Introduction
Cardiac myxoma (CM) is the most clinically common
primary cardiac benign tumor, its incidence accounts for more than
50% of cardiac tumors (1). CM
frequently occurs in the middle age and is more common in females
than in males but rarely seen in children, in whom it accounts for
15% of cardiac tumors (2,3). CM occurs anywhere in the heart, but
arises most commonly in the left atrium (4,5) with
occasionally on the right atrium, right ventricle and the left
ventricle (5,6). It can cause mild constitutional
symptoms, such as fever, weight loss, myalgia, skin flushing or
arthralgia, to serious hemodynamic derangement, depending on its
location and size, which can lead to disastrous embolic symptoms.
Therefore, early diagnosis and treatment are necessary, and once
the diagnosis is established, surgery should be performed without
delay. However, recurrence after surgical excision occurs in 2% of
the cases (7). CM can be easily
misdiagnosed and treatment can be delayed due to its complicated
clinical manifestations with no specific clinical performance.
Echocardiography is the most commonly used method for the detection
of CM. The diagnosis rate has been improved obviously since it was
widely used in the clinic. However, echocardiography has
limitations due to low sensitivity and specificity, lengthy
diagnostic time, and technical complexity. Therefore,
diagnostically sensitive and specific markers for early
uncomplicated CM detection are urgently needed.
A microRNA (miRNA/miR) is an endogenous small
single-stranded non-coding RNA molecule with a length of ~22
nucleotides. It can pair with the 3′UTR of a target gene's mRNA,
and inhibit the translation of the target genes or induce the
degradation of mRNA (8). miRNAs are
involved in numerous cancer-relevant processes, such as migration,
proliferation and apoptosis (9,10).
Aberrant expression of miRNA has been reported in various
malignancies such as gastric cancer (11) and breast cancer (12), and thus, their alteration can cause
malignant transformation. Recent evidence demonstrated that
circulating miRNAs can be regarded as reliable biomarkers for the
detection of various cancers and other diseases due to their
stability and ease of detection (13). However, miRNA in the serum of patients
with CM remains unexplored, and the pathogenesis of CM is not very
clear. Therefore, in the present study, we aimed to identify
differentially expressed miRNAs in the serum of patients with CM
and analyzed their potential role in CM development.
Materials and methods
Patients
Thirty patients with CM who were hospitalized in
Fujian Medical University Union Hospital (Fuzhou, China) from
October 2015 to May 2016 were recruited. Thirty healthy cases were
also recruited. Of the 30 patients with CM, 10 were male and 20
were female, and their age ranged from 17 to 88, with average age
of 58.5. In the control group, 10 were male and 20 were female, and
their age ranged from 18 to 74 with an average age of 55.6. No
significant difference was found (P>0.05) in the general
characteristics, such as gender and age, between the CM and healthy
groups. This study was approved by the Ethics Committee of Fujian
Medical University Union Hospital (Fuzhou, China) and informed
consent was provided by each patient.
Serum preparation and total RNA
isolation
Blood was collected from each subject into tubes
without anticoagulant and placed at 4°C for 2 h. The samples were
then centrifugated at 4,000 × g for 15 min, and the serum that was
separated was stored at −80°C until use. The 30 patients with CM
were randomly assigned into three equal groups of 10 subjects each,
then 100 µl of the serum form each group were pooled to generate a
serum pool (14) labeled as samples
A, B and E. Similarly, the samples from the healthy group were also
grouped, and the serum pools were labeled as samples a, b and e. We
named the groups randomly, these names have no meaning.
Total RNA from the serum was extracted and purified
using the miRNeasy Mini kit (Qiagen, Inc., Valencia, CA, USA). The
concentration and quality of the RNA were measured using a NanoDrop
1,000 Spectrophotometer (Thermo Fisher Scientific, Inc., Waltham,
MA, USA). RNA samples were subjected to electrophoresis with 1.4%
agarose-formaldehyde gels stained with ethidium bromide to verify
their integrity. The quality control criteria of RNA were 28S/18S
>1.5 and A260/A280 >1.8.
MiRNA expression profiling
The RNA with high purity (A260/A280 >1.8) and
high quality (28S/18S >1.5) was used for the microarray
experiments. The miRCURY Hy3/Hy5 Power Labeling kit (Exiqon,
Vedbaek, Denmark) was used according to the manufacturer's
guideline for miRNA labelling. One microgram of each sample was
3-end-labeled with Hy3TM fluorescent label by using T4 RNA ligase.
The mixture was incubated for 30 min at 37°C, and the reaction was
terminated by incubation for 5 min at 95°C. Then, 3.0 µl of
labeling buffer, 1.5 µl of fluorescent label (Hy3TM), 2.0 µl of
DMSO, and 2.0 µl of labeling enzyme were added into the mixture.
The labeling mixture was incubated for 1 h at 16°C, and the
reaction was terminated by incubation for 15 min at 65°C. After
stopping the labeling procedure, the Hy3-labeled samples were
hybridized to the Agilent miRNA microarray chip. Following the
hybridization, the slides were achieved, washed several times by
using a Wash Buffer kit (Exiqon), and finally dried by
centrifugation for 5 min at 400 × g. Then the microarray was
scanned using the GenePix 4000B microarray scanner (Axon
Instruments, Foster City, CA, USA). Data were extracted using the
Agilent Feature Extraction Software and analyzed using the
GeneSprint 13.0 software (Agilent Technologies, Inc., Santa Clara,
CA, USA). All raw data in control and CM group was processed with
‘percentile’ method for data normalization, and FC (fold-change)
was calculated using these normalized datas. The miRNAs with FC
≥2.0 and P<0.05 were considered significant, the choice of
cut-off was based on previous studies (15–18). This
section was accomplished by Beijing CapitalBio Corporation
(Beijing, China). The data in healthy group was used as control for
the comparisons.
Validation of the differentially
expressed miRNAs by quantitative reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Two selected candidate miRNAs identified from the
serum samples of the 30 patients with CM and 30 healthy controls
were validated by RT-qPCR. 1 µl of purified total RNA was reverse
transcribed using a miscript II RT kit (Qiagen, Courtaboeuf,
France) according to the manufacturer's instructions. The qPCR
assay was performed using reagents from the SYBR Green Real-Time
PCR kit (Takara Biotechnology Co., Ltd., Dalian, China), and the
reaction mix was composed of 10 µl SYBR Premix Ex Taq II, 0.8 µl
PCR forward primer (Table I), 0.8 µl
UnimiR qPCR primer, 2 µl cDNA, 0.4 µl ROX reference dye, and 6 µl
deionized H2O. The reactions were incubated in a 48-well
optical plate at 95°C for 30 sec, followed by 40 cycles of 95°C for
5 sec, and 60°C for 34 sec. The Cquantification cycle (Cq) was
determined by using the default threshold settings. All experiments
were performed in triplicate and repeated three times. RT-qPCR was
performed in Mx3000P (Stratagene, La Jolla, CA, USA). U6
small-nuclear RNA (U6 snRNA) was used as a reference gene to
normalize the expression of the miRNA. The mature sequences for all
miRNAs were acquired from miRBase (http://www.mirbase.org/). The primer sequences used
for the miRNA analysis are designed based on these mature sequences
and shown in Table I. Relative
expression was analyzed using the method of 2−ΔΔCq
(19).
 | Table I.miRNA-specific Primers Used in the
RT-qPCR. |
Table I.
miRNA-specific Primers Used in the
RT-qPCR.
| miRNA | Primer sequences
(5′-3′) |
|---|
| miR-320a | Forward:
5′-GCGAAAAGCTGGGTTGAGA-3′ |
|
| Reverse:
5′-AGTGCAGGGTCCGAGGTATT-3′ |
| miR-634 | Forward:
5′-CGAACCAGCACCCCAACT-3′ |
|
| Reverse:
5′-AGTGCAGGGTCCGAGGTATT-3′ |
| U6 | Forward:
5′-AGAGAAGATTAGCATGGCCCCTG-3′ |
|
| Reverse:
5′-AGTGCAGGGTCCGAGGTATT-3′ |
Bioinformatics analysis
The target genes of miR-320a were predicted using
three miRNA databases including miRanda (http://mirdb.org/miRDB/), TargetScan (http://www.targetscan.org/) and PicTar (http://pictar.mdc-berlin.de). The target genes that
were uniformly predicted by the three databases were selected. The
Gene Ontology (GO) database (http://www.geneontology.org/) was employed to enrich
the functions of miR-320a target genes. Fisher's exact test was
then employed to calculate the P-value for each function. A
function with P-value <0.05 was considered as significant. The
Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.kegg.jp/) was used to explore the relevant
signaling pathways of the target genes of miR-320a. Fisher's exact
test was then employed to calculate the P-value for each pathway. A
pathway with P-value <0.05 was considered as significant.
Statistical analysis. All data were shown as mean ±
standard deviation, and analyzed using the SPSS 20.0 software
(SPSS, Inc., Chicago, IL, USA). The differential expression levels
between two groups was analyzed using t-test for validation.
P<0.05 was considered to indicate a statistically significant
difference. P-values were corrected for multiple testing by false
discovery rate (FDR). Clustering analysis of miRNA based on
KK-means algorithm.
Results
Differentially expressed miRNAs in the
serum by miRNA microarray analysis
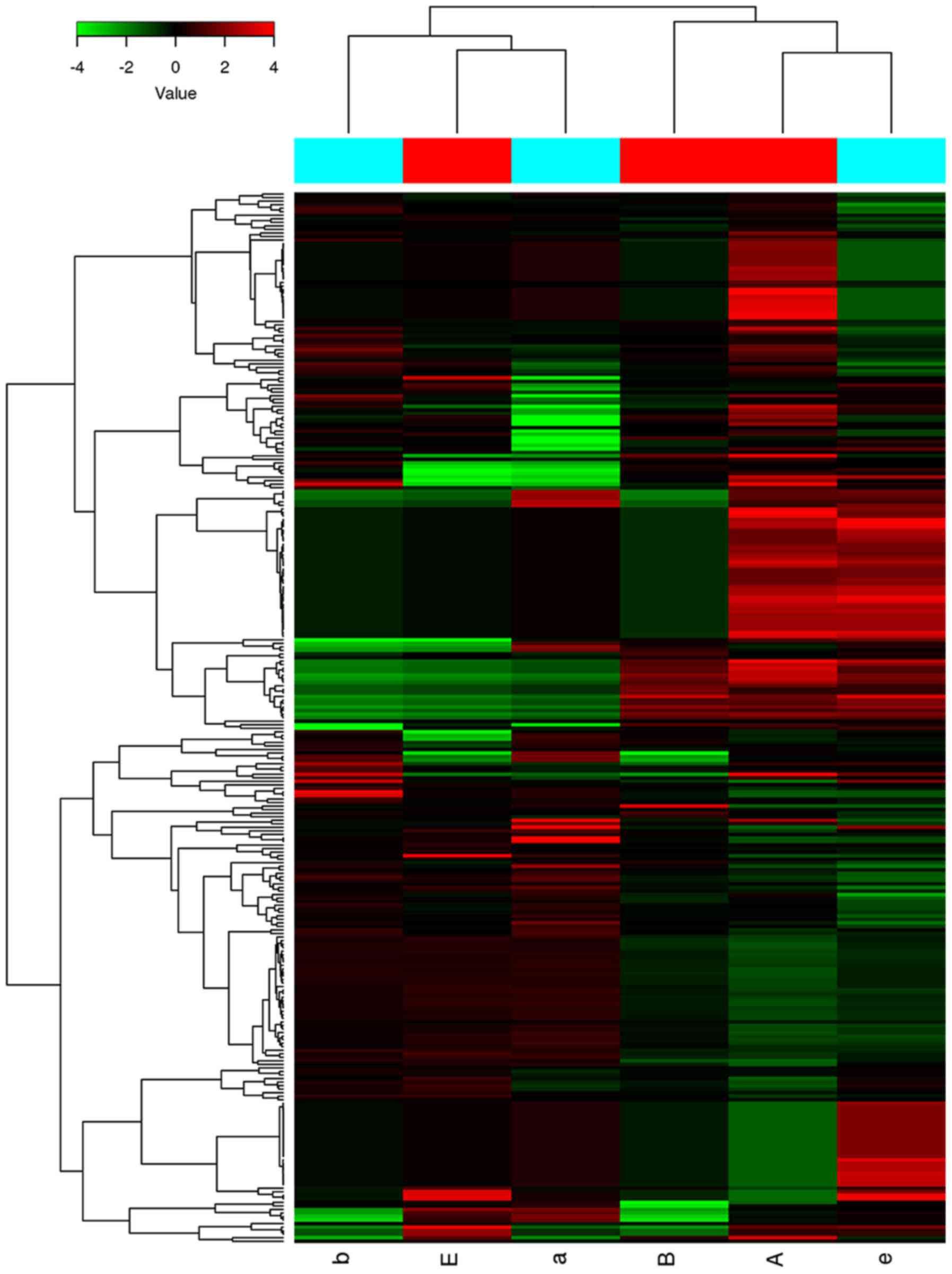
We performed a microarray analysis to identify the
miRNA expression patterns of the patients with CM. We made a heat
map to visualize the results of the two-way hierarchical clustering
of the miRNA (Fig. 1). The color
scale shown at the top illustrated the relative expression level of
a miRNA: Red represented a high relative expression level, whereas
green represented a low relative expression level (20).
Of the 207 miRNAs analyzed by miRNA microarray, only
four were identified as being differentially expressed by more than
two-fold. Of those, 2 (miR-320a and miR-1249-5p) were obviously
up-regulated and 2 (miR-634 and miR-6870-3p) were down-regulated in
the patients with CM in comparison with the healthy controls
(P<0.05; Table II).
 | Table II.Differentially expressed miRNA in the
serum of cardia myxoma patients. |
Table II.
Differentially expressed miRNA in the
serum of cardia myxoma patients.
| Systematic
name | P (Corr) | P-value | FC (abs) | Regulation |
|---|
| hsa-miR-320a | 0.96 | 0.032 | 2.05 | Up |
|
hsa-miR-1249-5p | 0.96 | 0.028 | 11.08 | Up |
| hsa-miR-634 | 0.96 | 0.047 | 5.47 | Down |
|
hsa-miR-6870-3p | 0.96 | 0.049 | 5.97 | Down |
Validation of the differentially
expressed miRNAs by RT-qPCR
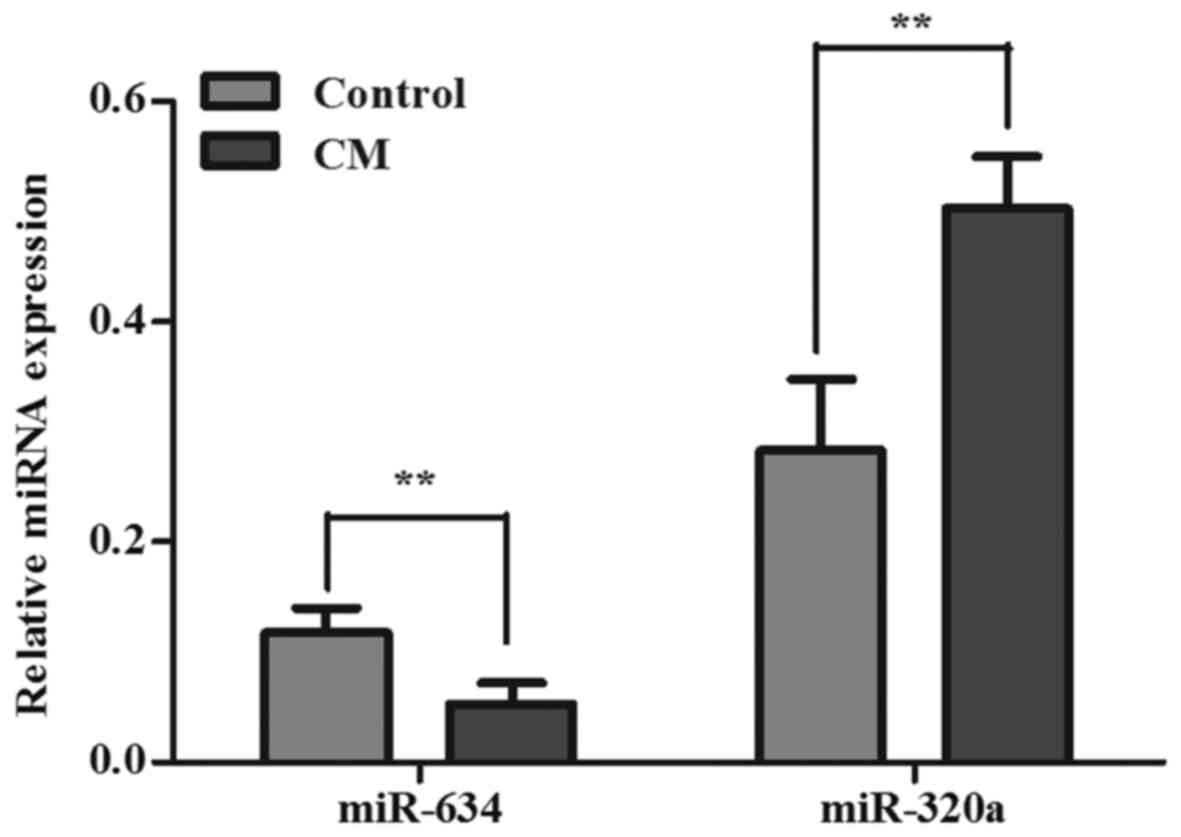
To validate the microarray results, miR-320a and
miR-634 were selected for further validation in the serum of the 30
CM patients and 30 healthy controls by using RT-qPCR due to no
target genes of miR-1249-5p and miR-6870-3p were predicted. Results
showed that the expression of miR-320a increased and the expression
of miR-634 decreased in the patients with CM in comparison with the
normal controls (P<0.05). These results were consistent with the
results of the microarray (Fig.
2).
Prediction of miR-320a target
genes
The miRNA target genes were predicted using three
databases including miRanda, TargetScan and PicTar. We identified
487 target genes from these three databases and only genes rank in
the top 21 that were predicted for miR-320a are shown in Table III, and the remaining 466 genes are
not shown in the table. However, following GO enrichment analysis
and pathway enrichment analysis are conducted on the basis of these
487 genes.
 | Table III.Some of genes that were predicted for
miR-320a. |
Table III.
Some of genes that were predicted for
miR-320a.
| miRNA | Target genes |
|---|
| miR-320a | CDH2, CPD, IGF1R,
MAT2A, PBX3, TAF5, NRP1, IGF2BP3, USP25, ZFP91, TMEM47, MTDH, DNER,
HECTD2, MIER3, ARF1, EREG, ESRRG, HOXA5, MAP1B, MN1 |
GO enrichment analysis for the target
genes of miR-320a
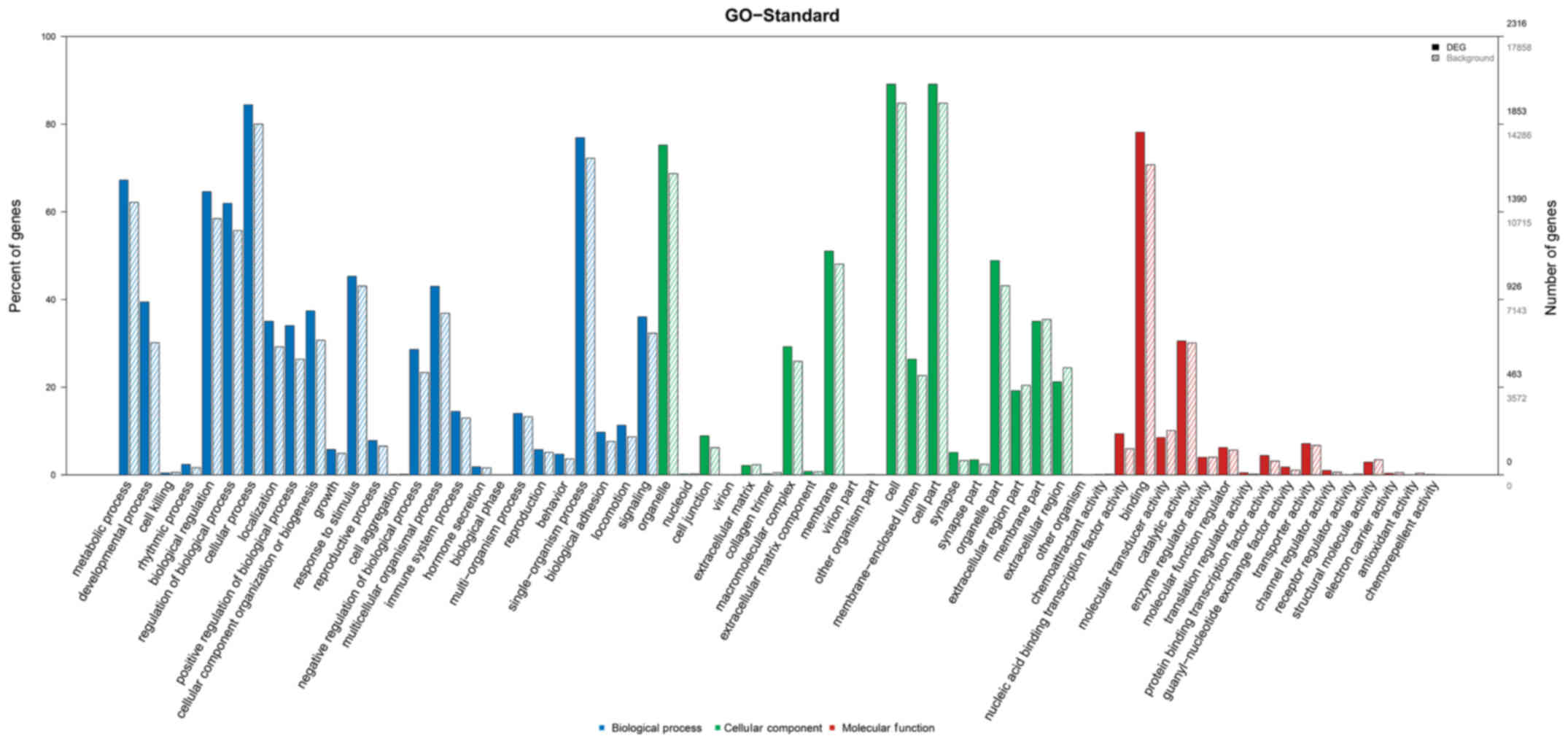
As shown in Fig. 3,
the GO functional enrichment results revealed that the miR-320a
target genes were enriched in numerous bioprocesses such as cell
adhesion, cellular process, multicellular organismal development,
and cellular component organization or biogenesis. The target genes
were also involved in the synthesis of many cellular components,
such as cell junction and extracellular matrix component. Moreover,
the target genes was closely related to many molecular functions,
such as necleic acid binding transcription factor activity.
Pathway enrichment analysis of the
miR-320a target genes
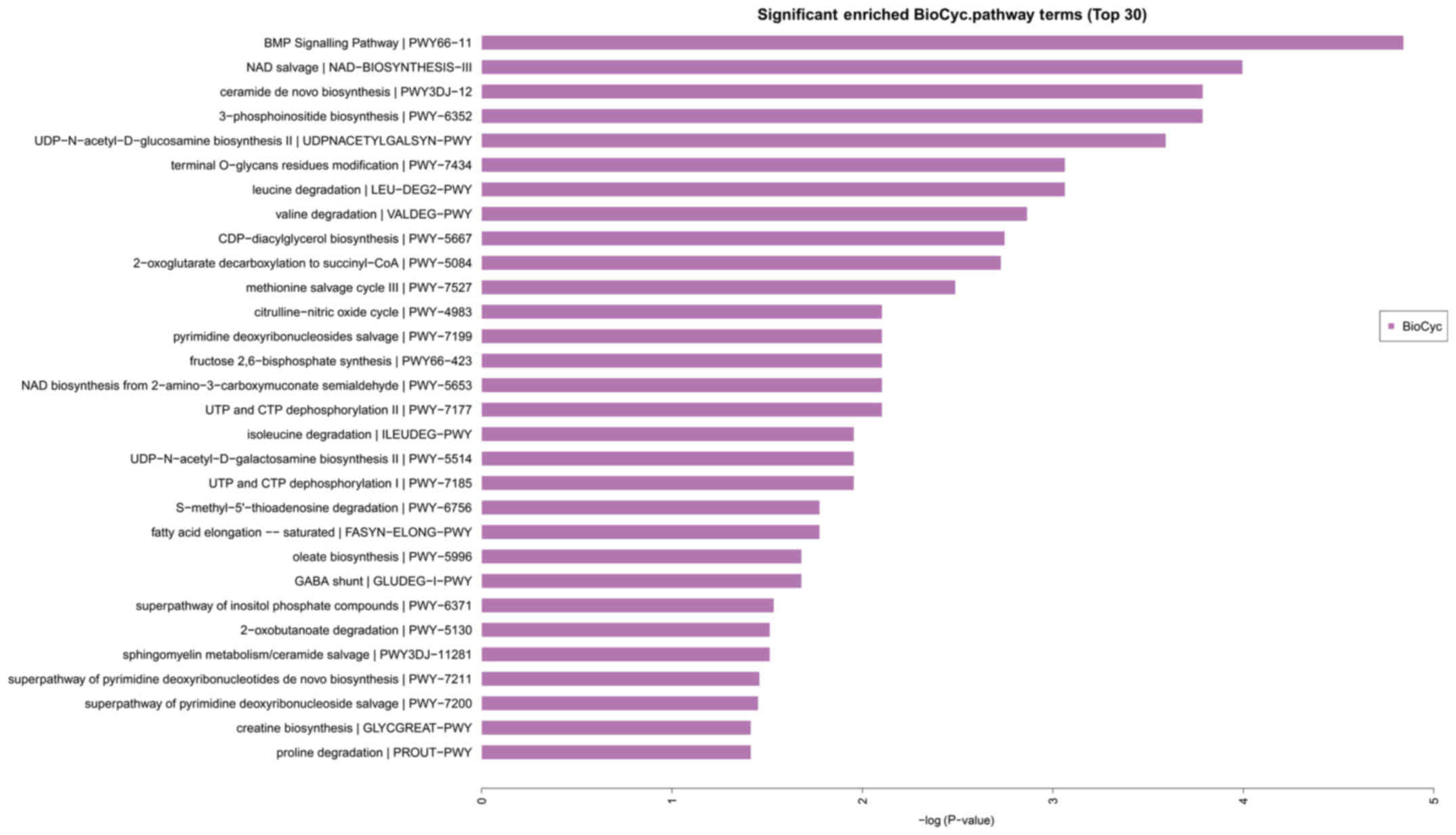
On the basis of the classification of the GO
annotation, the KEGG database was used to analyze the signaling
pathways modulated by the target genes. As shown in the Fig. 4, the miR-320a target genes were mainly
enriched in the bone morphogenetic protein (MBP) signaling pathway,
nicotinamide adenine dinucleotide (NAD) pathway, de novo ceramide
biosynthetic pathway and 3-phosphoinositide biosynthesis pathway
(Fig. 4).
Discussion
miRNAs are a series of short (typically 18–25
nucleotides), single-stranded and highly conserved non-coding RNA
molecules. They negatively regulate gene expression either through
inhibition of mRNA translation or by promoting mRNA degradation.
Emerging evidence suggests that miRNAs can be emerge as important
intervention targets and predictive tools for various human cancers
because of their stability and convenience of miRNA detection
(21,22). In particular, serum miRNAs may be used
as biomarkers in diagnosis (6). Zhang
et al (23) found that the
expression of miR-217 is downregulated in tissues of CM patients,
and overexpression of miR-217 inhibits the proliferation and
promotes the apoptosis of the primary CM cells. Cao et al
(24) reported that downregulation of
miR-218 promotes the proliferation of cells derived from patients
with CM. These findings suggested that miR-217 and miR-218 are
potential targets for CM prevention and therapy due to their tumor
suppression properties. However, the study on miRNA expression in
the serum of patients with CM has not yet been reported. The
present study reported for the first time the differential
expression profiles of miRNA in the serum of patients with CM. We
confirmed four differentially expressed miRNAs. Of these four
miRNAs, the expression levels of miR-320a and miR-1249-5p increased
and those of miR-634 and miR-6870-3p decreased in patients with CM
in comparison with the healthy controls. Only four differentially
expressed miRNAs were identified was due to large dispersion caused
by the small sample size of our study. In a study conducted by Liu
(25), sixteen patients were selected
and their serum miRNA profiles were determined using a microarray,
and the results showed that only four serum miRNAs were found to be
differentially altered, it is consistent with our experimental
results. Due to no target genes of miR-1249-5p and miR-6870-3p were
predicted, only miR-320a and miR-634 were selected for further
verification using RT-qPCR, and the results were consistent with
the microarray results, such finding indicated the high reliability
of the chip technology.
A newly found tumor suppressor, miR-634 plays an
important role in inducing the apoptosis of tumor cells. miR-634
inhibits the cell proliferation and induces apoptosis in cervical
cancer cells (26) and nasopharyngeal
carcinoma cells (27). Moreover,
Fujiwara et al (28) found
that miR-634 enhanced chemotherapy-induced cytotoxicity in a model
of esophageal squamous cell carcinoma. In the present study, the
expression of miR-634 was lower in the serum of CM patients,
suggesting that it may also be an important tumor suppressor in CM.
This finding provided the basis for the further study of the role
of miR-634 in CM.
In recent years, the differential expression of
miR-320a between tumor tissue and healthy tissue was identified
using qPCR, gene chips and Western blot. Previous studies have
revealed that miR-320a exhibits abnormal expression levels in
multiple malignancies and is involved in the formation, progression
and metastasis of cancer. miR-320a is down-regulated in various
cancers such as breast cancer (29),
bladder cancer (30), gastric cancer
(31), colorectal cancer (32), and nasopharyngeal cancer (33). The overexpression of miR-320a
suppresses the capability of cell migration and invasion and
induces G0/G1 growth arrest (32).
However, miR-320a does not exert an inhibition effect on all
cancers. Yao et al (34) found
that the overexpression of miR-320a promoted the migration and
invasion of the hepatocellular carcinoma (HCC) cell line SMMC-7721,
but decreased the expression of the endogenous miR-320a/c/d with
specific inhibitors significantly inhibiting the migration and
invasion of SK-Hep-1 cells. Wen et al (35) reported that miR-320a is markedly
overexpressed in the HCC patients and could be serve as preclinical
biomarkers. Xu et al (36)
reported that miR-320a is upregulated in prostate cancer cells, and
may exhibit an oncogenic function in prostate cancer. Recently, Xu
et al (37) found that
miR-320a is a potentially valuable biomarker for diagnosing older
females with gastric cancer. The difference expression levels in
different cancers are associated with tumor types, tumor size,
clinical stage and lymphatic metastasis (38).
Several studies on miR-320a in various cancers have
been conducted. However, no studies have investigated the
clinicopathological value of miR-320a expression in CM. In the
present study, the expression of miR-320a in CM patients was higher
than that in the healthy controls. This finding indicated that
miR-320a may be an oncogene in CM.
miR-320a is important in a number of cancer types
(39–41), and miR-320a was upregulated in this
study, hence, we predicted its target genes, and identified 487
target genes using miRanda, TargetScan and PicTar. Of these target
genes, myocyte enchancer factor 2D (MEF2D) is invovled in
regulating tumor biology of CM, it related to proliferation,
invasion and tumor size of myxoma (42). VEGFA, which possesses a highly
conserved binding site to miR-320a (43), probably induces angiogenesis for tumor
growth in CMs (44,45). Therefore, we assume that miR-320a is
probably involved in tumor growth in CM by targeting VEGFA and
MEF2D. Future research are needed to investigate this possibility.
Moreover, the GO functional enrichment results showed that the
major functional terms of the 487 target genes included cell
adhesion, cellular process, multicellular organismal development,
cellular component organization or biogenesis (biological process
category), cell junction, extracellular matrix component (cellular
component category) and nucleic acid binding transcription factor
activity (molecular function category), This result indicated that
some target genes regulated CM development in a joint manner.
The results of the pathway enrichment analysis
showed that the target genes of miR-320a are mainly involved in the
MBP signaling pathway, nicotinamide adenine dinucleotide (NAD)
pathway, de novo ceramide biosynthetic pathway and
3-phosphoinositide biosynthesis pathway. Among these, de novo
ceramide biosynthetic pathway is one of the most extensively
studied pathways in cancer. As a powerful tumor suppressor,
ceramide is a kind of sphingomyelin molecule that produced from
sphingolipid metabolism or de novo synthesis when cells encounter
extracellular signals and receptors such as TNF-α, radiation and
anticancer agents (46–51). It can act as the second messenger of
various signal transduction pathways involved in the regulation of
cell proliferation, differentiation and apoptosis through the
activation of c-Jun nh2-terminal kinase (JNK), mitogen-activated
protein kinase (MAPK), kinase suppressors of Ras (KSR) and other
signaling pathways, and also can through effector molecules such as
protein kinases, protein phosphatase 1 and protein phosphatase 2A
(51–54). However, deficiencies in the ceramide
production in cancer cells lead to the survival of tumour cells and
resistance to chemotherapy (55). The
apoptotic signaling pathways mediated by ceramide have been
considered as targets for anticancer therapies (51). Therefore, miR-320a may be involved in
CM development by through the ceramide signaling pathway. More of
the differentially expressed miRNAs we identified participated in
the ceramide pathway. Further studies are needed to elucidate the
key pathways for the development and maintenance of CM.
We should note that there are some limitations in
this study. First, this study has a relatively small sample size
and future studies with larger cohorts should be conducted in the
future. Second, due to limited funding for experiment, serum pools
were produced for microarray analysis. This method will be unable
to detect variations between individuals due to miRNA expression
varies greatly between individuals. Moreover, it is impossible to
perform a correlation analysis between the qPCR and microarray
results because of pooling the samples. Third, only miR-320a and
miR-634 were validated due to no target genes of miR-1249-5p and
miR-6870-3p were predicted. Target genes of miRNA prediction relys
on bioinfomatic algorithms. We used 11 softwares (miRWalk, Microt4,
miRanda, mirbridge, miRDB, Pictar, PITA, miRMap, RNA22, RNAhybrid,
Targetscan) to predict the targer genes of miR-1249-5p and
miR-6870-3p, and found less than 4 softwares successfully predicted
the targer genes of them. In our study, we chose the three most
commonly used predicting softwares (miRanda, TargetScan and PicTar)
to study the target genes of differently expressed miRNA, but no
targer genes of miR-1249-5p and miR-6870-3p were predicted by these
three softwares, this will not have impacts on the reliability of
our results. Fourth, there are few references on miRNA in tissue of
CM patients (23), and our study
reported for the first time the differential expression profiles of
miRNA in the serum of patients with CM, so it's not easy to write
incisive disccusion, unless the research goes further, and it is
also not enough to affirm the validity of the two differently
expressed miRNA detected in our study as markers for CM pathology,
the association only remain proved. Fifth, we did not found that CM
pathology associated to other miRNA serum markers related to miRNA
we have found. The meaning of our study is to provide an
experimental basis for the further exploration of exploring the
specific miRNA for the diagnosis and treatment of CM. Finally,
Common practice is to find differently expressed miRNAs and then
verified them between patients and healthy people. The miRNA
differential expression profile in CM patients remains unexplored,
so we only study differently expressed miRNAs between CM patients
and healthy people in our study. As the research moves along, we
will tempted a selective analysis between man and women.
In conclusion, we reported for the first time the
circulating miRNA profile of patients with CM and found that the
expression levels miR-320a and miR-1249-5p were up-regulated and
the expression levels of miR-634 and miR-6870-3p were
down-regulated in CM patients. miR-320a is mainly involved in the
cellular physiological reaction and possibly participates in CM
development through the ceramide signaling pathway. The results in
this study provided an experimental basis for the further
exploration of exploring the specific miRNA for the diagnosis and
treatment of CM.
Acknowledgements
The authors would like to thank Zhihao Yang for his
assistance with the data analysis in this study.
Funding
This study was supported by grants from the key
project of young and middle-aged backbone talents cultivation of of
Fujian Province Health System of China (No. 2015-ZQN-ZD-16).
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JL carried out sample preparation, and experimental
validation. QW and LY performed the experiments, analyzed the data,
and wrote the manuscript. LC conceived of the study and
participated in its design and coordination. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Ethical approval was given by the Ethics Committee
of The Union Hospital of Fujian Medical University (No.
2017KY027).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Debourdeau P, Gligorov J, Teixeira L,
Aletti M and Zammit C: Malignant cardiac tumors. Bull Cancer. 91
Suppl 3:S136–S146. 2004.(In French).
|
|
2
|
Lee KS, Kim GS, Jung Y, Jeong IS, Na KJ,
Oh BS, Ahn BH and Oh SG: Surgical resection of cardiac myxoma-a
30-year single institutional experience. J Cardiothorac Surg.
12:182017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mazzurin AV, Soboleva NI and Semenov BN:
Cardiac myxomas in children. Pediatriia. 46:50–53. 1967.(In
Russian). PubMed/NCBI
|
|
4
|
Butany J, Nair V, Naseemuddin A, Nair GM,
Catton C and Yau T: Cardiac tumors: Diagnosis and management.
Lancet Oncol. 6:219–228. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Reynen K: Cardiac myxomas. New Engl J Med.
333:1610–1617. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jain D, Maleszewski JJ and Halushka MK:
Benign cardiac tumors and tumorlike conditions. Ann of Diagn
Pathol. 14:215–230. 2010. View Article : Google Scholar
|
|
7
|
Rupp GM, Heyman RA, Martinez AJ, Sekhar LN
and Jungreis CA: The pathology of metastatic cardiac myxoma. Am J
Clin Pathol. 91:221–227. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Garzon R, Fabbri M, Cimmino A, Calin GA
and Croce CM: MicroRNA expression and function in cancer. Trends
Mol Med. 12:580–587. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jansson MD and Lund AH: MicroRNA and
cancer. Mol Onco. 6:590–610. 2012. View Article : Google Scholar
|
|
11
|
Chan SH, Wu CW, Li AF, Chi CW and Lin WC:
miR-21 microRNA expression in human gastric carcinomas and its
clinical association. Anticancer Res. 28:907–911. 2008.PubMed/NCBI
|
|
12
|
Iorio MV, Ferracin M, Liu CG, Veronese A,
Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M,
et al: MicroRNA gene expression deregulation in human breast
cancer. Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K,
Guo J, Zhang Y, Chen J, Guo X, et al: Characterization of microRNAs
in serum: A novel class of biomarkers for diagnosis of cancer and
other diseases. Cell Res. 18:997–1006. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhu Z, Qi Y, Ge A, Zhu Y, Xu K, Ji H, Shi
Z, Cui L and Zhou M: Comprehensive characterization of serum
microRNA profile in response to the emerging avian influenza A
(H7N9) virus infection in humans. Viruses. 6:1525–1539. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Krawczynski K, Najmula J, Bauersachs S and
Kaczmarek MM: MicroRNAome of porcine conceptuses and trophoblasts:
Expression profile of micrornas and their potential to regulate
genes crucial for establishment of pregnancy. Biol Reprod.
92:212015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shen J, Xiao Z, Wu WK, Wang MH, To KF,
Chen Y, Yang W, Li MS, Shin VY, Tong JH, et al: Epigenetic
silencing of miR-490-3p reactivates the chromatin remodeler SMARCD1
to promote Helicobacter pylori-induced gastric carcinogenesis.
Cancer Res. 75:754–765. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ho CY, Bar E, Giannini C, Marchionni L,
Karajannis MA, Zagzag D, Gutmann DH, Eberhart CG and Rodriguez FJ:
MicroRNA profiling in pediatric pilocytic astrocytoma reveals
biologically relevant targets, including PBX3, NFIB, and METAP2.
Neuro Oncol. 15:69–82. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li X, Peng B, Zhu X, Wang P, Xiong Y, Liu
H, Sun K, Wang H, Ou L, Wu Z, et al: Changes in related circular
RNAs following ERβ knockdown and the relationship to rBMSC
osteogenesis. Biochem Biophys Res Commun. 493:100–107. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Eisen MB, Spellman PT, Brown PO and
Botstein D: Cluster analysis and display of genome-wide expression
patterns. Proc Natl Acad Sci USA. 95:pp. 14863–14868. 1998;
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kong YW, Ferland-McCollough D, Jackson TJ
and Bushell M: microRNAs in cancer management. Lancet Oncol.
13:e249–e258. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Corsini LR, Bronte G, Terrasi M, Amodeo V,
Fanale D, Fiorentino E, Cicero G, Bazan V and Russo A: The role of
microRNAs in cancer: Diagnostic and prognostic biomarkers and
targets of therapies. Expert Opin Ther Targets. 16 Suppl
2:S103–S109. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang J, Wang C and Xu H: miR-217
suppresses proliferation and promotes apoptosis in cardiac myxoma
by targeting Interleukin-6. Biochem Biophys Res Commun.
490:713–718. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cao Q, Dong P and Wang Y, Zhang J, Shi X
and Wang Y: miR-218 suppresses cardiac myxoma proliferation by
targeting myocyte enhancer factor 2D. Oncol Rep. 33:2606–2612.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu N, Cui RX, Sun Y, Guo R, Mao YP, Tang
LL, Jiang W, Liu X, Cheng YK, He QM, et al: A four-miRNA signature
identified from genome-wide serum miRNA profiling predicts survival
in patients with nasopharyngeal carcinoma. Int J Cancer.
134:1359–1368. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cong J, Liu R, Wang X, Jiang H and Zhang
Y: MiR-634 decreases cell proliferation and induces apoptosis by
targeting mTOR signaling pathway in cervical cancer cells. Artif
Cells Nanomed Biotechnol. 44:1694–1701. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Peng X, Cao P, He D, Han S, Zhou J, Tan G,
Li W, Yu F, Yu J, Li Z and Cao K: MiR-634 sensitizes nasopharyngeal
carcinoma cells to paclitaxel and inhibits cell growth both in
vitro and in vivo. Int J Clin Exp Pathol. 7:6784–6791.
2014.PubMed/NCBI
|
|
28
|
Fujiwara N, Inoue J, Kawano T, Tanimoto K,
Kozaki K and Inazawa J: miR-634 activates the mitochondrial
apoptosis pathway and enhances chemotherapy-induced cytotoxicity.
Cancer Res. 75:3890–3901. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang B, Yang Z, Wang H, Cao Z, Zhao Y,
Gong C, Ma L, Wang X, Hu X and Chen S: MicroRNA-320a inhibits
proliferation and invasion of breast cancer cells by targeting
RAB11A. Am J Cancer Res. 5:2719–2729. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shang C, Zhang H, Guo Y, Hong Y, Liu Y and
Xue Y: MiR-320a down-regulation mediates bladder carcinoma invasion
by targeting ITGB3. Mol Biol Rep. 41:2521–2527. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhu Y, Zhang Y, Sui Z, Zhang Y, Liu M and
Tang H: USP14 de-ubiquitinates vimentin and miR-320a modulates
USP14 and vimentin to contribute to malignancy in gastric cancer
cells. Oncotarget. 8:48725–48736. 2017.PubMed/NCBI
|
|
32
|
Zhao H, Dong T, Zhou H, Wang L, Huang A,
Feng B, Quan Y, Jin R, Zhang W, Sun J, et al: miR-320a suppresses
colorectal cancer progression by targeting Rac1. Carcinogenesis.
35:886–895. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Qi X, Li J, Zhou C, Lv C and Tian M:
MicroRNA-320a inhibits cell proliferation, migration and invasion
by targeting BMI-1 in nasopharyngeal carcinoma. FEBS Lett.
588:3732–3738. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yao J, Liang LH, Zhang Y, Ding J, Tian Q,
Li JJ and He XH: GNAI1 suppresses tumor cell migration and invasion
and is post-transcriptionally regulated by Mir-320a/c/d in
hepatocellular carcinoma. Cancer Biol Med. 9:234–241.
2012.PubMed/NCBI
|
|
35
|
Wen Y, Han J, Chen J, Dong J, Xia Y, Liu
J, Jiang Y, Dai J, Lu J, Jin G, et al: Plasma miRNAs as early
biomarkers for detecting hepatocellular carcinoma. Int J Cancer.
137:1679–1690. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xu G, Wu J, Zhou L, Chen B, Sun Z, Zhao F
and Tao Z: Characterization of the small RNA transcriptomes of
androgen dependent and independent prostate cancer cell line by
deep sequencing. PLoS One. 5:e155192010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xu Q, Dong QG, Sun LP, He CY and Yuan Y:
Expression of serum miR-20a-5p, let-7a, and miR-320a and their
correlations with pepsinogen in atrophic gastritis and gastric
cancer: A case-control study. BMC Clin Pathol. 13:112013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yang HP, Yu J, Wang L, Ding D, Zhang L,
Chu C, Chen Q, Xu Z, Zou Q and Liu X: miR-320a is an independent
prognostic biomarker for invasive breast cancer. Oncol Lett.
8:1043–1050. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Diakos C, Zhong S, Xiao Y, Zhou M,
Vasconcelos GM, Krapf G, Yeh RF, Zheng S, Kang M, Wiencke JK, et
al: TEL-AML1 regulation of survivin and apoptosis via miRNA-494 and
miRNA-320a. Blood. 116:4885–4893. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hummel R, Wang T, Watson DI, Michael MZ,
Van der Hoek M, Haier J and Hussey DJ: Chemotherapy-induced
modification of microRNA expression in esophageal cancer. Oncol
Rep. 26:1011–1017. 2011.PubMed/NCBI
|
|
41
|
Salendo J, Spitzner M, Kramer F, Zhang X,
Jo P, Wolff HA, Kitz J, Kaulfuß S, Beißbarth T, Dobbelstein M, et
al: Identification of a microRNA expression signature for
chemoradiosensitivity of colorectal cancer cells, involving
miRNAs-320a, −224, −132 and let7g. Radiother Oncol. 108:451–457.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Huo Y, Zhao Q, Wang C, Zhao F, Du Y and
Sun W: The involvement of myocyte enhancer factor 2D in regulating
tumor biology of cardiac myxoma. Tumour Biol. 37:5405–5411. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yin Z, Zhao Y, Li H, Yan M, Zhou L, Chen C
and Wang DW: miR-320a mediates doxorubicin-induced cardiotoxicity
by targeting VEGF signal pathway. Aging (Albany NY). 8:192–207.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kono T, Koide N, Hama Y, Kitahara H,
Nakano H, Suzuki J, Isobe M and Amano J: Expression of vascular
endothelial growth factor and angiogenesis in cardiac myxoma: A
study of fifteen patients. J Thorac Cardiovasc Surg. 119:101–107.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Amano J, Kono T, Wada Y, Zhang T, Koide N,
Fujimori M and Ito K: Cardiac myxoma: Its origin and tumor
characteristics. Ann Thorac Cardiovasc Surg. 9:215–221.
2003.PubMed/NCBI
|
|
46
|
Hannun YA and Obeid LM: Ceramide: An
intracellular signal for apoptosis. Trends Biochem Sci. 20:73–77.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Gulbins E: Regulation of death receptor
signaling and apoptosis by ceramide. Pharmacol Res. 47:393–399.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kolesnick R and Fuks Z: Radiation and
ceramide-induced apoptosis. Oncogene. 22:5897–5906. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ogretmen B and Hannun YA: Biologically
active sphingolipids in cancer pathogenesis and treatment. Nat Rev
Cancer. 4:604–616. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Modrak DE, Gold DV and Goldenberg DM:
Sphingolipid targets in cancer therapy. Mol Cancer Ther. 5:200–208.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Lin CF, Chen CL and Lin YS: Ceramide in
apoptotic signaling and anticancer therapy. Curr Med Chem.
13:1609–1616. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Mathias S, Peña LA and Kolesnick RN:
Signal transduction of stress via ceramide. Biochem J. 335:465–480.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hannun YA and Obeid LM: Ceramide and the
eukaryotic stress response. Biochem Soc Trans. 25:1171–1175. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Stoica BA, Movsesyan VA, Lea PM IV and
Faden AI: Ceramide-induced neuronal apoptosis is associated with
dephosphorylation of Akt, BAD, FKHR, GSK-3beta, and induction of
the mitochondrial-dependent intrinsic caspase pathway. Mol Cell
Neurosci. 22:365–382. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Morad SAF and Cabot MC:
Ceramide-orchestrated signalling in cancer cells. Nat Rev Cancer.
13:51–65. 2013. View Article : Google Scholar : PubMed/NCBI
|