANCR Is a positive
regulator of proliferation and chondrogenic differentiation in
human Synovium-derived stem cells. DNA Cell Biol. 36:136–142. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yuan SX, Wang J, Yang F, Tao QF, Zhang J,
Wang LL, Yang Y, Liu H, Wang ZG, Xu QG, et al: Long noncoding RNA
DANCR increases stemness features of hepatocellular carcinoma by
derepression of CTNNB1. Hepatology. 63:499–511. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jin L, Fu H, Quan J, Pan X, He T, Hu J, Li
Y, Li H, Yang Y, Ye J, et al: Overexpression of long non-coding RNA
differentiation antagonizing non-protein coding RNA inhibits the
proliferation, migration and invasion and promotes apoptosis of
renal cell carcinoma. Mol Med Rep. 16:4463–4468. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang L, Sun X, Chen S, Yang C, Shi B,
Zhou L and Zhao J: Long noncoding RNA DANCR regulates miR-1305-Smad
4 axis to promote chondrogenic differentiation of human
synovium-derived mesenchymal stem cells. Biosci Rep. 37:pii:
BSR20170347. 2017. View Article : Google Scholar
|
|
26
|
Chen L, Song Z, Huang S, Wang R, Qin W,
Guo J and Lin Z: lncRNA DANCR suppresses odontoblast-like
differentiation of human dental pulp cells by inhibiting
wnt/β-catenin pathway. Cell Tissue Res. 364:309–318. 2016.
View Article : Google Scholar : PubMed/NCBI
|