Introduction
Colorectal cancer (CRC) is the third most common
human malignancy and the fourth most common cause of
cancer-associated mortality globally (1). In China, the morbidity of CRC has been
increasing and was the fourth most common cause of
cancer-associated mortality in 2011 (2). The lack of sensitivity to chemotherapy
and metastasis are the most common reasons for treatment failure
(3) and for the mortality of
patients. Therefore, it is important to elucidate the underlying
molecular mechanisms of migration and invasion of CRC cells.
Long non-coding RNAs (lncRNAs) are a type of
non-coding RNA with >200 nucleotides in length and do not encode
for proteins (4,5). lncRNAs serve a function in tumorigenesis
(3,6–12) and have
been identified in various types of cancer (3,7–11,13–21).
lncRNA H19 is one type of lncRNA and is located on chromosome 11 in
humans and serves an important function in mammalian development
(22). According to previous studies,
H19 is upregulated in numerous types of cancer, including CRC
(23,24), hepatocellular carcinoma (25,26),
esophageal (27), bladder (28) and breast cancer (29,30). The
finding that H19 is overexpressed in cancer tissues suggests that
it may serve an oncogenic function. However, the exact underlying
molecular mechanism of H19 remains unclear.
The RAS/mitogen-activated protein kinase (MAPK)
signaling pathway is an important pathway in human cancer.
Tumorigenic mutations of RAS occur in ~30% of tumors. In this
pathway, activated RAS triggers the activation of RAF and
subsequently activated RAF phosphorylates and activates
mitogen-activated protein kinase kinase (MEK), which phosphorylates
and activates MAPK/extracellular signal-related kinase (ERK)
(31). However, the association
between H19 and the RAS-MAPK signaling pathway in colorectal cancer
remains unresolved. In the present study, it was identified that
H19 increases the migration and invasion of CRC cells by activating
the RAS-MAPK signaling pathway.
Materials and methods
Cell culture and reagents
The human CRC cell lines SW480 and HCT116 were
obtained from the Institute of Biochemistry and Cell Biology
(Shanghai, China). The SW480 and HCT116 cells were cultured in
RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham,
MA, USA), 2% penicillin-streptomycin (10 U/ml) at 37°C in a 5%
CO2 atmosphere. MAPK inhibitor (PD098059) was purchased
from Cell Signaling Technology, Inc. (Danvers, MA, USA). The SW480
and HCT116 cells were cultured with 45 µmol/l PD098059 when
required.
Transfection
pWPXL lentiviral vectors were employed to
overexpress H19 in SW480 and HCT116 cells (32). The full-length sequence of H19
(forward, 5′-GGATCCAGTTAGAAAAAGCCCGGGCT-3′ and reverse,
5′-ACGCGTGCTGTAACAGTGTTTATTGA-3′) was amplified and inserted into
the pWPXL vector (cat. no. 12257; Addgene, Inc., Cambridge, MA,
USA). A small interfering RNA (siRNA) against H19 (siRNA sequence,
5′-GCAGGACAUGACAUGGUCC-3′) and a non-targeted sequence (negative
control, NC, 5′-UCCGCUGACGACAAGGAUG-3′) were synthesized by
Shanghai GenePharma Co., Ltd. (Shanghai, China). The transfection
of plasmid and siRNAs was conducted using Lipofectamine
2000® (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cultured cells
following transfection by TRIzol reagent (Takara Biotechnology Co.,
Ltd., Dalian, China), according to the manufacturer's protocol. A
total of 2 µg RNA was reverse transcribed into cDNA using a
PrimeScript RT Master Mix Perfect Real Time kit (Takara
Biotechnology Co., Ltd.). RT-qPCR was performed using an ABI 7900
RT-PCR system with a SYBR Premix Ex Taq kit (Takara Biotechnology
Co., Ltd.). The H19 and reference gene α-tubulin primers were
synthesized by GenePharma (Shanghai GenePharma Co., Ltd., Shanghai,
China). The RT-qPCR primers were as follows: H19 forward,
5′-CTGGGCAACGGAGGTGTA-3′ and reverse, 5′-CTGGGAGGGTGTCTGCTTC-3′;
α-tubulin forward, 5′-ACCTTAACCGCCTTATTAGCCA-3′ and reverse,
5′-ACATTCAGGGCTCCATCAAATC-3′. The thermocycling conditions were as
follows: 95°C for 10 min, followed by 39 cycles at 95°C for 10 sec
and 60°C for 40 sec. The analysis of RT-qPCR results was performed
using the 2−ΔΔCq method (33).
Ras activity assay
Active Ras Pull-Down and Detection kit (Thermo
Fisher Scientific, Inc.) was utilized according to the
manufacturer's protocol. After 48 h of transfection, human CRC
cells were harvested and prepared in accordance with the
manufacturer's protocol. The cell samples were loaded onto a 10%
SDS-PAGE gel followed by western blotting on nitrocellulose
membrane according to the manufacturer's protocol, and the
polyvinylidene difluoride membrane (PVDF) membranes were incubated
with anti-Ras antibody.
Western blot analysis
Total protein was extracted from the human CRC cells
using radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology, Haimen, China). A total of 50 µg protein was loaded
into each lane on a 10% SDS-PAGE gel and transferred to PVDF
membranes. The membranes were incubated in 5% skimmed milk
dissolved in TBST for 1 h at room temperature, followed by
incubation with anti-Raf (1:1,200; cat. no. ab137435), anti-p-Raf
(1:1,500; cat. no. ab173539), anti-ERK (1:2,000; cat. no. ab54230),
anti-p-ERK (1:1,500; cat. no. ab50011), anti-MEK (1:1,000; cat. no.
ab32091), anti-p-MEK (1:2,000; cat. no. ab96379) and anti-GAPDH
(1:2,000; cat. no. ab9485; all from Abcam, Cambridge, MA, USA)
overnight at 4°C. The membranes were incubated with secondary
peroxidase-conjugated antibody (1:1,500; cat. no.
ab205718/ab205719; Abcam) for 1 h following washing with TBS. The
membranes were assessed using an enhanced chemiluminescence kit
(Beyotime Institute of Biotechnology). The relative level of each
band was measured using ImageJ K.1.45 software (National Institutes
of Health, Bethesda, MD, USA).
Transwell migration assay
A migration assay was conducted using Transwell
chambers (pore size, 8 µm; Costar, Cambridge, MA, USA). Human
colorectal cancer cells that overexpressed H19 or control human
colorectal cancer cells (2×104) were trypsinized and
re-suspended in 200 µl serum-free RPMI-1640 medium. The cells were
seeded into the upper Transwell chamber. The lower chambers were
filled with 800 µl complete RPMI-1640 medium with 10% fetal bovine
serum (Gibco; Thermo Fisher Scientific, Inc.). Following incubation
for 24 h at 37°C, the cells on the upper side of the chamber were
carefully scraped off with cotton swab. The cells that had migrated
to the lower side of the chamber were fixed using 1% crystal violet
for 15 min at room temperature before being counted. The images
were captured using an inverted microscope (magnification, ×200;
Olympus Corporation, Tokyo, Japan). The number of cells that
migrated were counted using ImageJ software.
Transwell invasion assay
Transwell invasion assay was performed using
Transwell chambers with BD Matrigel coating the upper side of the
Transwell insert (Costar). The cells were seeded, and the number of
cells that invaded through the Matrigel insert were counted using
the aforementioned method for the Transwell migration assay.
Statistical analysis
All statistical analyses were performed using SPSS
software (version 17; SPSS, Inc., Chicago, IL, USA). The
significance of the differences between two groups was assessed
using Student's t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Overexpression of H19 promotes the
migration and invasion of CRC cells
In order to elucidate the function of H19 in the
migration and invasion of CRC cells, the pWPXL lentiviral vector
was utilized to overexpress H19 in SW480 and HCT116 cells. In order
to detect H19 expression in CRC cells, RT-qPCR was conducted.
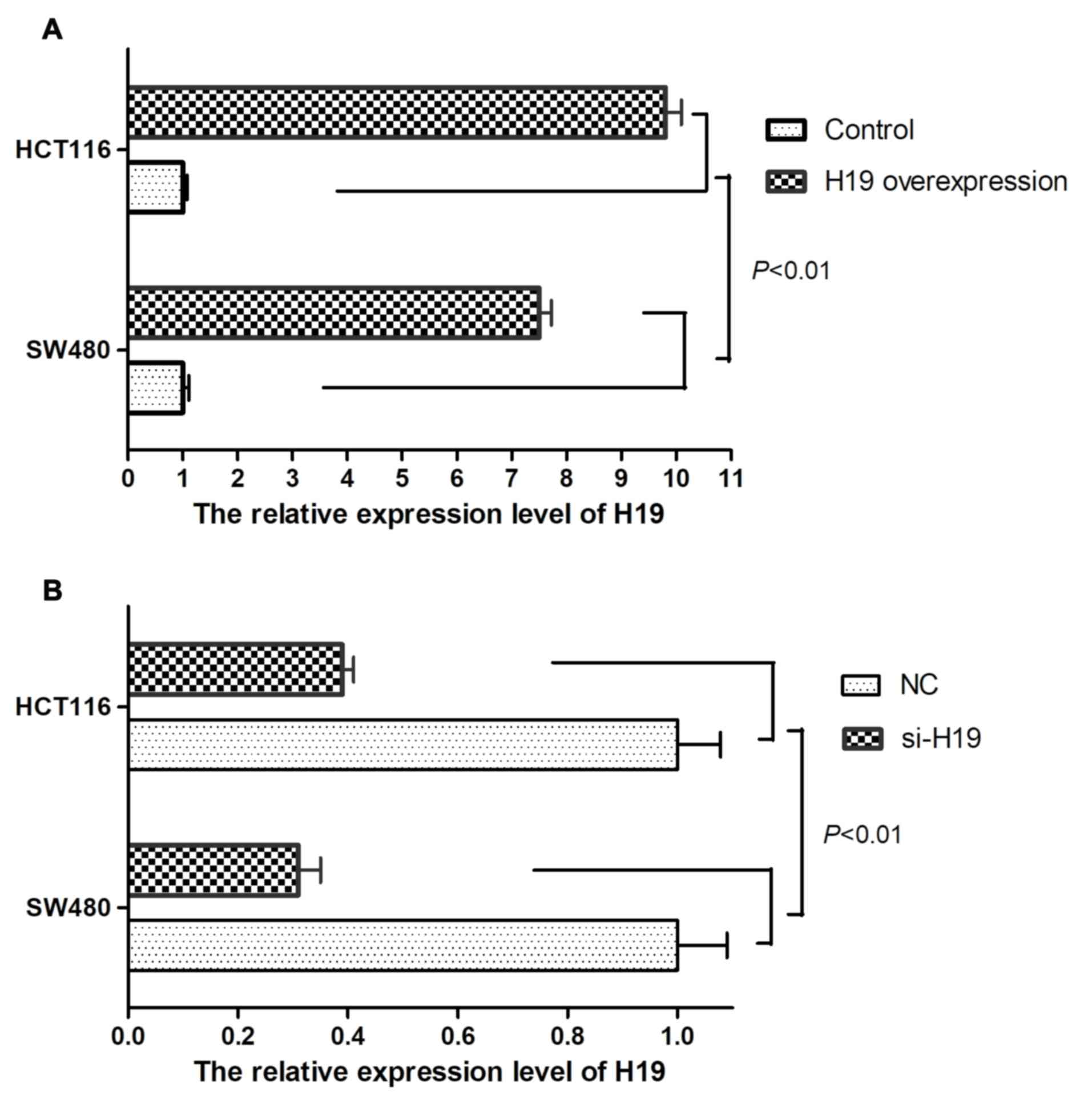
Compared with control CRC cells, overexpression of H19 in SW480 and
HCT116 cells significantly elevated H19 expression levels (Fig. 1A; P<0.01). Whereas, knockdown of
H19 in SW480 and HCT116 cells reduced H19 expression levels
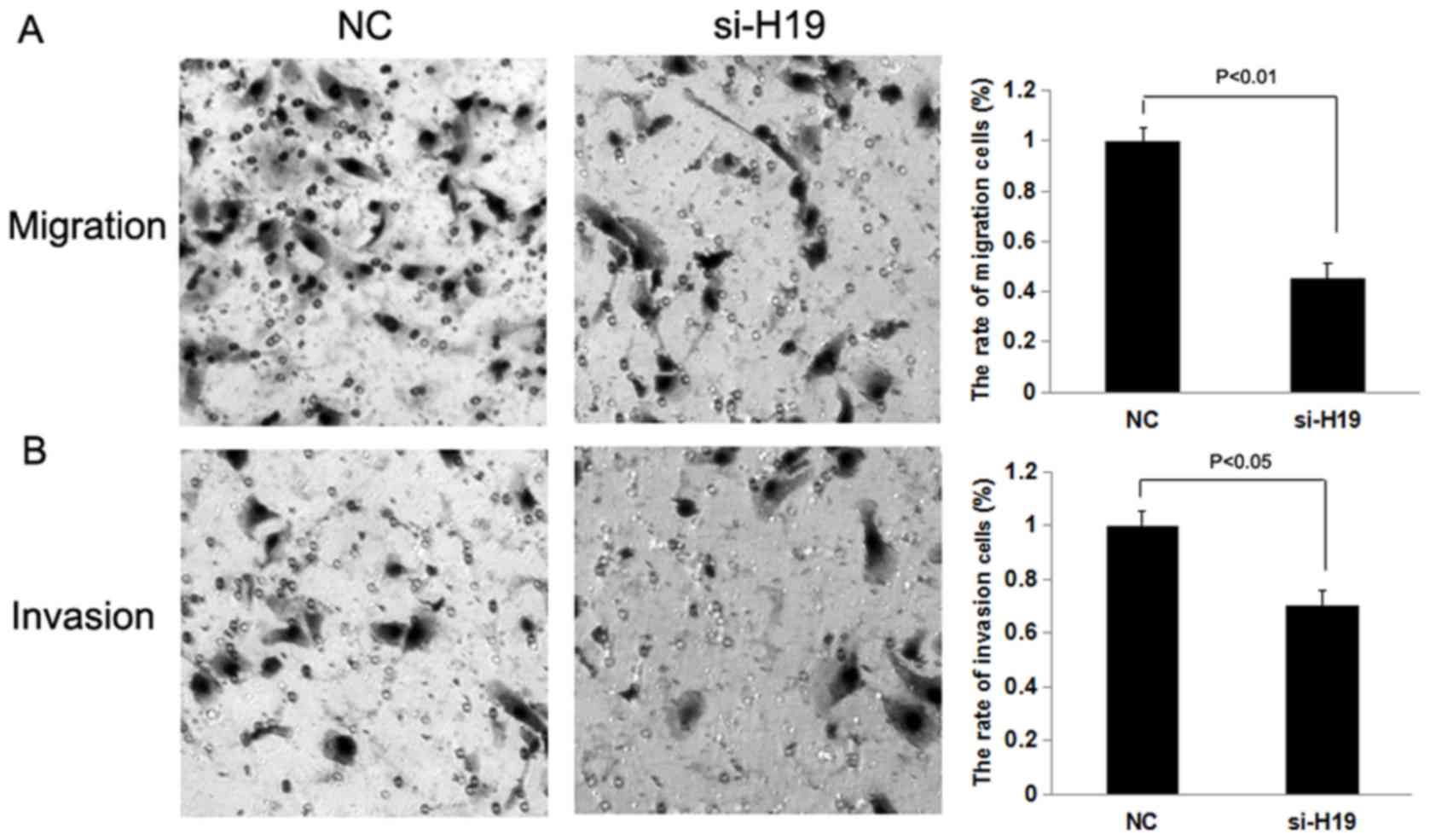
compared with CRC negative control cells (Fig. 1B; P<0.01). Transwell migration and
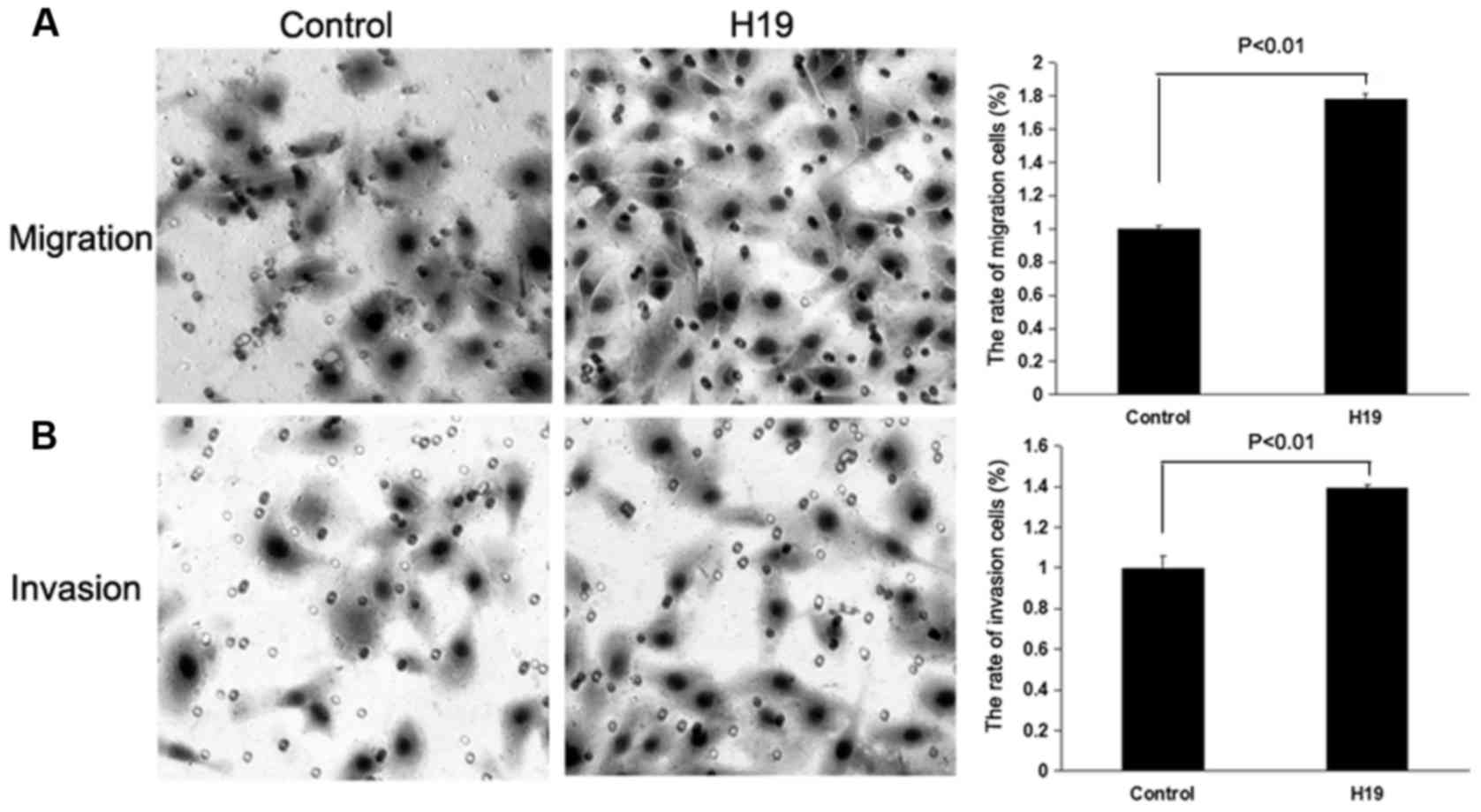
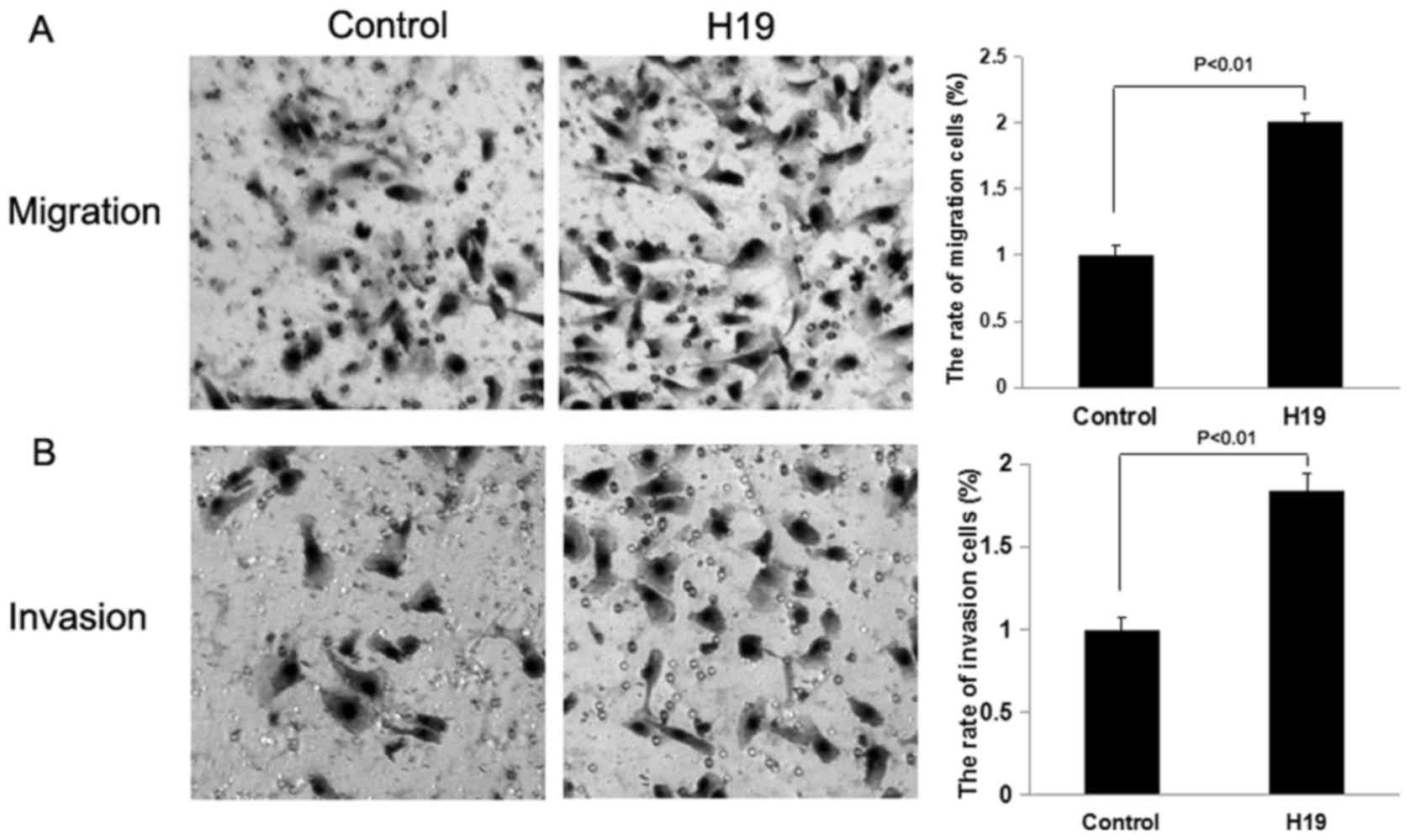
invasion assays were performed to evaluate the migratory and
invasive capacity of the CRC cells. The Transwell migration and
invasion assays demonstrated that the overexpression of H19 in
SW480 and HCT116 cells was able to promote the migration and
invasion of CRC cells compared with the control treatment
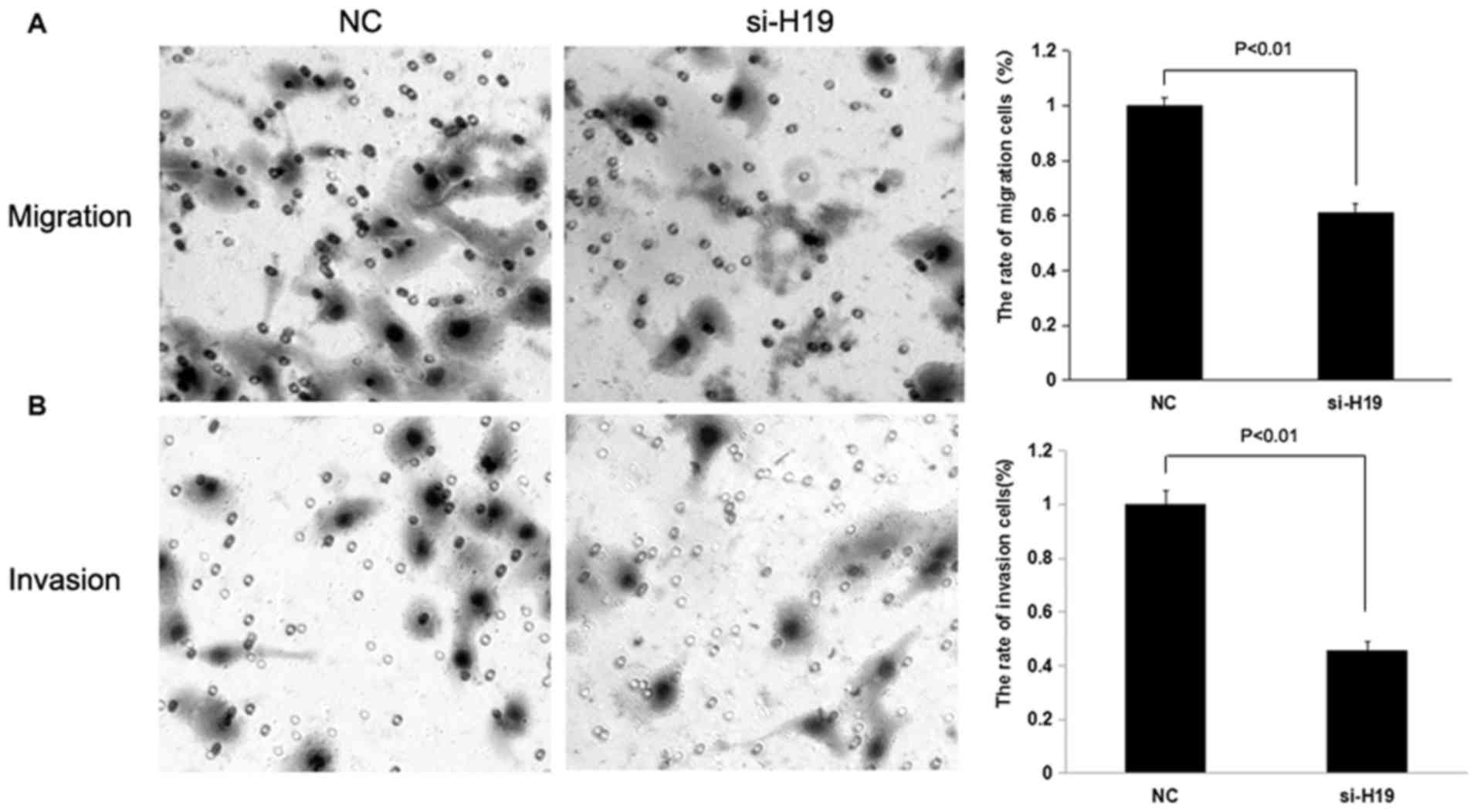
(P<0.01; Figs. 2 and 3), whereas the knockdown of H19
significantly decreased the migration and invasion of SW480 and
HCT116 CRC cells compared with the control treatment group
(P<0.01 and P<0.05; Figs. 4 and
5).
H19 regulates the activation of
Ras
The underlying molecular mechanism of H19 in
promoting cell migration and invasion was investigated. A potential
signaling pathway was identified which may be regulated by H19 and
is associated with migration and invasion of human CRC cells.
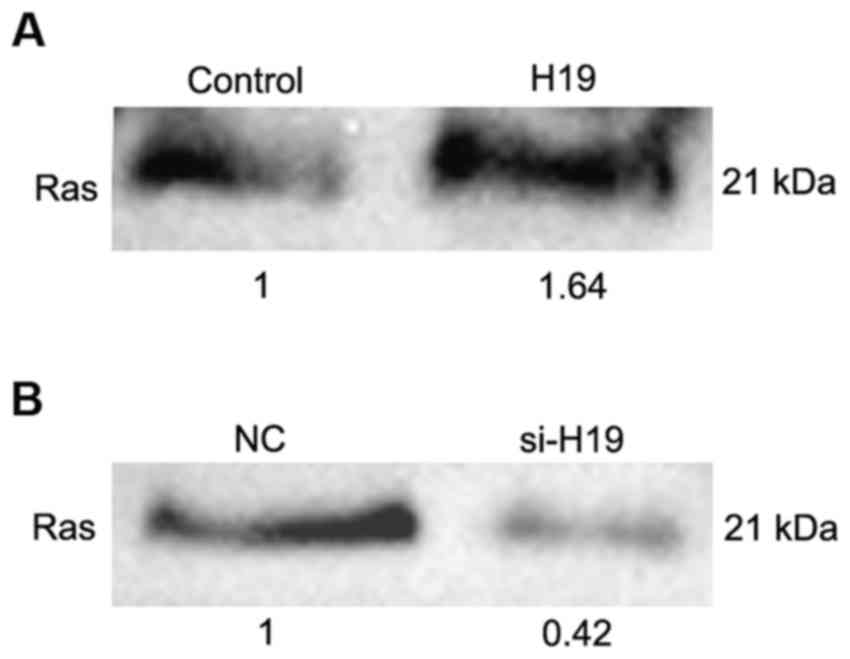
The RAS-MAPK signaling pathway is one of the most
frequently dysregulated pathways in human cancer (31). In order to evaluate the association
between H19 and the RAS-MAPK signaling pathway, a Ras activity
assay was performed following transfection with H19 full-length
sequence and siRNA against H19 in SW480 and HCT116 cells. The
overexpression of H19 was able to upregulate the expression level
of active Ras (Fig. 6A), whereas the
knockdown of H19 was able to downregulate the expression
level of active Ras (Fig. 6B). These
results indicate that H19 may affect the expression level of active
Ras.
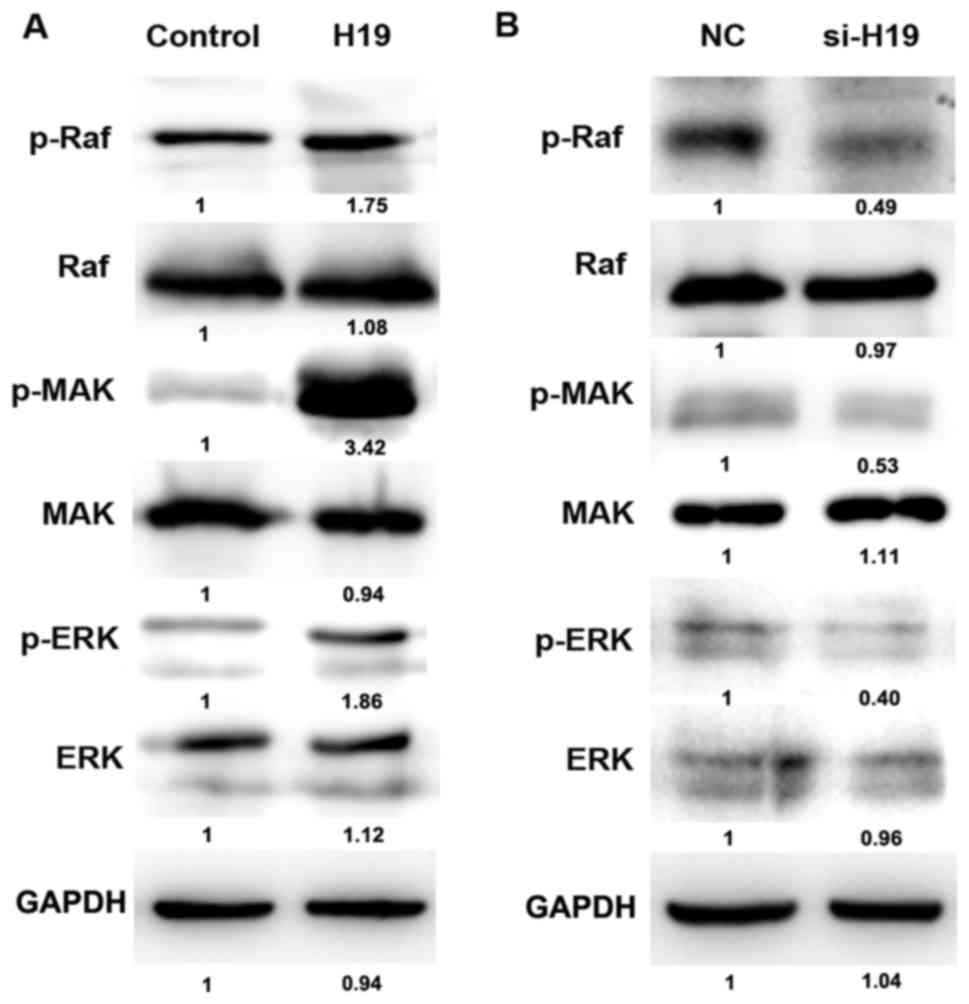
H19 affects the level of p-Raf, p-ERK
and p-MEK
The underlying molecular mechanism of H19 in
promoting cell migration and invasion, and the association between
H19 and the RAS-MAPK signaling pathway were evaluated. The
phosphorylation of Raf, ERK and MEK is an important element of the
RAS-MAPK signaling pathway. The level of p-Raf, p-ERK and p-MEK was
analyzed using western blotting. The overexpression of H19 was able
to increase the phosphorylation level of Raf, ERK and MEK (Fig. 7A), whereas H19 knockdown decreased the
level of p-Raf, p-ERK and p-MEK (Fig.
7B). The results suggest that H19 may be able to regulate the
level of p-Raf, p-ERK and p-MEK.
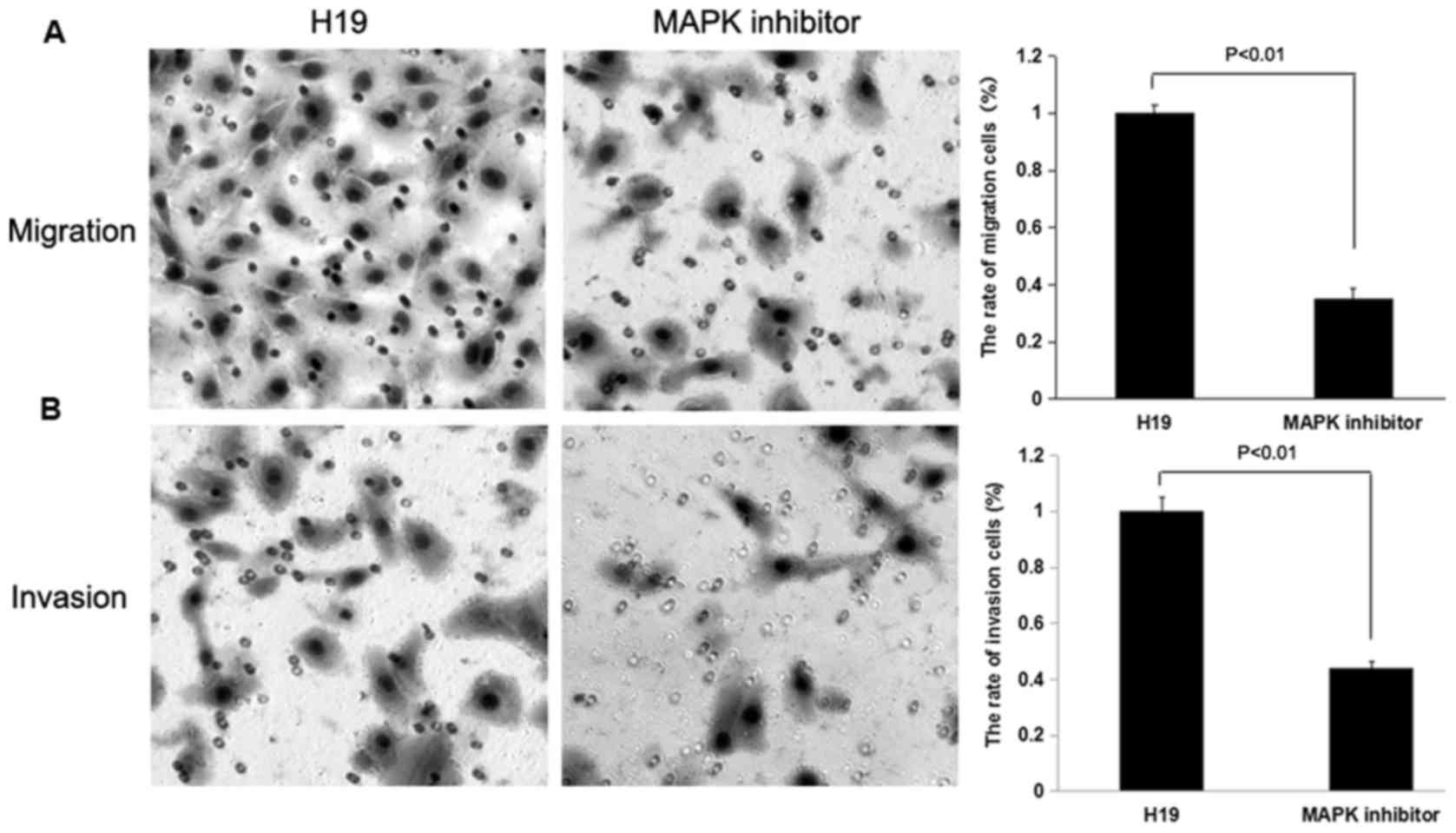
Effect of H19 on cell migration and
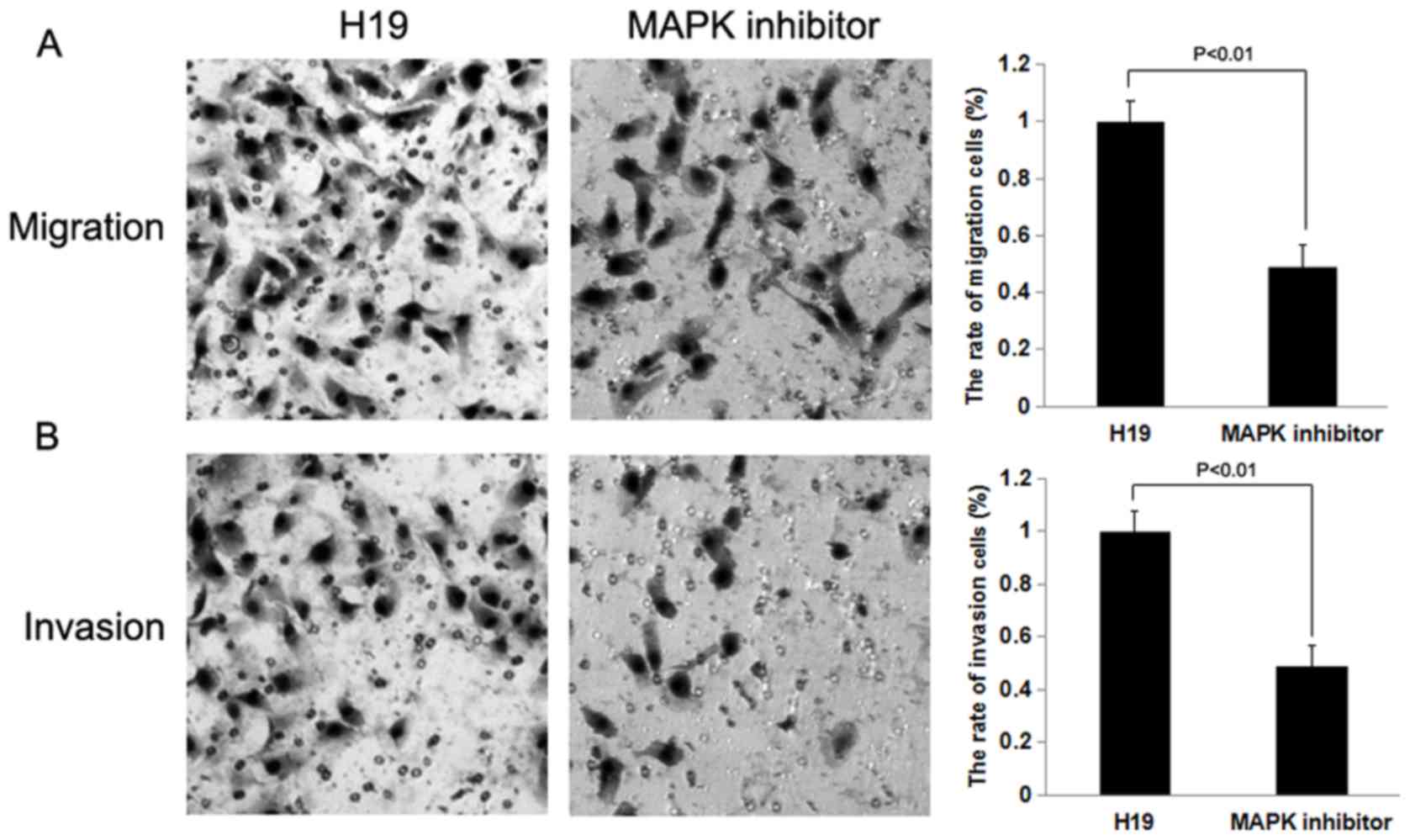
invasion may be reversed by treatment with a MAPK inhibitor
To determine whether H19 promotes cell migration and
invasion through the RAS-MAPK signaling pathway, H19-overexpressed
SW480 and HCT116 cells were treated with a MAPK inhibitor, and
subsequently Transwell migration and invasion assays were
performed. The rates of cell migration and invasion were decreased
following treatment with the MAPK inhibitor (P<0.01; Figs. 8 and 9).
The results indicated that the inhibition of MAPK was able to
decrease the effect of H19 on cell migration and invasion on SW480
and HCT116 cells.
Discussion
lncRNAs serve important functions in a number of
biological processes, including cell cycle, proliferation,
apoptosis, differentiation and invasion (34–36).
Numerous studies have demonstrated that lncRNAs are important
molecules in human malignancies (19–21,37–39).
However, the functions of lncRNAs and the underlying molecular
mechanism of the oncogenicity of lncRNAs are not well understood.
Previous studies have suggested that lncRNA H19 is overexpressed in
a variety of types of cancer and that H19 promotes cancer
progression (22,40,41). For
example, it was previously demonstrated that H19 promoted the
proliferation of colorectal cancer cells via a competing endogenous
RNA mechanism (42). By binding to
miR-200a, H19 increased the level of CTNNB1, which is a target of
miR-200a, and therefore facilitated the proliferation of CRC cells
(42). Similarly, it has been
identified that H19 may promote epithelial to mesenchymal
transition by acting as a ‘miRNA sponge’ in colorectal cancer
(43). Previous research has
demonstrated that H19 is an oncogene and promotes the proliferation
and metastasis of CRC cells (44).
With regard to the underlying molecular mechanisms of H19 in
promoting the progression of CRC, a number of studies have focused
on the functions of H19 as a competing endogenous RNA or ‘miRNA
sponge’ (43,45). In the present study, it has been
demonstrated that H19 may promote the migration and invasion of
colorectal cancer cells via a novel mechanism.
The Ras/Raf/MEK/ERK cascade couples signals from
cell-surface receptors to transcription factors, and they may be
able to regulate gene expression (31). This signaling pathway may also be
activated in certain tumors via the overexpression of wild-type or
mutated receptor, including epidermal growth factor receptor,
chromosomal translocations of breakpoint cluster region-c-abl
oncogene 1 (BCR-ABL) and cytokine receptor mutations e.g., Fms
related tyrosine kinase 3 (31).
Ras, a small guanosine triphosphate-binding protein,
is one of the most common upstream molecules of certain signaling
pathways, including Raf/MEK/ERK (46). In total, four types of Ras have been
identified: Ha-Ras, N-Ras, Ki-Ras 4A and Ki-Ras 4B. Ki-Ras 4A and
Ki-Ras 4B are generated from the same gene by alternative splicing.
Ki-Ras is a strong inducer of the Raf/MEK/ERK signaling cascade
compared with Ha-Ras and is more frequently mutated in human cancer
(47). The overexpression of the
Ras/Raf/MEK/ERK signaling pathway has been implicated in advanced
prostate cancer and has been associated with poor patient prognosis
(48). The molecules in the
Ras/Raf/MEK/ERK signaling pathway, which act as growth factors may
be induced by autocrine and paracrine signaling (49). However, a number of studies assessed
the function of the RAS-MAPK signaling pathway in human colorectal
cancer, particularly the combined functions of H19 and the RAS-MAPK
signaling pathway. In the present study, in order to investigate
whether H19 promotes invasion and migration via activation of MAPK
signaling, MAPK was inhibited and the effect of H19 on migration
and invasion of cancer cells was assessed. The results revealed
that the inhibition of MAPK was able to suppress the pro-metastatic
function of H19, which supports the hypothesis that H19 promotes
the migration and invasion of cancer cells via activation of MAPK
signaling. However, the present study has only identified the
association between H19 and RAS-MAPK signaling pathway in CRC using
in vitro techniques. Therefore, in vivo experiments
should be carried out to corroborate the in vitro data.
In conclusion, H19 may promote the migration and
invasion of CRC cells, by activating Ras protein and upregulating
the levels of p-Raf, p-MEK and p-ERK. H19 may facilitate the
migration and invasion of CRC cells via the RAS-MAPK signaling
pathway.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Fundamental
Research Funds for the Provincial Universities (grant no.
2017-KYYWF-0289), the National Natural Science Foundation of China
(grant no. 81201688) and The Key Program of Pecking University
International Hospital Scientific Research Foundation (grant no.
YN2016ZD04).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
NN conceived and designed the experiments. WY, RER
and CZ performed the experiments and analyzed the data. WY and CZ
wrote the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Karsa LV, Lignini TA, Patnick J, Lambert R
and Sauvaget C: The dimensions of the CRC problem. Best Pract Res
Clin Gastroenterol. 24:381–396. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Zeng H, Zhang S and He J:
Annual report on status of cancer in China, 2011. Chin J Cancer
Res. 27:2–12. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhou M, Zhao H, Wang Z, Cheng L, Yang L,
Shi H, Yang H and Sun J: Identification and validation of potential
prognostic lncRNA biomarkers for predicting survival in patients
with multiple myeloma. J Exp Clin Cancer Res. 34:1022015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
St Laurent G, Wahlestedt C and Kapranov P:
The Landscape of long noncoding RNA classification. Trends Genet.
31:239–251. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shang C, Guo Y, Zhang H and Xue YX: Long
noncoding RNA HOTAIR is a prognostic biomarker and inhibits
chemosensitivity to doxorubicin in bladder transitional cell
carcinoma. Cancer Chemother Pharmacol. 77:507–513. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sun J, Shi H, Wang Z, Zhang C, Liu L, Wang
L, He W, Hao D, Liu S and Zhou M: Inferring novel lncRNA-disease
associations based on a random walk model of a lncRNA functional
similarity network. Mol Biosyst. 10:2074–2081. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou M, Wang X, Li J, Hao D, Wang Z, Shi
H, Han L, Zhou H and Sun J: Prioritizing candidate disease-related
long non-coding RNAs by walking on the heterogeneous lncRNA and
disease network. Mol Biosyst. 11:760–769. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhou M, Xu W, Yue X, Zhao H, Wang Z, Shi
H, Cheng L and Sun J: Relapse-related long non-coding RNA signature
to improve prognosis prediction of lung adenocarcinoma. Oncotarget.
7:29720–29738. 2016.PubMed/NCBI
|
|
10
|
Sun J, Chen X, Wang Z, Guo M, Shi H, Wang
X, Cheng L and Zhou M: A potential prognostic long non-coding RNA
signature to predict metastasis-free survival of breast cancer
patients. Sci Rep. 5:165532015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhou M, Zhao H, Xu W, Bao S, Cheng L and
Sun J: Discovery and validation of immune-associated long
non-coding RNA biomarkers associated with clinically molecular
subtype and prognosis in diffuse large B cell lymphoma. Mol Cancer.
16:162017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhou M, Zhang Z, Zhao H, Bao S, Cheng L
and Sun J: An immune-related six-lncRNA signature to improve
prognosis prediction of glioblastoma multiforme. Mol Neurobiol.
55:3684–3697. 2017.PubMed/NCBI
|
|
13
|
Morris KV: Long antisense non-coding RNAs
function to direct epigenetic complexes that regulate transcription
in human cells. Epigenetics. 4:296–301. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ricciuti B, Mencaroni C, Paglialunga L,
Paciullo F, Crinò L, Chiari R and Metro G: Long noncoding RNAs: New
insights into non-small cell lung cancer biology, diagnosis and
therapy. Med Oncol. 33:182016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Guo Q, Cheng Y, Liang T, He Y, Ren C, Sun
L and Zhang G: Comprehensive analysis of lncRNA-mRNA co-expression
patterns identifies immune-associated lncRNA biomarkers in ovarian
cancer malignant progression. Sci Rep. 5:176832015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Qiu M, Feng D, Zhang H, Xia W, Xu Y, Wang
J, Dong G, Zhang Y, Yin R and Xu L: Comprehensive analysis of
lncRNA expression profiles and identification of functional lncRNAs
in lung adenocarcinoma. Oncotarget. 7:16012–16022. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sun J, Cheng L, Shi H, Zhang Z, Zhao H,
Wang Z and Zhou M: A potential panel of six-long non-coding RNA
signature to improve survival prediction of diffuse large-B-cell
lymphoma. Sci Rep. 6:278422016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang L, Yi K, Wang H, Zhao Y and Xi M:
Comprehensive analysis of lncRNAs microarray profile and
mRNA-lncRNA co-expression in oncogenic HPV-positive cervical cancer
cell lines. Oncotarget. 7:49917–49929. 2016.PubMed/NCBI
|
|
19
|
Zhou M, Guo M, He D, Wang X, Cui Y, Yang
H, Hao D and Sun J: A potential signature of eight long non-coding
RNAs predicts survival in patients with non-small cell lung cancer.
J Transl Med. 13:2312015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou M, Sun Y, Sun Y, Xu W, Zhang Z, Zhao
H, Zhong Z and Sun J: Comprehensive analysis of lncRNA expression
profiles reveals a novel lncRNA signature to discriminate
nonequivalent outcomes in patients with ovarian cancer. Oncotarget.
7:32433–32448. 2016.PubMed/NCBI
|
|
21
|
Zhou M, Wang X, Shi H, Cheng L, Wang Z,
Zhao H, Yang L and Sun J: Characterization of long non-coding
RNA-associated ceRNA network to reveal potential prognostic lncRNA
biomarkers in human ovarian cancer. Oncotarget. 7:12598–12611.
2016.PubMed/NCBI
|
|
22
|
Keniry A, Oxley D, Monnier P, Kyba M,
Dandolo L, Smits G and Reik W: The H19 lincRNA is a developmental
reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell
Biol. 14:659–665. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cui H, Onyango P, Brandenburg S, Wu Y,
Hsieh CL and Feinberg AP: Loss of imprinting in colorectal cancer
linked to hypomethylation of H19 and IGF2. Cancer Res.
62:6442–6446. 2002.PubMed/NCBI
|
|
24
|
Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung
JJ and Kwok TT: Oncofetal H19-derived miR-675 regulates tumor
suppressor RB in human colorectal cancer. Carcinogenesis.
31:350–358. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Matouk IJ, DeGroot N, Mezan S, Ayesh S,
Abu-lail R, Hochberg A and Galun E: The H19 non-coding RNA is
essential for human tumor growth. PLoS One. 2:e8452007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ariel I, Miao HQ, Ji XR, Schneider T, Roll
D, de Groot N, Hochberg A and Ayesh S: Imprinted H19 oncofetal RNA
is a candidate tumour marker for hepatocellular carcinoma. Mol
Pathol. 51:21–25. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hibi K, Nakamura H, Hirai A, Fujikake Y,
Kasai Y, Akiyama S, Ito K and Takagi H: Loss of H19 imprinting in
esophageal cancer. Cancer Res. 56:480–482. 1996.PubMed/NCBI
|
|
28
|
Byun HM, Wong HL, Birnstein EA, Wolff EM,
Liang G and Yang AS: Examination of IGF2 and H19 loss of imprinting
in bladder cancer. Cancer Res. 67:10753–10758. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Berteaux N, Lottin S, Monté D, Pinte S,
Quatannens B, Coll J, Hondermarck H, Curgy JJ, Dugimont T and
Adriaenssens E: H19 mRNA-like noncoding RNA promotes breast cancer
cell proliferation through positive control by E2F1. J Biol Chem.
280:29625–29636. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lottin S, Adriaenssens E, Dupressoir T,
Berteaux N, Montpellier C, Coll J, Dugimont T and Curgy JJ:
Overexpression of an ectopic H19 gene enhances the tumorigenic
properties of breast cancer cells. Carcinogenesis. 23:1885–1895.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
McCubrey JA, Steelman LS, Chappell WH,
Abrams SL, Wong EW, Chang F, Lehmann B, Terrian DM, Milella M,
Tafuri A, et al: Roles of the Raf/MEK/ERK pathway in cell growth,
malignant transformation and drug resistance. Biochim Biophys Acta.
1773:1263–1284. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li X, Zhang J, Gao L, McClellan S, Finan
MA, Butler TW, Owen LB, Piazza GA and Xi Y: MiR-181 mediates cell
differentiation by interrupting the Lin28 and let-7 feedback
circuit. Cell Death Differ. 19:378–386. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Khaitan D, Dinger ME, Mazar J, Crawford J,
Smith MA, Mattick JS and Perera RJ: The melanoma-upregulated long
noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer
Res. 71:3852–3862. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Calin GA, Liu CG, Ferracin M, Hyslop T,
Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, et
al: Ultraconserved regions encoding ncRNAs are altered in human
leukemias and carcinomas. Cancer Cell. 12:215–229. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Braconi C, Valeri N, Kogure T, Gasparini
P, Huang N, Nuovo GJ, Terracciano L, Croce CM and Patel T:
Expression and functional role of a transcribed noncoding RNA with
an ultraconserved element in hepatocellular carcinoma. Proc Natl
Acad Sci USA. 108:786–791. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Matouk IJ, Halle D, Raveh E, Gilon M,
Sorin V and Hochberg A: The role of the oncofetal H19 lncRNA in
tumor metastasis: Orchestrating the EMT-MET decision. Oncotarget.
7:3748–3765. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xia T, Liao Q, Jiang X, Shao Y, Xiao B, Xi
Y and Guo J: Long noncoding RNA associated-competing endogenous
RNAs in gastric cancer. Sci Rep. 4:60882014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang W, Ning N and Jin X: The lncRNA H19
promotes cell proliferation by competitively binding to miR-200a
and derepressing β-catenin expression in colorectal cancer. Biomed
Res Int. 2017:27674842017.PubMed/NCBI
|
|
43
|
Liang WC, Fu WM, Wong CW, Wang Y, Wang WM,
Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF and Waye MM: The lncRNA
H19 promotes epithelial to mesenchymal transition by functioning as
miRNA sponges in colorectal cancer. Oncotarget. 6:22513–22525.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liang W, Zou Y, Qin F, Chen J, Xu J, Huang
S, Chen J and Dai S: sTLR4/MD-2 complex inhibits colorectal cancer
migration and invasiveness in vitro and in vivo by lncRNA H19
down-regulation. Acta Biochim Biophys Sin (Shanghai). 49:1035–1041.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ohtsuka M, Ling H, Ivan C, Pichler M,
Matsushita D, Goblirsch M, Stiegelbauer V, Shigeyasu K, Zhang X,
Chen M, et al: H19 Noncoding RNA, an independent prognostic factor,
regulates essential Rb-E2F and CDK8-β-catenin signaling in
colorectal cancer. EBioMedicine. 13:113–124. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Peyssonnaux C, Provot S,
Felder-Schmittbuhl MP, Calothy G and Eychène A: Induction of
postmitotic neuroretina cell proliferation by distinct Ras
downstream signaling pathways. Mol Cell Biol. 20:7068–7079. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yan J, Roy S, Apolloni A, Lane A and
Hancock JF: Ras isoforms vary in their ability to activate Raf-1
and phosphoinositide 3-kinase. J Biol Chem. 273:24052–24056. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gioeli D, Mandell JW, Petroni GR, Frierson
HF Jr and Weber MJ: Activation of mitogen-activated protein kinase
associated with prostate cancer progression. Cancer Res.
59:279–284. 1999.PubMed/NCBI
|
|
49
|
Abreu-Martin MT, Chari A, Palladino AA,
Craft NA and Sawyers CL: Mitogen-activated protein kinase kinase
kinase 1 activates androgen receptor-dependent transcription and
apoptosis in prostate cancer. Mol Cell Biol. 19:5143–5154. 1999.
View Article : Google Scholar : PubMed/NCBI
|