Introduction
MicroRNAs (miRNAs) are a class of small non-coding
RNA of 18–24 nucleotides, which post-transcriptionally regulate the
translation or degradation of protein-coding genes through
imperfect base pairing with the 3′-untranslated region of target
mRNAs (1). miRNAs are believed to be
linked to tissue morphogenesis and cellular processes, e.g.
apoptosis and major signaling pathways (2). For example, the miR-23a/24-2/27a cluster
promotes cell invasion and metastasis by targeting Sprouty2 in
breast cancer (3). It is reported
that >60% of human protein-coding genes are regulated by miRNAs
(4). As miRNAs may regulate the
expression of tumor-associated genes in multiple tumor types, they
are considered to be potential therapeutic targets (5,6).
MicroRNA-137 (miR-137) is located on chromosome 1p22
and functions as a tumor suppressor by targeting a number of
oncogenic mRNAs (7–10). Recent studies have demonstrated that
miR-137 is downregulated in various types of tumor, suggesting that
it can be regarded as a negative regulator of certain
tumor-associated genes. For example, miR-137 acts as a tumor
suppressor by directly targeting carboxyl-terminal binding protein
1 to inhibit the epithelial-mesenchymal transition and induce the
apoptosis of melanoma cells (11).
miR-137 inhibits the invasive ability of melanoma cells through the
downregulation of melanogenesis associated transcription factor,
enhancer of zeste 2 polycomb repressive complex 2 subunit, c-Met
and Y-box binding protein 1 (12). To
the best of our knowledge, only astrocyte elevated gene-1 has been
reported as a target of miR-137 in human ovarian cancer (13). The function of miR-137 in ovarian
cancer remains to be elucidated. The miR-137-knockout cell model
established in the present study using a CRISPR/Cas9 editing
approach will provide a useful platform for the functional study of
ovarian cancer.
Clustered regularly interspaced short palindromic
repeats (CRISPR) are a distinctive feature of the genomes of
bacteria and archaea (14). Together
with CRISPR-associated system (Cas) genes, they are hypothesized to
be involved in resistance to bacteriophages, and resistance
specificity is determined by spacer-phage sequence similarity
(15). The strategy was first
documented in 1987 when Ishino et al (16) were studying the isozyme conversion of
alkaline phosphatase in Escherichia coli. The CRISPR/Cas9
system, consisting of Cas9 protein and guide RNA (gRNA), utilizes
the type II prokaryotic CRISPR/Cas9 adaptive immune system and
targets the Cas9 nuclease by a 20 nucleotide guide sequence cloned
upstream of a 5′-NGG protospacer adjacent motif (17).
Genome editing using CRISPR/Cas9 is hypothesized to
be a more efficient strategy than designer nuclease technologies,
including zinc-finger nucleases and transcription activator-like
effectors, for the generation of genetically modified cells and
organisms (18,19). These easily engineered systems are
increasingly being employed and have successfully knocked out or
edited protein-coding genes in several model organisms and a
variety of cell lines (20–25). Zhao et al (26) have reported the silencing of
non-coding miRNA genes using CRISPR/Cas9. This suggested that
miR-30a-5p and miR-21 could be edited in cells, and stimulated the
production of the present study, in which an miR-137 gene knockout
A2789 cell model was constructed using this technology. This may
provide the basis for the functional study of miR-137 in ovarian
cancer.
Materials and methods
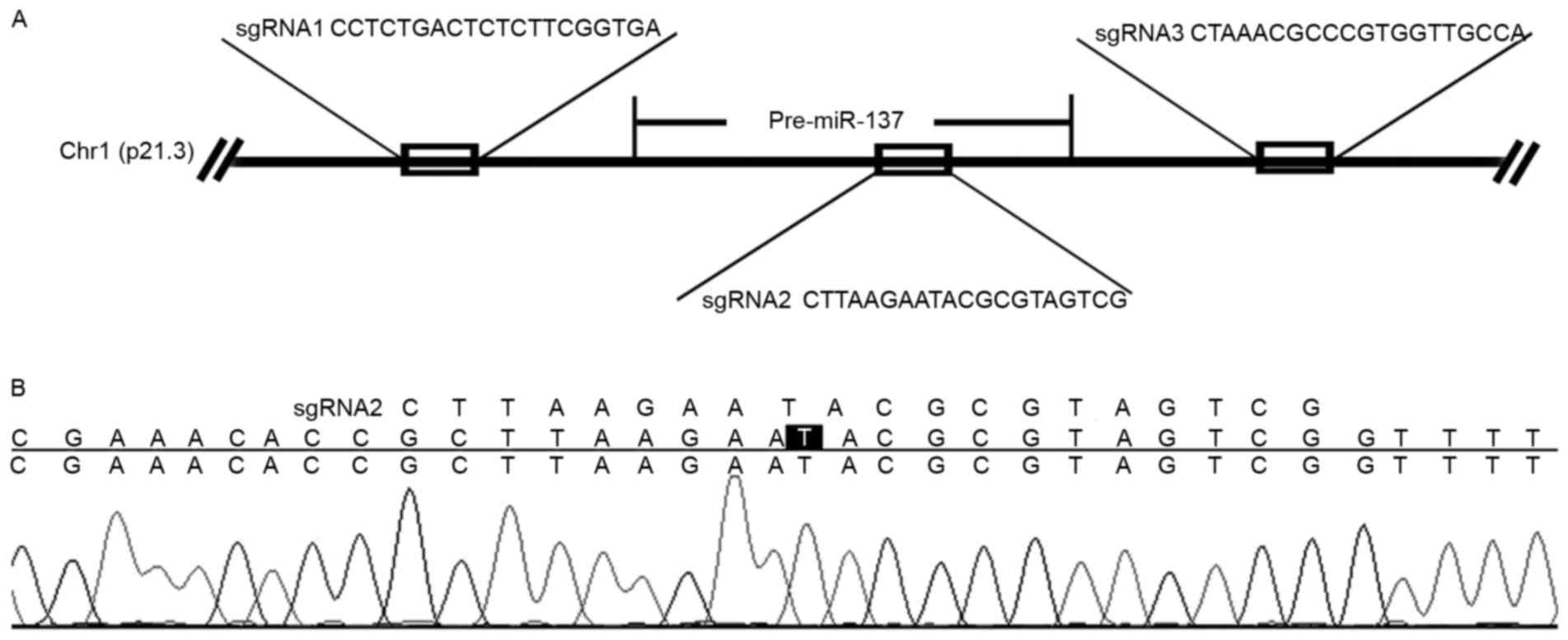
Design of the sgRNAs
The sgRNAs targeting miR-137 were designed using
CRISPR DESIGN (http://crispr.mit.edu/). A total of
320-bp sequences upstream of 5′-NGG were chosen as sgRNAs, and
BsmBIsticky ends were attached to flank the sgRNAs. The
pairs of oligonucleotides designed for the miR-137 site sequences
(Fig. 1A) were sgRNA1, forward,
5′-CACCGCCTCTGACTCTCTTCGGTGA-3′ and reverse,
5′-AAACTCACCGAAGAGAGTCAGAGGC-3′; sgRNA2, forward,
5′-CACCGCTTAAGAATACGCGTAGTC-3′ and reverse,
5′-AAACCGACTACGCGTATTCTTAAGC-3′; sgRNA3, forward,
5′-CACCGCTAAACGCCCGTGGTTGCCA-3′, and reverse,
5′-AAACTGGCAACCACGGGCGTTTAGC-3′.
Construction of sgRNA expression
vectors
Each oligonucleotide was diluted to 10 µM, and each
pair of oligonucleotides were mixed in equal volumes. Next, 1 µl
combined oligonucleotides, 1 µl annealing buffer and 6 µl water
were mixed in a total reaction volume of 8 µl, placed in boiling
water for 10 min and then decreased to 25°C. The annealed
oligonucleotides were added 1 µl ligation buffer and ligated with
BsmB1-digested linearized pXPR001 plasmids (1 µl). The
ligation mix was transformed into competent Dh5α E. coli
cells (Shanghai DingGuo Biotech., Co., Ltd., Shanghai, China) and
the constructs were confirmed by sequencing.
Cell lines and cell culture
293T and A2780 cells from the American Type Culture
Collection (Manassas, VA, USA) were cultured in Dulbecco's modified
Eagle's medium or RPMI, respectively, supplemented with 10% fetal
bovine serum and penicillin/streptomycin (100 U/ml; Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA), and maintained
in an incubator at 37°C with 5% CO2.
Virus generation and infection
293T cells (8×105) were seeded in a 6-cm
plate 1day prior to transfection. Co-transfection of the 1.8 µg
pPAX2 packaging plasmid, 0.8 µg pMD2.G envelope plasmid and 2.5 µg
pXPR001-sgRNA vector was performed using Fugene HD transfection
reagent (Roche Diagnostics, Basel, Switzerland). A total of 48 h
after transfection, viral supernatants were harvested and stored at
−80°C.
Single-cell culture and selection of
miR-137-knockout clones
A2780 cells were seeded in a 6-well plate at
4×105 cells/well and 100 µl viral supernatants were
added 24 h later. Puromycin (800 ng/ml)was added 1 day after
transduction for a 2-day period of selection. The remaining cells
were sorted into 96-well plates by flow cytometry for single-cell
culture. When the cells reached a certain density, they were then
transferred to 24-well plates. In total, ~500 cells were used for
direct identification using PCR. These cells were directly added to
25 µl PCR system. The PCR system consisted of 12.5 µl 2× PCR Buffer
for KOD FX, 5 µl 2 mm dNTP, 1.5 µl positive and 5 µM reverse
primers, 2 µl PCR template cells and 4 µl ddH2O.
Polymerase chain reaction (PCR) was performed using KOD FX DNA
polymerase (Toyobo Life Science, Osaka, Japan), using primers
covering the 3 miR-137 targeting sites to identify the cell clones
with miR-137 gene disruption. The primer sequences used were as
follows: forward, 5′-TGGATCCTTCTTTAGGGAAATCGAGT-3′, reverse,
5′-GAAAACACCCGAGGAAATGAAAAGAAC-3′. Thermocycling took place as
follows: denaturation at 95°C for 5 min, followed by 35 cycles of
denaturation at 98°C for 10 sec, annealing and extension at 68°C
for 40 sec. The PCR products were separated in a 1.5% agarose gel,
and were extracted from the gel using a gel purification kit (Omega
Bio-Tek, Inc., Norcross, GA, USA). The sequence of the sgRNA2
construct is illustrated in Fig.
1B.
RNA extraction & reverse
transcription-quantitative (RT-q)PCR
Total RNA was extracted from the cells with TRIzol
(Invitrogen; Thermo Fisher Scientific, Inc.) and was reverse
transcribed by ReverTraAce-α-Transcriptase (Toyobo Life Science),
according to the manufacturers' protocol. The expression of miR-137
in cell lines was quantified using a SYBR® Premix Ex
Taq™ II (TliRNaseH Plus) kit (Takara Bio, Inc., Otsu, Japan); U6
was used as an internal control. Thermocycling was performed using
a Light Cycler 480 Real-Time PCR system (Roche Diagnostics, Basel,
Switzerland) with the following conditions: denaturation at 95°C
for 5 min, followed by 40 cycles of denaturation at 95°C for 10
sec, annealing at 60°C for 20 sec, and extension at 72°C for 30
sec. The data were analyzed using LightCycler 480 software version
1.5 (Roche Diagnostics, Basel, Switzerland). The relative fold
changes of miR-137 expression in the miR-137-knockout cells vs.
control cells were calculated using the 2−ΔΔCq method
(27). The primers used were as
follows: miR-137RT:
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCTACGCGT-3′, miR-137
forward, 5′-CCAGCTGGGTTAAGAATACGCGTAG-3′, miR-137 reverse,
5′-CTGGTGTCGTGGAGTCGGCAATT-3′; U6 RT,
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAAAAATATG-3′, U6 forward,
5′-CTCGCTTCGGCAGCACA-3′, U6 reverse,
5′-AACGCTTCACGAATTTGCGT-3′.
MTT assay
A2780 cells and miR-137-knockout single-cell clones
derived from A2780 cells were seeded in a 96-well plate at
1×104 cells/well. At 72 h, 5 mg/ml MTT was added to the
medium, and the plate was incubated in a 37°C, 5% CO2
incubator for 4 h. DMSO was added at a volume of 100 ml/well, and
the plate was thoroughly mixed for 10 min. The absorbance of the
samples was measured at 490 nm by a microplate reader (BioTek
Instruments, Inc., Winooski, VT, USA).
Statistical analysis
GraphPad Prism (version 5.0; GraphPad Software,
Inc., La Jolla, CA, USA) was used for data analysis. The data are
presented as the mean ± standard deviation of ≥3 separate
experiments and were analyzed using one-way analysis of variance
and Dunnett's post-hoc test). P<0.05 was considered to indicate
a statistically significant difference. P<0.05, 0.01, 0.001 or
0.0001 are indicated by asterisks *, **, ***, or ****,
respectively.
Results
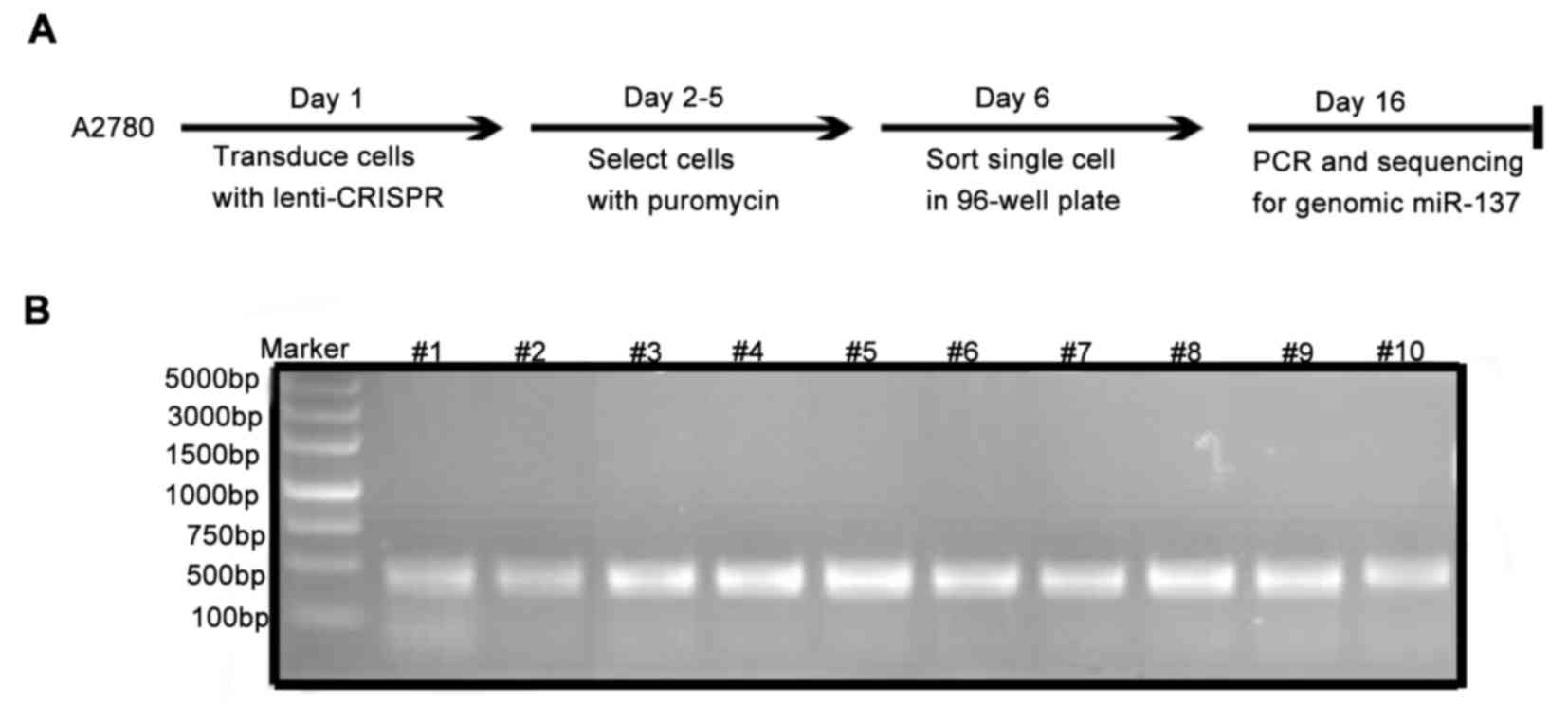
Selection of the single-cell cultures
with miR-137-knockout
The expression level of miR-137 in A2780 was the
highest in a panel of 7 ovarian cancer cell lines (data not shown).
Therefore, A2780 cells were selected for the establishment of the
miR-137-knockout cell model. Single-cell cultures
exhibitingmiR-137-knockout were selected according to the process
illustrated in Fig. 2A. The
transduced cells were selected in 800 nM puromycin for 2 days, and
the single-cell cultures with miR-137 disruption were identified
via sequencing the PCR products (Fig.
2B) amplified from single-cell cultures using primers extending
across the 3 miR-137-targeting sites.
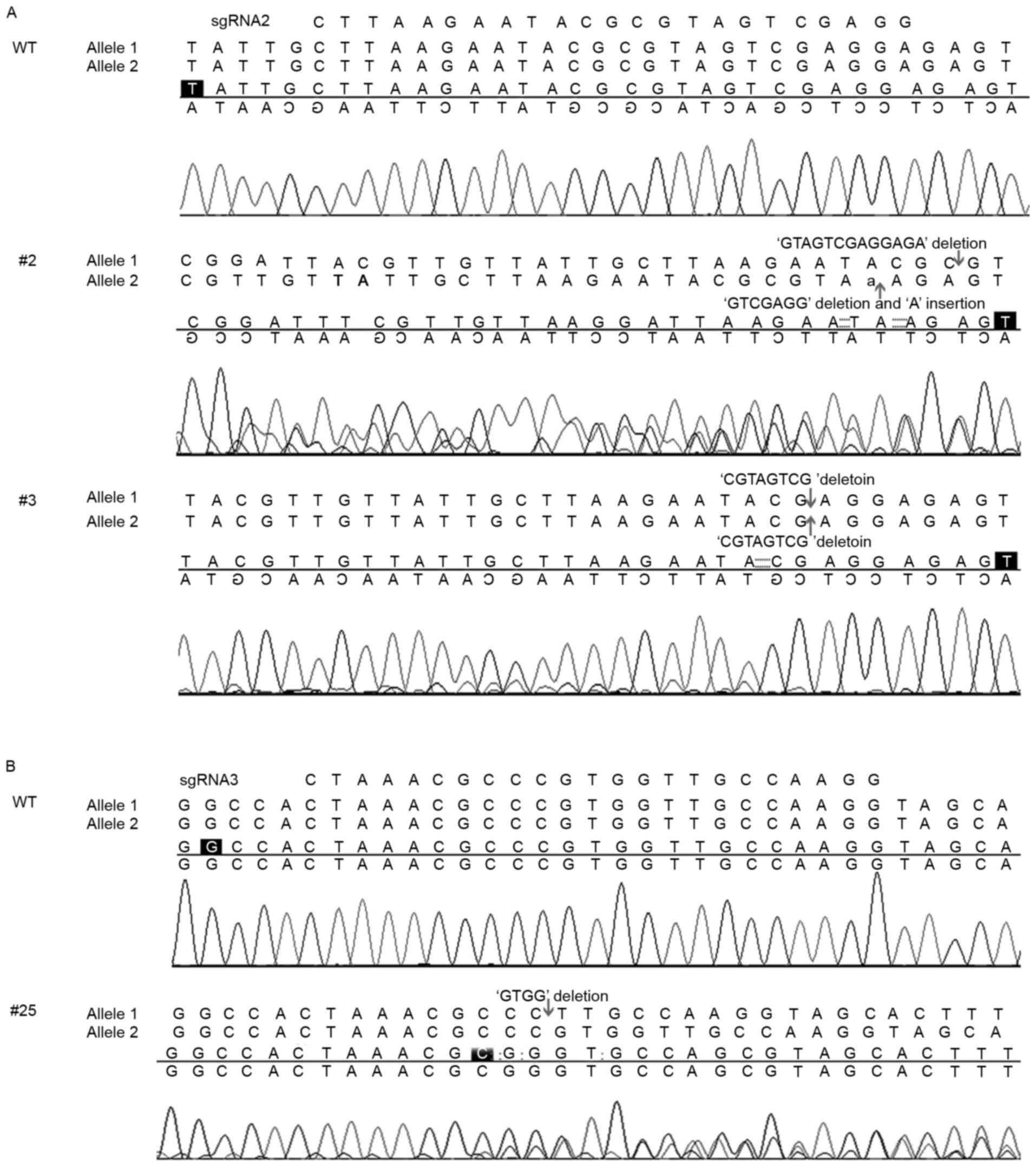
A total of 25 single-cell cultures transduced with
each sgRNA were selected for miR-137-knockout identification. None
of the sgRNA1 cultures were positive for miR-137-knockout; however,
18/25 single-cell cultures of sgRNA2 and 1/25 single-cell cultures
of sgRNA3 were positive for miR-137-knockout. This demonstrated a
higher gene knockout efficiency of sgRNA2. Further analysis
demonstrated that 32% of the sgRNA2 single-cell cultures had
1allele of miR-137 edited, and 40% had 2alleles edited. For
example, in the target site region of clone #2, a 2-allele knockout
single-cell culture, there were 17 and 7-base deletions for each
allele. In the target site region of clone #3, another 2-allele
knockout single-cell culture, there were 8-base deletions for both
alleles (Fig. 3A). For comparison,
the sequencing result of a miR-137-knockout single-cell culture of
sgRNA3 is included in Fig. 3B, which
did not disrupt the pre-miR-137 sequence. Therefore, clones #2 and
#3 derived from sgRNA2 were selected for further experiments.
Establishment of the miR-137-knockout
cell model
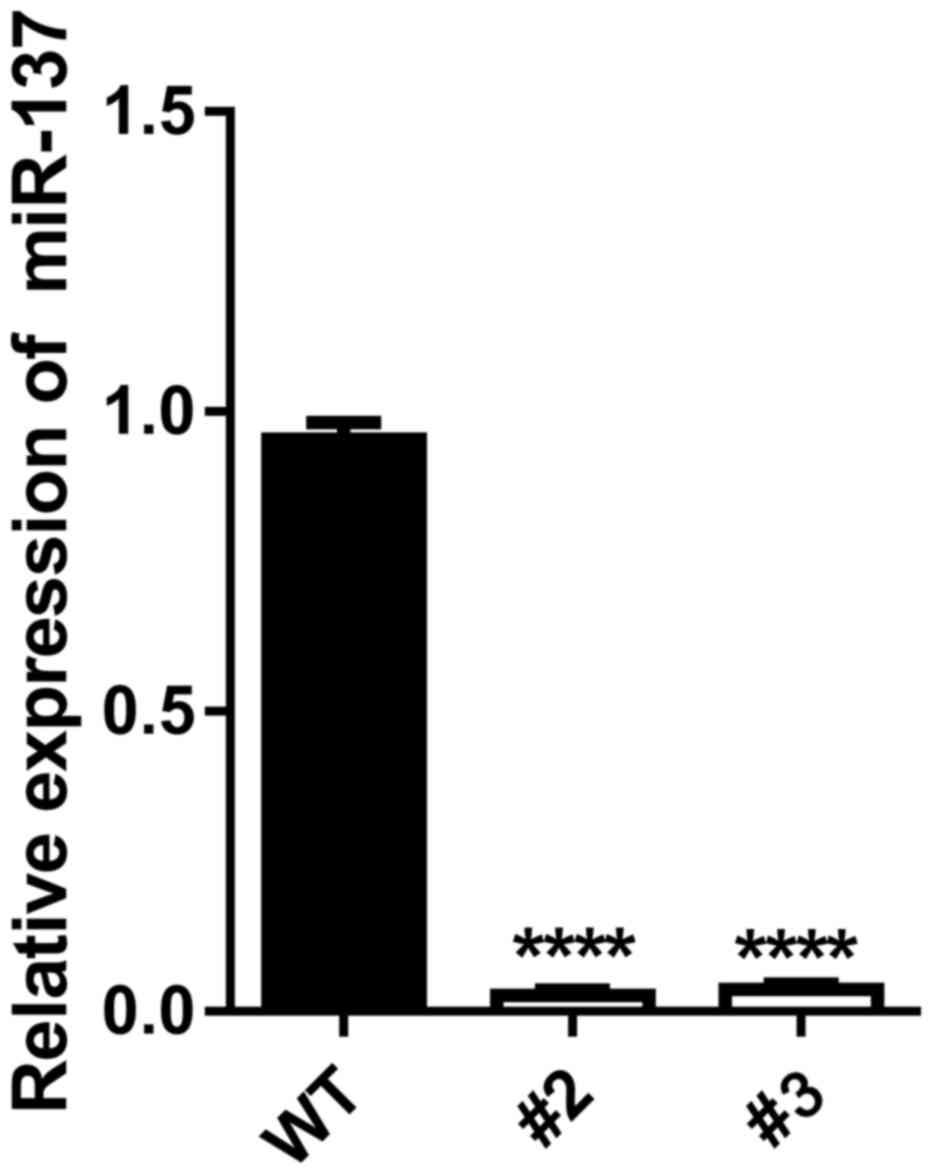
RT-qPCR was performed to detect the expression
levels of miR-137 in the miR-137-knockout clones. The RT-qPCR
results of clones #2 and #3 indicated the low expression levels of
miR-137 compared with wild-type cells (P<0.0001; Fig. 4). This suggested that the cell model
with miR-137-knockout had been successfully established using
CRISPR/Cas9 technology.
miR-137-knockout promotes
proliferation in A2780 cells
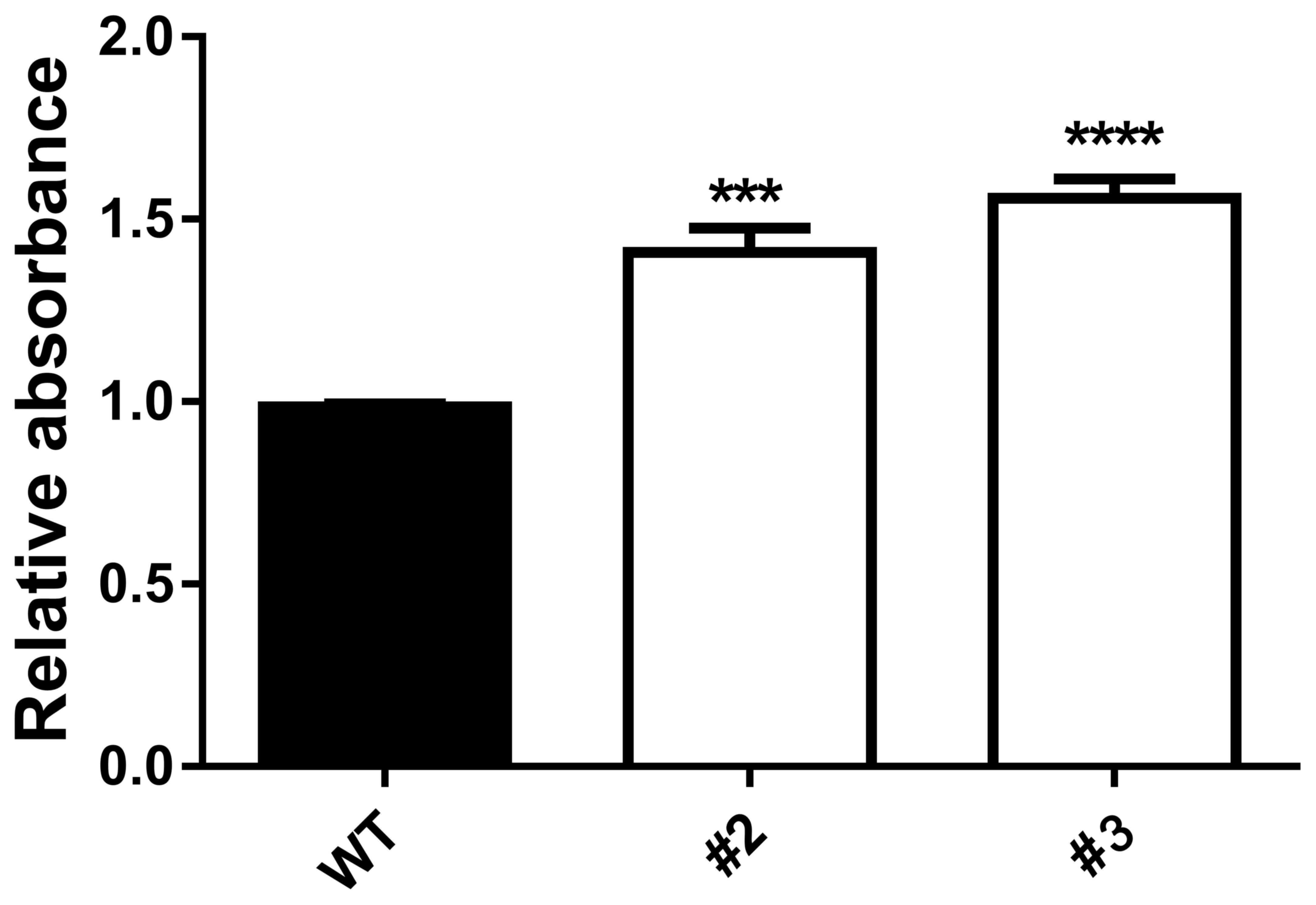
To investigate the effect of miR-137-knockout in
A2780 ovarian cancer cells, the rate of proliferation was assessed
using an MTT assay. A total of 72 h after seeding, the relative
absorbance values of the miR-137-knockout clones (#2 and #3) were
significantly increased compared with A2780 cells expressing the
CRISPR/cas9 empty vector (WT; P<0.001 for #2 and P<0.0001 for
#3; Fig. 5). These results suggest
that the miR-137-knockout promoted the proliferation of A2780
cells.
Discussion
Precise and efficient genome-targeting technologies
allowing selective perturbation of individual genetic elements can
facilitate the reverse engineering or reconstruction of useful
biological systems. Although genome-editing technologies, including
designer zinc fingers and transcription activator-like effectors,
have begun to enable targeted genome modifications (19), each of these platforms has limitations
(28). Genome editing using
CRISPR/Cas9 is a more affordable and efficient strategy for the
generation of gene-modified cells and organisms. It is now the most
popular tool for the precise alteration of the genomes of diverse
species. Its application in genome-wide studies will enable
large-scale screening for drug targets and enhance the development
of human gene therapy (29).
Mali et al (17) obtained targeting rates of 10–25% of
the endogenous AAVS1 locus in 293T cells, 13–38% in K562 cells, and
2–4% in induced pluripotent stem cells. In the present study,
miR-137 gene knockout efficiency was 56% for sgRNA2 and 2% for
sgRNA3, whereas sgRNA1 demonstrated no editing efficiency,
indicating that differences in target sequences affected the
efficiency of knockout.
miRNAs can regulate gene expression in diverse
biological processes, including immune modulation, metabolic
control, neuronal development, cell cycle, muscle differentiation
and stem cell differentiation (2).
Previous studies have indicated that miR-137 serves critical roles
in gastric cancer, glioblastoma, non-small cell lung cancer,
colorectal cancer, ovarian cancer and neuroblastoma (9,25–29). In the present study, sgRNAs were
designed and synthesized to target miR-137 in A2780 cells. The
delivering Cas9 nuclease and miR-137-specific sgRNAs were
transformed into lentiviruses, which were used to infect the A2780
cells. These cells were selected with puromycin and sorted into
96-well plates for single-cell culture. The RT-qPCR results
demonstrated that miR-137 was minimally detected in the
miR-137-knockout cells. An MTT assay indicated that
miR-137-knockout promoted proliferation in A2780 cells. Thus, the
miR-137-knockout A2780 cell model was successfully established
using CRISPR/Cas9 technology and exhibited a functional effect.
Acknowledgements
The present study was supported by funding from the
Health and Family Planning Commission of Guangdong Province (grant
no., A2015314).
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li X, Liu X, Xu W, Zhou P, Gao P, Jiang S,
Lobie PE and Zhu T: c-MYC-regulated miR-23a/24-2/27a cluster
promotes mammary carcinoma cell invasion and hepatic metastasis by
targeting Sprouty2. J Biol Chem. 288:18121–18133. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Christopher AF, Kaur RP, Kaur G, Kaur A,
Gupta V and Bansal P: MicroRNA therapeutics: Discovering novel
targets and developing specific therapy. Perspect Clin Res.
7:68–74. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Feng Q, Wu Q, Liu X, Xiong Y and Li H:
MicroRNA-137 acts as a tumor suppressor in osteosarcoma by
targeting enhancer of zeste homolog 2. Exp Ther Med. 13:3167–3174.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li X, Chen W, Zeng W, Wan C, Duan S and
Jiang S: microRNA-137 promotes apoptosis in ovarian cancer cells
via the regulation of XIAP. Br J Cancer. 116:66–76. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun L, Liang J, Wang Q, Li Z, Du Y and Xu
X: MicroRNA-137 suppresses tongue squamous carcinoma cell
proliferation, migration and invasion. Cell Prolif. 49:628–635.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu L, Chen J, Ding C, Wei S, Zhu Y, Yang
W, Zhang X, Wei X and Han D: MicroRNA-137 contributes to dampened
tumorigenesis in human gastric cancer by targeting AKT2. PLoS One.
10:e01301242015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Deng Y, Deng H, Bi F, Liu J, Bemis LT,
Norris D, Wang XJ and Zhang Q: MicroRNA-137 targets
carboxyl-terminal binding protein 1 in melanoma cell lines. Int J
Biol Sci. 7:133–137. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Luo C, Tetteh PW, Merz PR, Dickes E,
Abukiwan A, Hotz-Wagenblatt A, Holland-Cunz S, Sinnberg T, Schittek
B, Schadendorf D, et al: miR-137 inhibits the invasion of melanoma
cells through downregulation of multiple oncogenic target genes. J
Invest Dermatol. 133:768–775. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guo J, Xia B, Meng F and Lou G: miR-137
suppresses cell growth in ovarian cancer by targeting AEG-1.
Biochem Biophys Res Commun. 441:357–363. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jansen R, Embden JD, Gaastra W and Schouls
LM: Identification of genes that are associated with DNA repeats in
prokaryotes. Mol Microbiol. 43:1565–1575. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Barrangou R, Fremaux C, Deveau H, Richards
M, Boyaval P, Moineau S, Romero DA and Horvath P: CRISPR provides
acquired resistance against viruses in prokaryotes. Science.
315:1709–1712. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ishino Y, Shinagawa H, Makino K, Amemura M
and Nakata A: Nucleotide sequence of the iap gene, responsible for
alkaline phosphatase isozyme conversion in Escherichia coli, and
identification of the gene product. J Bacteriol. 169:5429–5433.
1987. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mali P, Yang L, Esvelt KM, Aach J, Guell
M, DiCarlo JE, Norville JE and Church GM: RNA-guided human genome
engineering via Cas9. Science. 339:823–826. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cong L, Ran FA, Cox D, Lin S, Barretto R,
Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA and Zhang F:
Multiplex genome engineer using CRISPR/Cas systems. Science.
339:819–823. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
LaFountaine JS, Fathe K and Smyth HD:
Delivery and therapeutic applications of gene editing technologies
ZFNs, TALENs, and CRISPR/Cas9. Int J Pharm. 494:180–194. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sakuma T, Nishikawa A, Kume S, Chayama K
and Yamamoto T: Multiplex genome engineering in human cells using
all-in-one CRISPR/Cas9 vector system. Sci Rep. 4:54002014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schwank G, Koo BK, Sasselli V, Dekkers JF,
Heo I, Demircan T, Sasaki N, Boymans S, Cuppen E, van der Ent CK,
et al: Functional repair of CFTR by CRISPR/Cas9 in intestinal stem
cell organoids of cystic fibrosis patients. Cell Stem Cell.
13:653–658. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu Y, Liang D, Wang Y, Bai M, Tang W, Bao
S, Yan Z, Li D and Li J: Correction of a genetic disease in mouse
via use of CRISPR-Cas9. Cell Stem Cell. 13:659–662. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hruscha A, Krawitz P, Rechenberg A,
Heinrich V, Hecht J, Haass C and Schmid B: Efficient CRISPR/Cas9
genome editing with low off-target effects in zebrafish.
Development. 140:4982–4987. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Friedland AE, Tzur YB, Esvelt KM,
Colaiacovo MP, Church GM and Calarco JA: Heritable genome editing
in C. Elegans via a CRISPR-Cas9 system. Nat Methods. 10:741–743.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yu Z, Ren M, Wang Z, Zhang B, Rong YS,
Jiao R and Gao G: Highly efficient genome modifications mediated by
CRISPR/Cas9 in Drosophila. Genetics. 195:289–291. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhao Y, Dai Z, Liang Y, Yin M, Ma K, He M,
Ouyang H and Teng CB: Sequence-specific inhibition of microRNA via
CRISPR/CRISPRi system. Sci Rep. 4:39432014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hsu PD, Lander ES and Zhang F: Development
and applications of CRISPR-Cas9 for genome engineering. Cell.
157:1262–1278. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Doudna JA and Charpentier E: Genome
editing. The new frontier of genome engineering with CRISPR-Cas9.
Science. 346:92014. View Article : Google Scholar
|