Introduction
Gastric cancer (GC) represents the fourth most
common malignant neoplasm and the second leading cause of
cancer-related death worldwide. More than 40% of all GC cases
diagnosed annually occur in China, of whom almost 50% are diagnosed
at an advanced stage and cannot be cured. Treatment of advanced GC
is challenging. Epirubicin, cisplatin, and fluorouracil (ECF) and
its modified regimens, such as EOF (epirubicin, oxaliplatin, and
fluorouracil) and EOX (epirubicin, oxaliplatin, and capecitabine),
are widely used to treat GC patients based on the results of phase
III clinical trials. However, although these regimens are common
first-line treatments, their response rates remain <50%.
Biomarkers able to predict chemotherapeutic efficacy are therefore
urgently needed.
Both membrane transporters and metabolic enzymes
affect cytotoxic-drug metabolism. Solute carrier (SLC) superfamily
proteins and ATP-binding cassette (ABC)-transporters are vital
membrane transporters. SLCO1B1 encodes the transporter
protein, organic anion-transporting polypeptide-1 (OATP1B1), which
mediates liver uptake of a wide variety of drugs, and its role in
the efficacy of cytotoxic drugs, including 5-fluorouracil (5FU)
(1,2),
methotrexate (MTX) (3–5), irinotecan (6,7), and
paclitaxel (8) has been widely
reported. MTX was the first cytotoxic drug reported to be
associated with SLCO1B1, and SLCO1B1 SNPs were shown
to affect MTX pharmacokinetics in children with acute lymphoblastic
leukemia, particularly in terms of deposition and toxicity
(5). These findings have been
validated in several later studies (9–11). The
breast cancer resistance protein (BCRP/ABCG2) has been reported to
affect drug resistance in many cancer types, including colorectal
cancer, lymphoblastic leukemia, and breast cancer (12–15), and
ABCG2 polymorphisms are prognostic factors in breast cancer
patients treated with anthracycline-based neoadjuvant chemotherapy
(16). Expression levels of glucose
transporters (GLUT/SLC1A) were also shown to be related to response
to 5FU chemotherapy in GC cells (17), and glucose transporters were reported
to be independent prognostic factors in patients with GC (18,19).
Among metabolic enzymes, the cytochrome-P450 (CYP)
enzyme family plays an important role in the metabolism of various
anticancer drugs (20). Several
studies have shown that SNPs in CYP2C9 influence
disease-free survival in breast cancer patients treated with
tamoxifen (21,22). Moreover, CYP2C9 polymorphism
was related to response to fluorouracil-based neoadjuvant
chemotherapy in breast cancer (23).
CYP2C19 is involved in the metabolism of cyclophosphamide (24,25) and
tamoxifen (26,27), and P450 enzymes in human liver
microsomes, including CYP1A2 have also been reported to catalyze
tegafur into 5FU (28).
Metabolism-related genes, including SLCO1B1,
ABCG2, SLC2A9, CYP2C9, CYP2C19, and CYP1A2, might thus
influence the efficacy of EOF regimens. In the present study, we
investigated the associations between metabolism-related genes and
the clinical outcomes of GC patients treated with first-line EOF
regimens, in terms of disease-control rate (DCR), progression-free
survival (PFS), and overall survival (OS).
Materials and methods
Study population
This retrospective study enrolled 108 consecutive
Chinese Han patients with metastatic GC (MGC) treated with EOF
regimens as first-line chemotherapy at Fudan University Shanghai
Cancer Center (Shanghai, China) between May 2009 and June 2012.
Their diagnoses were pathologically confirmed as gastric
adenocarcinoma. The study was approved by the Ethics Committee of
Fudan University Shanghai Cancer and complied with the principles
of the Helsinki Accord. This was a retrospective study and patient
consent was therefore deemed unnecessary. However, blood samples
were collected from all subjects before treatment, with patient
consent, and stored in the tissue bank at Fudan University Shanghai
Cancer Center.
Treatment
All patients in this study were treated with
first-line chemotherapy using an EOF regimen, consisting of
epirubicin infusion (50 mg/m2) combined with an intravenous
infusion of oxaliplatin (130 mg/m2) for 2 h on day 1, following a
24-h continuous infusion of 5FU (375–425 mg/m2/day) for 5 days,
over a 21-day treatment cycle. Tumor responses were evaluated every
6 weeks in accordance to the Response Evaluation Criteria in Solid
Tumors 1.0 (RECIST 1.0). Treatment was terminated in the event of
disease progression or unacceptable toxicity. If the lesions
continued to shrink after six cycles, with no unacceptable
toxicity, a further one or two EOF cycles were recommended;
otherwise, oral FU was recommended as follow-up treatment.
SNP selection and genotyping
We selected and genotyped 13 drug-metabolism-related
genetic polymorphisms located at SLCO1B1 (rs4149056),
SLC2A9 (rs16890979, rs6449213, rs734553), ABCG2
(rs2231142), CYP2C9 (rs1057910, rs1799853), CYP2C19
(rs72552267, rs28399504, rs56337013, rs41291556), and CYP1A2
(rs12720461, rs56107638), respectively, from the Hapmap project
(www.hapmap.org) and dbSNP databases (www.ncbi.nlm.nih.gov/SLP) (Table I). Genomic DNA was extracted from
venous blood leukocytes using a standard phenol-chloroform method.
The selected SNPs were genotyped using the TaqMan assay method and
an ABI 7900 DNA detection system (Applied Biosystems, Foster City,
CA, USA). All the probes and primers were designed using the
Assay-on-Design service from Applied Biosystems. The experiments
were repeated for 15% of the samples. The genotype error rate was
<0.03%.
 | Table I.SNPs in the SLCO1B1, SLC2A9, SLC17A1,
ABCG2, CYP2C9, CYP2C19 and CYP1A2 genes analyzed in the
article. |
Table I.
SNPs in the SLCO1B1, SLC2A9, SLC17A1,
ABCG2, CYP2C9, CYP2C19 and CYP1A2 genes analyzed in the
article.
| Gene | SNP ID | Chromosome | Function | Allele | HWE test
P-value |
|---|
| SLCO1B1 | rs4149056 | 12:21178615 | Missense | T/C | 0.608 |
| SLC2A9 | rs16890979 | 4:9920543 | Missense | C/T | 0.903 |
| SLC2A9 | rs6449213 | 4:9992591 | Intron | C/T | 0.903 |
| SLC2A9 | rs734553 | 4:9921380 | Intron | A/C | 0.903 |
| ABGC2 | rs2231142 | 4:88131171 | Missense | A/C | 0.047 |
| CYP2C9 | rs1057910 | 10:94981296 | Missense | A/C | 0.808 |
| CYP2C9 | rs1799853 | 10:94942290 | Missense | C/T | 1.000 |
| CYP2C19 | rs72552267 | 10:94775453 | Missense | A/G | 1.000 |
| CYP2C19 | rs28399504 | 10:94762706 | Missense | A/G/T | 1.000 |
| CTP2C19 | rs56337013 | 10:94852738 | Missense | C/T | 1.000 |
| CYP2C19 | rs41291556 | 10:94775416 | Missense | C/T | 1.000 |
| CYP1A2 | rs12720461 | 15:74749010 | Intron | C/T | 1.000 |
| CYP1A2 | rs56107638 | 15:74753271 | Splice donor | A/C/G | 1.000 |
Statistical analysis
The allelic and genotypic distributions and
Hardy-Weinberg equilibrium were analyzed using the online analysis
tool, SHEsis. Differences in clinical characteristics among the 108
GC patients were analyzed by χ2 tests, and differences in allelic
and genotypic frequencies between the controlled-disease and
progressive-disease groups were compared with χ2 or Fisher's exact
probability tests. Genetic power was calculated using the G*Power
program (29). Survival curves were
analyzed using the Kaplan-Meier method. Differences in PFS and OS
between genotype groups were estimated by log-rank tests.
Multivariate analysis of prognostic predictors was carried out
using a Cox proportional hazards model. A two-sided P-value
<0.05 was considered significant. P-values for association
analysis were corrected by a false-discovery rate (FDR) procedure
(30). To be detailed, FDR p=p*n/a.
In this formula, n is the number of SNPs in the same gene, and a is
the rank of the P-value among the SNPs in the same gene.
Results
Patient characteristics
Most of the 108 GC patients enrolled in the study
had ≥3 tumor sites, with the most common metastatic organs being
the liver and retroperitoneal lymph nodes. Among the
clinicopathological features, liver metastasis and pleural effusion
were significantly associated with OS, whereas histological grade,
number of tumor sites, retroperitoneal lymph node involvement, and
ascites were significantly associated with PFS (Table II).
 | Table II.Associations between patient
characteristics and survival. |
Table II.
Associations between patient
characteristics and survival.
|
Characteristics | Number of patients
(%) | Median OS | P-value | Median PFS | P-value |
|---|
| Age (years) |
|
≤60 | 80 (74.1) | 465 | 0.916 | 159 | 0.176 |
|
>60 | 28 (25.9) | 403 |
| 187 |
|
| Sex |
|
Male | 64 (59.3) | 534 | 0.359 | 166 | 0.163 |
|
Female | 44 (40.7) | 372 |
| 182 |
|
| ECOG performance
status |
| 0 | 14 (13.0) | 704 | 0.153 | 240 | 0.768 |
| 1 | 89 (82.4) | 367 |
| 167 |
|
| 2 | 5 (4.6) | 299 |
| 237 |
|
| Histological
grade |
|
Moderate and high | 13 (12.0) | 403 | 0.226 | 156 | 0.004 |
|
Low/undifferentiated | 66 (61.2) | 875 |
| 380 |
|
|
Unclassified | 29 (26.8) | 367 |
| 180 |
|
| Number of tumor
sites |
| 1 | 5 (4.6) | 984 | 0.076 | 545 | 0.038 |
| 2 | 8 (7.4) | 570 |
| 411 |
|
| ≥3 | 95 (88.0) | 372 |
| 167 |
|
| Metastasis
sites |
|
Liver |
|
Yes | 36 (33.3) | 281 | 0.004 | 169 | 0.232 |
|
No | 72 (66.7) | 570 |
| 178 |
|
|
Lung |
|
Yes | 7 (6.5) | 252 | 0.997 | 169 | 0.885 |
|
No | 101 (93.5) | 444 |
| 187 |
|
| Retroperitoneal
lymph node |
|
Yes | 45 | 372 | 0.710 | 156 | 0.033 |
| No | 63 | 534 |
| 192 |
|
| Ascites |
|
Yes | 32 | 312 | 0.101 | 126 | 0.015 |
| No | 76 | 534 |
| 192 |
|
| Pleural
effusion |
|
Yes | 9 | 211 | 0.001 | 144 | 0.054 |
| No | 99 | 475 |
| 178 |
|
The responses to EOF chemotherapy were CR, n=1; PR,
n=41; SD, n=47; and PD, n=19. The 89 patients with CR, PR, or SD
were classified as the disease-control group and the 19 patients
with PD were classified as the disease-progression group.
Genotype frequency and disease
control
There was no significant relationship between
genotype frequency and disease-control rate for any of the 13 SNPs
analyzed in the present study (Table
III). There was only one genotype distribution each for
CYP2C19 (rs72552267, rs28399504, rs56337013, rs41291556) and
CYP1A2 (rs12720461, rs56107638) in the 108 patients (data
not shown).
 | Table III.Allelic and genotypic distribution of
the 15 SNPs in the disease control (CR, PR and SD) and the disease
progressive (PD) to chemotherapy. |
Table III.
Allelic and genotypic distribution of
the 15 SNPs in the disease control (CR, PR and SD) and the disease
progressive (PD) to chemotherapy.
| Gene | SNP | Allele
frequency | Chi2 value | P-value | Odds ratio 95%
CI | Genotype
frequency | P-value |
|---|
| SLCO1B1 | Rs4149056 | C | T |
|
|
| CC | CT | TT |
|
|
| CR+PR+SD | 4 (0.105) | 34 (0.895) | 0.372 | 0.541 | 0.705
(0.229–2.168) | 0 (0.000) | 4 (0.211) | 15 (0.789) | 0.700 |
|
| PD | 24 (0.143) | 144 (0.857) |
|
|
|
|
|
|
|
| SLC2A9 | rs16890979 | C | T |
|
|
| CC | CT |
|
|
|
| CR+PR+SD | 35 (0.972) | 1 (0.028) | 0.185 | 0.666 | 0.606
(0.061–6.006) | 17 (0.944) | 1 (0.056) |
| 0.663 |
|
| PD | 173 (0.983) | 3 (0.017) |
|
|
| 85 (0.966) | 3 (0.034) |
|
|
| SLC2A9 | rs6449213 | C | T |
|
|
| CT | TT |
|
|
|
| CR+PR+SD | 1 (0.028) | 35 (0.972) | 0.036 | 0.847 | 1.242
(0.134–11.457) | 1 (0.056) | 17 (0.944) |
| 0.845 |
|
| PD | 4 (0.022) | 174 (0.978) |
|
|
| 4 (0.045) | 85 (0.955) |
|
|
| SLC2A9 | rs734553 | A | C |
|
|
| AA | AC |
|
|
|
| CR+PR+SD | 35 (0.972) | 1 (0.028) | 0.168 | 0.681 | 0.621
(0.062–6.149) | 17 (0.944) | 1 (0.056) |
| 0.678 |
|
| PD | 169 (0.983) | 3 (0.017) |
|
|
| 83 (0.965) | 3 (0.035) |
|
|
| ABGC2 | Rs2231142 | A | C |
|
|
| AA | AC | CC |
|
|
| CR+PR+SD | 7 (0.194) | 29 (0.806) | 2.081 | 0.149 | 0.525
(0.217–1.272) | 2 (0.111) | 3 (0.167) | 13 (0.722) | 0.175 |
|
| PD | 56 (0.315) | 122 (0.685) |
|
|
| 11 (0.124) | 34 (0.382) | 44 (0.494) |
|
| CYP2C9 | Rs1057910 | A | C |
|
|
| AA | AC |
|
|
|
| CR+PR+SD | 36 (0.947) | 2 (0.053) | 2.952 | 0.085 | 0.204
(0.027–1.500) | 17 (0.895) | 2 (0.105) |
| 0.082 |
|
| PD | 176 (0.989) | 2 (0.011) |
|
|
| 87 (0.978) | 2 (0.022) |
|
|
| CYP2C9 | rs1799853 | C | T |
|
|
| CC | CT |
|
|
|
| CR+PR+SD | 36 (1.000) | 0 (0.000) | 0.203 | 0.652 |
| 18 (1.000) | 0 (0.000) |
| 0.651 |
|
| PD | 177 (0.994) | 1 (0.006) |
|
|
| 88 (0.989) | 1 (0.011) |
|
|
Genotype frequency and survival
analysis
Univariate analysis of the 13 SNPs in relation to
survival (Table IV) showed that
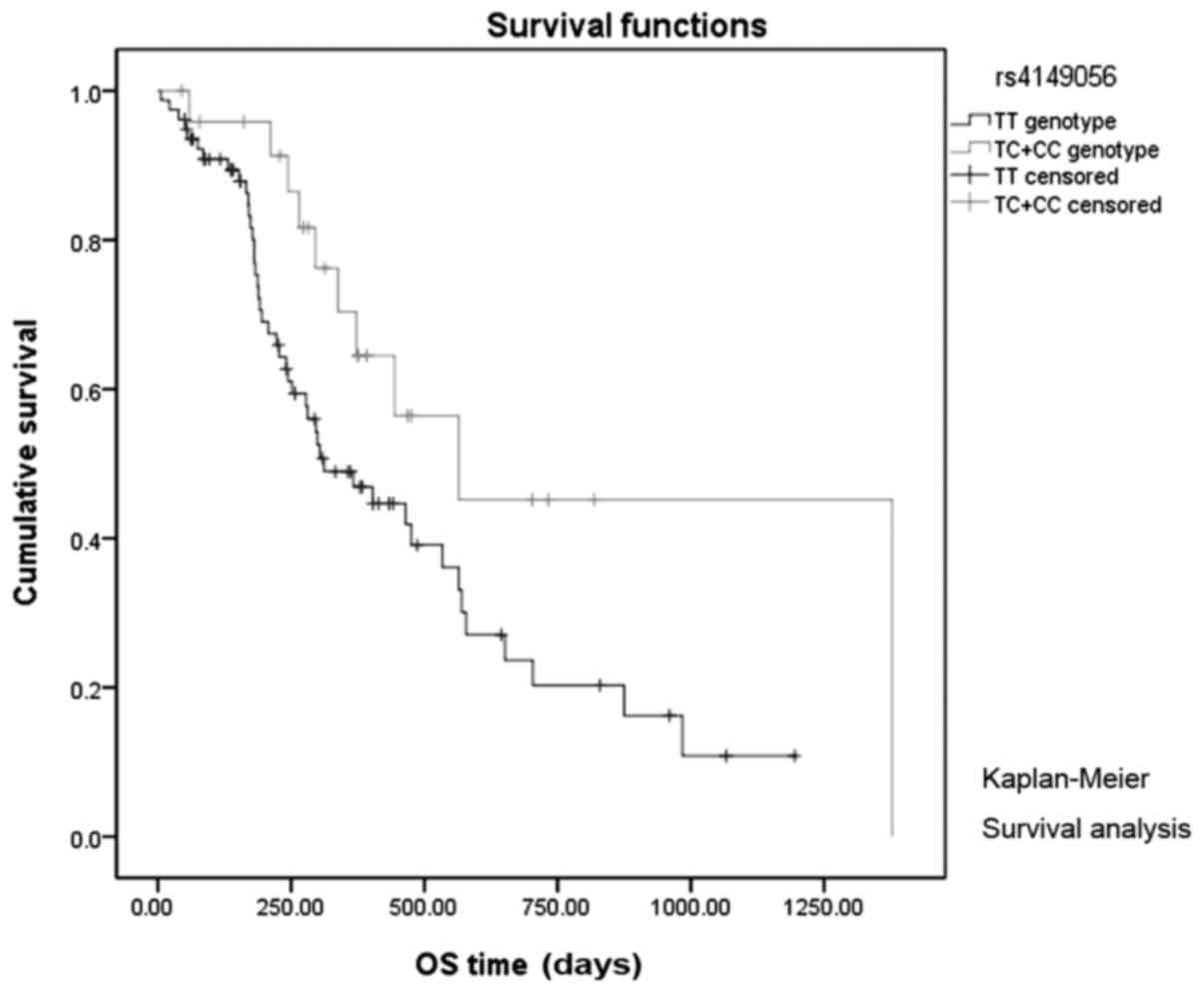
patients with SLCO1B1 rs4149056 CC and CT genotypes had
significantly longer median OS than patients with the TT genotype
(565 vs. 312 days, log-rank P=0.039; Fig.
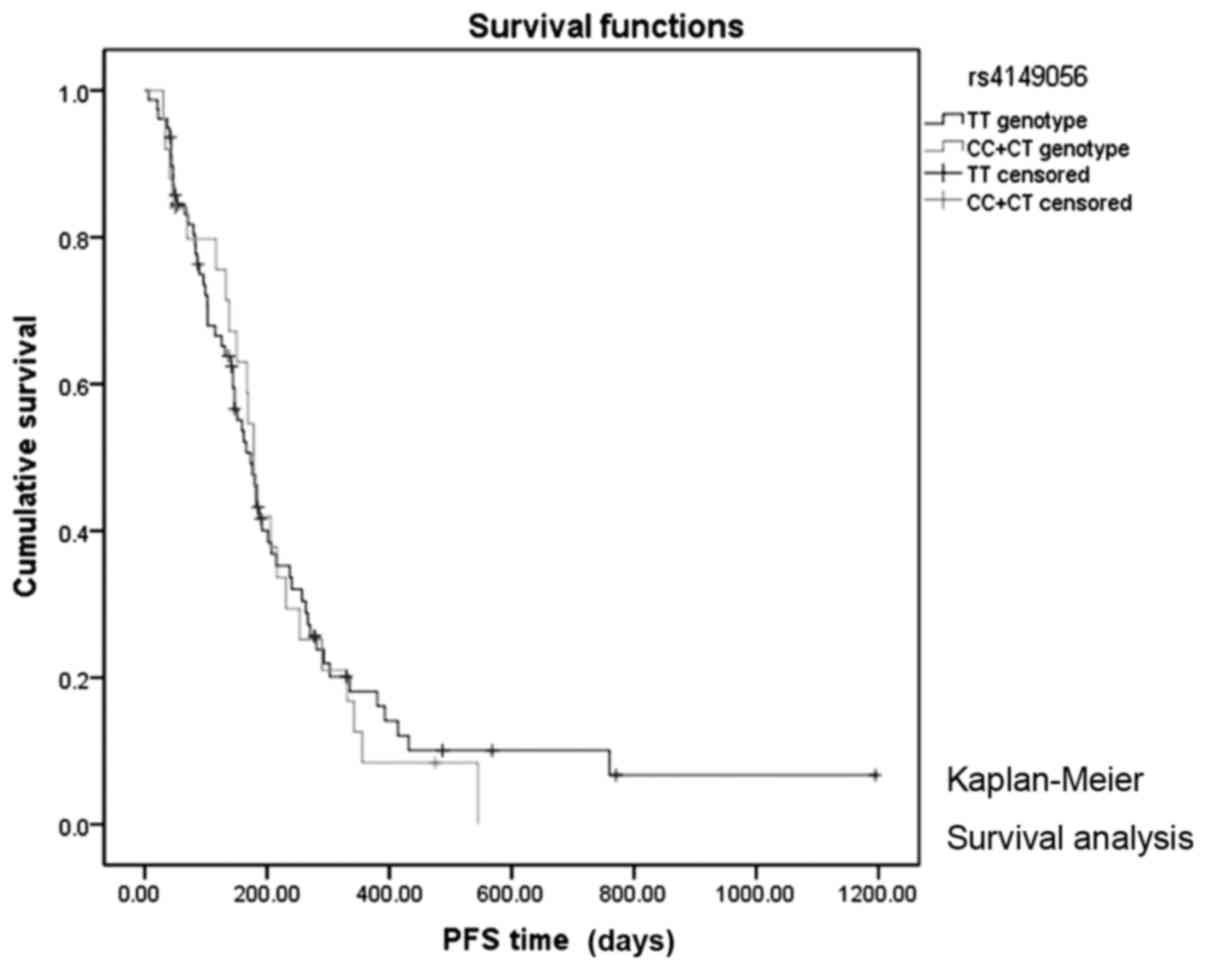
1). However, there was no significant association between PFS
and SLCO1B1 rs4149056 (log-rank P=0.956; Fig. 2), or between any of the other 12 SNPs
and OS or PFS (Table IV).
 | Table IV.Univariate survival analysis of SNPs
and overall survival/progression free survival time. |
Table IV.
Univariate survival analysis of SNPs
and overall survival/progression free survival time.
| SNPs | Median OS | 95% CI | P-value | Median PFS | 95% CI | P-value |
|---|
| SLCO1B1
rs4149056 |
| TT | 312.0 | 178.0–445.9 | 0.039 | 173.0 | 142.7–203.2 | 0.760 |
|
CC+CT | 565.0 | 108.9–1021.0 |
| 179.0 | 157.6–200.3 |
|
| SLC2A9
rs16890979 |
| CC | 465.0 | 303.1–626.8 | 0.301 | 176.0 | 156.7–195.2 | 0.427 |
| TC | 189.0 | 0.0–383.6 |
| 162.0 | 0.0–353.6 |
|
| SLC2A9
rs6449213 |
| CT | 265.0 | 94.0–435.9 | 0.161 | 162.0 | 55.8–268.1 | 0.257 |
| TT | 465.0 | 304.2–625.7 |
| 176.0 | 161.4–190.5 |
|
| SLC2A9
rs734553 |
| AA | 444.0 | 307.7–580.2 | 0.282 | 173.0 | 154.2–191.7 | 0.489 |
| CA | 189.0 | 0.0–383.6 |
| 162.0 | 0-353.6 |
|
| ABGC2
rs2231142 |
| CC | 444.0 | 293.2–594.7 | 0.750 | 167.0 | 144.1–189.8 | 0.083 |
|
AA+CA | 475.0 | 128.5–821.4 |
| 182.0 | 138.1–225.8 |
|
| CYP2C9
rs1057910 |
| AA | 444.0 | 289.8–598.1 | 0.382 | 176 | 156.8–195.1 | 0.311 |
| CA | 295.0 | 0.0–727.6 |
| 41 | 0.0–202.7 |
|
| CYP2C9
rs1799853 |
| CC | 465.0 | 272.9–657.0 | 0.711 | 173.0 | 153.6–192.3 | 0.750 |
| CT | 444.0 | – |
| 178.0 | – |
|
Multivariate PFS and OS analysis with
cox regression
Multivariate analysis showed that SLCO1B1
rs4149056 genotype, liver metastasis, ascites, pleural effusion,
and number of tumor sites were significantly or
borderline-significantly associated with OS (Table V). In addition, histological grade,
retroperitoneal lymph node involvement, ascites, pleural effusion,
and number of tumor sites were significantly or
borderline-significantly associated with PFS. All the above factors
were included in a stepwise multivariate Cox regression model,
which confirmed that SLCO1B1 rs4149056 was an independent
prognostic factor for shorter OS (P=0.014), but not PFS
(P=0.533).
 | Table V.Multi-factorial analysis of
prognostic factors for PFS and OS. |
Table V.
Multi-factorial analysis of
prognostic factors for PFS and OS.
|
| OS | PFS |
|---|
|
|
|
|
|---|
| Clinical
factor | P-value | HR 95% CI | P-value | HR 95% CI |
|---|
| rs4149056 TT | 0.014 | 2.565
(1.215–5.415) | – | – |
| Grade | – | – | 0.221 | – |
| PLN | – | – | 0.003 | 2.041
(1.271–3.278) |
| Ascites | 0.050 | 0.508
(0.258–0.999) | 0.436 | 0.818
(0.494–1.355) |
| Pleural
effusion | 0.016 | 0.363
(0.159–0.827) | 0.071 | 0.497
(0.232–1.061) |
| Liver | 0.002 | 0.372
(0.179–0.703) | – | – |
| Number of
sites | – | – | 0.200 | – |
| rs2231142 CC | – | – | 0.342 | 1.239
(0.796–1.929) |
Linkage disequilibrium analysis
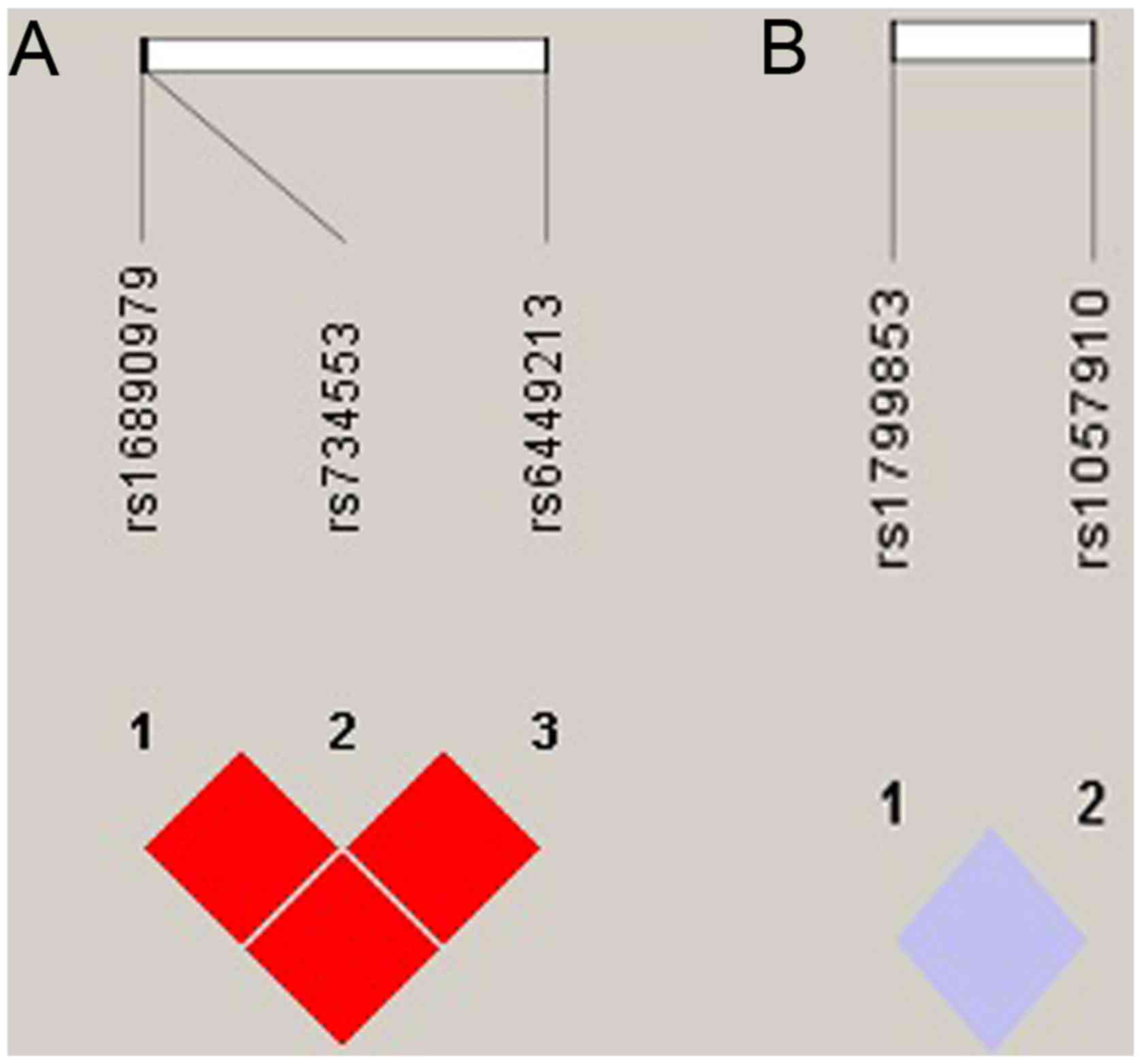
We analyzed linkage disequilibrium (LD) for the
SLC2A9 and CYP2C9 SNPs in AGC patients HaploView software. The
SLC2A9 SNPs rs16890979 and rs6449213 (D'=1.000, r2=0.796),
SLC2A9 SNPs rs16890979 and rs734553 (D'=1.000, r2=1.000) and
SLC2A9 SNPs rs6449213 and rs734553 (D'=1.000,
r2=0.796) all showed strong LD (Fig.
3A). CYP2C9 SNPs rs1057910 and rs1799853 showed no LD
(D'=1.000, r2=0.000) (Fig. 3B). Since
the most samples showed the same allele and only less than 5
samples showed different allele in the SCL2A9 SNPs, linkage
disequilibrium and haplotype analysis cannot find clinically
meaningful result.
Discussion
We investigated the association between SNPs of
metabolism-related genes and chemotherapy response in patients with
MGC. Patients with SLCO1B1 rs4149056 CC and CT genotypes had
longer OS than patients with TT genotype (312 vs. 565 days,
P=0.039), and SLCO1B1 rs4149056 TT was confirmed as an
independent prognostic factor for shorter OS in GC patients treated
with EOF by Cox regression analysis (hazard ratio: 2.565, 95%
confidence interval: 1.215–5.415, P=0.014).
SLCO1B1 encodes the SLC family member
OATP1B1, which is highly expressed in hepatocytes and associated
with hepatic drug uptake and elimination (31). Among single-nucleotide variants of
SLCO1B1, c.521T>C rs4149056 had the greatest effect on
OATP1B1 activity in this study. SLCO1B1 c.521T>C
decreases OATP1B1 transporting activity, thus increasing plasma
concentrations of drugs, including cytotoxic drugs. SLCO1B1
rs4149056 has been significantly associated with exposure to SN-38,
the active metabolite of irinotecan (7). In another study, OATP1B1 was responsible
for SN-38 uptake from plasma into hepatocytes (6). Innocenti et al also revealed that
SLCO1B1 rs4149056 increased patient exposure to CPT-11 and
was associated with an increased risk of severe neutropenia
(6). OATP1B1 was shown to transport
paclitaxel in an in vitro ovarian cancer study, implying
that it contributed to the disposition of paclitaxel (8). Huang et al reported that another
SLCO1B1 variant, rs2306283, was an independent prognostic
factor for longer PFS in patients with metastatic colorectal cancer
treated with a fluoropyrimidine plus irinotecan (1). Overall, these studies suggest that
SLCO1B1 affects the metabolism of fluoropyrimidines, CPT-11,
and docetaxel, thus influencing their chemotherapeutic efficacy,
consistent with the results of the current study.
To the best of our knowledge, the present study
provides the first evidence for an association between
SLCO1B1 and GC. A literature search of MEDLINE via PubMed
found no similar reports with respect to the efficacy of first-line
chemotherapy, GC risk, or prognosis.
Notably, although we identified SLCO1B1
rs4149056 as a prognostic factor for OS in MGC patients treated
with first-line EOF, we failed to detect any correlation between
SLCO1B1 rs4149056 and PFS. There are two possible reasons
for this apparent discrepancy. First, SLCO1B1 rs4149056 may
only be a prognostic factor in GC patients, and may not affect the
short-term efficacy of first-line chemotherapy. Second, CPT-11 or
paclitaxel were administered as the main second- or third-line
chemotherapy regimens in the present study; as noted above,
SLCO1B1 rs4149056 has been associated with the metabolism of
CPT-11 and paclitaxel, and rs4149056 may thus affect OS by
influencing the efficacy of the second- or third-line regimens.
In our study, ABCG2 (BCRP2) C421A rs2231142
was borderline-significantly associated with PFS (P=0.083), though
multivariate analysis did not identify it as an independent
predictor in GC patients treated with EOF regimen. However, several
previous studies reported significant relationships between
ABCG2 (BCRP2) C421A rs2231142 and the toxicity or efficacy
of oxaliplatin or anthracyclines. Custodio et al revealed
that ABCG2 rs3114018 was associated with oxaliplatin-induced
peripheral neuropathy in patients with stage II–III colon cancer
(32), while breast cancer patients
with an ABCG2 rs2231142 A allele reportedly showed better
responses to anthracycline-based neoadjuvant chemotherapy (16). Ghafouri et al also showed that
the ABCG2 rs22311442 A allele was associated with stronger
responses to anthracyclines and paclitaxel (33). Overall, these three studies indicated
that patients with an ABCG2 rs22311442 A allele showed
better responses to anthracyclines and paclitaxel. Although the
current univariate analysis failed to find an association between
ABCG rs22311442 and PFS, there was a nonsignificant tendency
towards longer PFS among rs22311442 AA/AC compared with rs22311442
CC patients. There are three possible reasons for this apparent
discrepancy. First, it is possible that the sample-size was too
small to demonstrate any significant difference between the
survival trends of patients with rs22311442 A and C alleles,
respectively. Second, the previous studies involved patients with
colon cancer or breast cancer, rather than GC, and the results may
thus have been affected by this tumor heterogeneity. Third, the EOF
regimen contains epirubicin, oxaliplatin, and 5FU. Custodio showed
an association between ABCG2 rs22311442 and oxaliplatin
toxicity rather than oxaliplatin response, which differed from our
current study.
Evidence suggests that some CYP450 enzyme family
members affect 5FU metabolism (34–37). 5FU
was reported to reduce the ability to metabolize a CYP2C9 probe
drug (38). Moreover, several studies
have indicated that 5FU can down-regulate the expression of CYP
enzymes, including CYP2C9 and CYP2C19 (39,40).
However, although CYP2C9 and CYP2C19 have been shown to influence
5FU metabolism in vitro, these results have not yet been
verified in clinical studies. Furthermore, no studies have reported
on the relationship between 5FU efficacy and SNPs in CYP2C9
or CYP2C19. Our results showed that all 108 blood samples
had the same genotypic distributions of CYP2C9 and
CYP2C19 SNPs, except for CYP2C9 rs1057910, which was
not significantly associated with PFS or OS. The roles of CYP2C9
and CYP2C19 in 5FU metabolism thus currently remain unclear.
There were several limitations to the present study.
First, we only analyzed some of the genes related to the CYP, SLC,
and ABC families, and any associations between the remaining
untested genes and GC remain unknown. Second, we only detected
selected polymorphisms for each target agent, and other potentially
related polymorphisms may have been missed. Third, the sample size
was relatively small. The conclusions of this study therefore
require verification in further studies.
In conclusion, the present study identified an
association between the SLCO1B1 rs4149056 SNP and clinical
outcomes of MGC patients treated with EOF chemotherapy. The
resulting risk model successfully divided patients into low-risk
and high-risk groups, with a significant difference in OS, but not
PFS. SLCO1B1 rs4149056 is therefore a prognostic, but not a
predictive factor in MGC patients treated with EOF. Further studies
are required to validate our results and to detect other potential
prognostic sites in SLCO1B1.
Acknowledgements
Not applicable.
Funding
This study was funded by The National Key Research
and Development Program of China (grant no. 2017YFC1308900).
Availability of data and materials
All data generated or analysed during this study are
included in this published article.
Authors' contributions
WF and XL performed the statistical analysis and
wrote the article. XZhu and ZC designed and managed the research.
XZha and MH performed the experiments. WG and JY assisted with the
research design.
Ethics approval and consent to
participant
Informed consent was been obtained from all
participants. The study was approved by the Ethics Committee of
Fudan University Shanghai Cancer.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Huang L, Zhang T, Xie C, Liao X, Yu Q,
Feng J, Ma H, Dai J, Li M, Chen J, et al: SLCO1B1 and SLC19A1 gene
variants and irinotecan-induced rapid response and survival: A
prospective multicenter pharmacogenetics study of metastatic
colorectal cancer. PLoS One. 8:e772232013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
De Mattia E, Toffoli G, Polesel J,
D'Andrea M, Corona G, Zagonel V, Buonadonna A, Dreussi E and
Cecchin E: Pharmacogenetics of ABC and SLC transporters in
metastatic colorectal cancer patients receiving first-line FOLFIRI
treatment. Pharmacogenet Genomics. 23:549–557. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ramsey LB, Panetta JC, Smith C, Yang W,
Fan Y, Winick NJ, Martin PL, Cheng C, Devidas M, Pui CH, et al:
Genome-wide study of methotrexate clearance replicates SLCO1B1.
Blood. 121:898–904. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Radtke S, Zolk O, Renner B, Paulides M,
Zimmermann M, Möricke A, Stanulla M, Schrappe M and Langer T:
Germline genetic variations in methotrexate candidate genes are
associated with pharmacokinetics, toxicity, and outcome in
childhood acute lymphoblastic leukemia. Blood. 121:5145–5153. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Treviño LR, Shimasaki N, Yang W, Panetta
JC, Cheng C, Pei D, Chan D, Sparreboom A, Giacomini KM, Pui CH, et
al: Germline genetic variation in an organic anion transporter
polypeptide associated with methotrexate pharmacokinetics and
clinical effects. J Clin Oncol. 27:5972–5978. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Innocenti F, Kroetz DL, Schuetz E, Dolan
ME, Ramírez J, Relling M, Chen P, Das S, Rosner GL and Ratain MJ:
Comprehensive pharmacogenetic analysis of irinotecan neutropenia
and pharmacokinetics. J Clin Oncol. 27:2604–2614. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Teft WA, Welch S, Lenehan J, Parfitt J,
Choi YH, Winquist E and Kim RB: OATP1B1 and tumour OATP1B3 modulate
exposure, toxicity, and survival after irinotecan-based
chemotherapy. Br J Cancer. 112:857–865. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Svoboda M, Wlcek K, Taferner B, Hering S,
Stieger B, Tong D, Zeillinger R, Thalhammer T and Jäger W:
Expression of organic anion-transporting polypeptides 1B1 and 1B3
in ovarian cancer cells: Relevance for paclitaxel transport. Biomed
Pharmacother. 65:417–426. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang HN, He XL, Wang C, Wang Y, Chen YJ,
Li JX, Niu CH and Gao P: Impact of SLCO1B1 521T>C variant on
leucovorin rescue and risk of relapse in childhood acute
lymphoblastic leukemia treated with high-dose methotrexate. Pediatr
Blood Cancer. 61:2203–2207. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ramsey LB, Bruun GH, Yang W, Treviño LR,
Vattathil S, Scheet P, Cheng C, Rosner GL, Giacomini KM, Fan Y, et
al: Rare versus common variants in pharmacogenetics: SLCO1B1
variation and methotrexate disposition. Genome Res. 22:1–8. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lima A, Bernardes M, Azevedo R, Monteiro
J, Sousa H, Medeiros R and Seabra V: SLC19A1, SLC46A1 and SLCO1B1
polymorphisms as predictors of methotrexate-related toxicity in
Portuguese rheumatoid arthritis patients. Toxicol Sci. 142:196–209.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yuan SQ, Zhou ZW, Liang YJ, Fu LW, Chen G,
Qiu HB and Zhang LY: Correlation of chemosensitivity tested using
histoculture drug response assay to expression of multidrug
resistance genes and proteins in colorectal cancer tissues. Ai
Zheng. 28:932–938. 2009.PubMed/NCBI
|
|
13
|
Cortez MA, Scrideli CA, Yunes JA, Valera
ET, Toledo SR, Pavoni-Ferreira PC, Lee ML, Petrilli AS, Brandalise
SR and Tone LG: mRNA expression profile of multidrug resistance
genes in childhood acute lymphoblastic leukemia. Low expression
levels associated with a higher risk of toxic death. Pediatr Blood
Cancer. 53:996–1004. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou S, Liao L, Chen C, Zeng W, Liu S, Su
J, Zhao S, Chen M, Kuang Y, Chen X and Li J: CD147 mediates
chemoresistance in breast cancer via ABCG2 by affecting its
cellular localization and dimerization. Cancer Lett. 337:285–292.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang Z, Wang N, Li W, Liu P, Chen Q, Situ
H, Zhong S, Guo L, Lin Y, Shen J and Chen J: Caveolin-1 mediates
chemoresistance in breast cancer stem cells via β-catenin/ABCG2
signaling pathway. Carcinogenesis. 35:2346–2356. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu H, Liu Y, Kang H, Xiao Q, Yao W, Zhao
H, Wang E and Wei M: Genetic variations in ABCG2 gene predict
breast carcinoma susceptibility and clinical outcomes after
treatment with anthracycline-based chemotherapy. Biomed Res Int
2015. 2791092015.
|
|
17
|
Won HJ, Ha TK, Kwon SJ, Cho HY, Hur SJ,
Baik HH, Suh SI, Ha E and Kim YH: Differential effects of
5-fluorouracil on glucose transport and expressions of glucose
transporter proteins in gastric cancer cells. Anticancer Drugs.
21:270–276. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Berlth F, Mönig S, Pinther B, Grimminger
P, Maus M, Schlösser H, Plum P, Warnecke-Eberz U, Harismendy O,
Drebber U, et al: Both GLUT-1 and GLUT-14 are independent
prognostic factors in gastric adenocarcinoma. Ann Surg Oncol. 22
Suppl 3:S822–S831. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schlößer HA, Drebber U, Urbanski A, Haase
S, Baltin C, Berlth F, Neiß S, von Bergwelt-Baildon M, Fetzner UK,
Warnecke-Eberz U, et al: Glucose transporters 1, 3, 6, and 10 are
expressed in gastric cancer and glucose transporter 3 is associated
with UICC stage and survival. Gastric Cancer. 20:83–91. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dees EC and Watkins PB: Role of cytochrome
P450 phenotyping in cancer treatment. J Clin Oncol. 23:1053–1055.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jernström H, Bågeman E, Rose C, Jönsson PE
and Ingvar C: CYP2C8 and CYP2C9 polymorphisms in relation to tumour
characteristics and early breast cancer related events among 652
breast cancer patients. Br J Cancer. 101:1817–1823. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Boruban MC, Yasar U, Babaoglu MO, Sencan O
and Bozkurt A: Tamoxifen inhibits cytochrome P450 2C9 activity in
breast cancer patients. J Chemother. 18:421–424. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Seredina TA, Goreva OB, Talaban VO,
Grishanova AY and Lyakhovich VV: Association of cytochrome P450
genetic polymorphisms with neoadjuvant chemotherapy efficacy in
breast cancer patients. BMC Med Genet. 13(45)2012.PubMed/NCBI
|
|
24
|
Jamieson D, Lee J, Cresti N, Jackson R,
Griffin M, Sludden J, Verrill M and Boddy AV: Pharmacogenetics of
adjuvant breast cancer treatment with cyclophosphamide, epirubicin
and 5-fluorouracil. Cancer Chemother Pharmacol. 74:667–674. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Melanson SE, Stevenson K, Kim H, Antin JH,
Court MH, Ho VT, Ritz J, Soiffer RJ, Kuo FC, Longtine JA and
Jarolim P: Allelic variations in CYP2B6 and CYP2C19 and survival of
patients receiving cyclophosphamide prior to myeloablative
hematopoietic stem cell transplantation. Am J Hematol. 85:967–971.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Beelen K, Opdam M, Severson TM, Koornstra
RH, Vincent AD, Hauptmann M, van Schaik RH, Berns EM, Vermorken JB,
van Diest PJ and Linn SC: CYP2C19 2 predicts substantial tamoxifen
benefit in postmenopausal breast cancer patients randomized between
adjuvant tamoxifen and no systemic treatment. Breast Cancer Res
Treat. 139:649–655. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
van Schaik RH, Kok M, Sweep FC, van Vliet
M, van Fessem M, Meijer-van Gelder ME, Seynaeve C, Lindemans J,
Wesseling J, Van't Veer LJ, et al: The CYP2C19*2 genotype predicts
tamoxifen treatment outcome in advanced breast cancer patients.
Pharmacogenomics. 12:1137–1146. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Komatsu T, Yamazaki H, Shimada N, Nakajima
M and Yokoi T: Roles of cytochromes P450 1A2, 2A6, and 2C8 in
5-fluorouracil formation from tegafur, an anticancer prodrug, in
human liver microsomes. Drug Metab Dispos. 28:1457–1463.
2000.PubMed/NCBI
|
|
29
|
Faul F, Erdfelder E, Lang AG and Buchner
A: G*Power 3: A flexible statistical power analysis program for the
social, behavioral, and biomedical sciences. Behav Res Methods.
39:175–191. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Benjamini Y, Drai D, Elmer G, Kafkafi N
and Golani I: Controlling the false discovery rate in behavior
genetics research. Behav Brain Res. 125:279–284. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Niemi M, Pasanen MK and Neuvonen PJ:
Organic anion transporting polypeptide 1B1: A genetically
polymorphic transporter of major importance for hepatic drug
uptake. Pharmacol Rev. 63:157–181. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Custodio A, Moreno-Rubio J, Aparicio J,
Gallego-Plazas J, Yaya R, Maurel J, Higuera O, Burgos E, Ramos D,
Calatrava A, et al: Pharmacogenetic predictors of severe peripheral
neuropathy in colon cancer patients treated with oxaliplatin-based
adjuvant chemotherapy: A GEMCAD group study. Ann Oncol. 25:398–403.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ghafouri H, Ghaderi B, Amini S, Nikkhoo B,
Abdi M and Hoseini A: Association of ABCB1 and ABCG2 single
nucleotide polymorphisms with clinical findings and response to
chemotherapy treatments in Kurdish patients with breast cancer.
Tumour Biol. 37:7901–7906. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Park SR, Kong SY, Nam BH, Choi IJ, Kim CG,
Lee JY, Cho SJ, Kim YW, Ryu KW, Lee JH, et al: CYP2A6 and ERCC1
polymorphisms correlate with efficacy of S-1 plus cisplatin in
metastatic gastric cancer patients. Br J Cancer. 104:1126–1134.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Park SR, Hong YS, Lim HS, Seong MW, Kong
SY, Kim SY, Park YI and Jung KH: Phase I clinical and
pharmacokinetic/pharmacogenetic study of a triplet regimen of
S-1/irinotecan/oxaliplatin in patients with metastatic colorectal
or gastric cancer. Cancer Chemother Pharmacol. 72:953–964. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kim YW, Kim MJ, Ryu KW, Lim HS, Lee JH,
Kong SY, Lee JS, Choi IJ, Kim CG, Lee JY, et al: A phase II study
of perioperative S-1 combined with weekly docetaxel in patients
with locally advanced gastric carcinoma: Clinical outcomes and
clinicopathological and pharmacogenetic predictors for survival.
Gastric Cancer. 19:586–596. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kong SY, Lim HS, Nam BH, Kook MC, Kim YW,
Ryu KW, Lee JH, Choi IJ, Lee JS, Park YI, et al: Association of
CYP2A6 polymorphisms with S-1 plus docetaxel therapy outcomes in
metastatic gastric cancer. Pharmacogenomics. 10:1147–1155. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Karadag O, Babaoglu MO, Altundag K,
Elkiran T, Yasar U and Bozkurt A: 5-Fluorouracil-induced coronary
spasm: May inhibition of hyperpolarization factors produced by
CYP2C enzymes be the cause? Oncology. 66:510–511. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Helsby NA, Lo WY, Thompson P and Laking
GR: Do 5-fluorouracil therapies alter CYP2C19 metaboliser status?
Cancer Chemother Pharmacol. 66:405–407. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Afsar A, Lee C and Riddick DS: Modulation
of the expression on constitutive rat hepatic cytochrome P450
isozymes by 5-fluorouracil. Can J Physiol Pharmacol. 74:150–156.
1996. View Article : Google Scholar : PubMed/NCBI
|