Introduction
A total of >85% lung cancer is classified as
non-small cell lung cancer (NSCLC) (1). Although early diagnosis and novel
therapeutic approaches have markedly prolonged the overall survival
of patients with NSCLC, the survival rate for patients with drug
resistance remains low (2).
Long non-coding RNAs (lncRNAs) are transcripts
consisting of >200 nucleotides with no ability to code protein
(3). Previous studies have indicated
that lncRNAs were involved in the pathogenesis of various diseases,
including cancer (4).
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) has
been identified to regulate cancer cell proliferation, migration
and invasion in lung, hepatocellular, ovarian and colorectal cancer
(5–7).
MALAT1 was also demonstrated to induce temozolomide resistance in
glioblastoma (8). However, whether
and how MALAT1 contributes to cisplatin resistance in NSCLC is
unknown.
MicroRNAs (miRNAs) are small non-coding RNAs that
function in RNA silencing through complementary binding. miR-145 is
a well-known tumor suppressor that inhibits oncogene expression in
numerous types of cancer (9,10). Additionally, increased miR-145
expression levels increased the sensitivity of hepatocellular
carcinoma cells to chemotherapy via the downregulation of Mothers
against decapentaplegic homolog 3 (11). In lung cancer, miR-145 was suggested
to inhibit cancer cell proliferation, migration and invasion, but
its role in chemoresistance was not identified (12).
Cancer stem cells have drawn much attention for
their pivotal role in promoting relapse and drug resistance,
including cisplatin resistance (13).
With the ability to maintain cancer cell stemness, Kruppel-like
factor 4 (KLF4) serves as an oncogene in a number of types of
cancer, and has been demonstrated to be associated with
chemoresistance (14,15). For example, the interaction between
KLF4 and High mobility group box 1 conferred resistance in
osteosarcoma cells to multiple chemotherapy agents, including
cisplatin (16). In lung cancer, an
increased expression of KLF4 was observed in high-grade NSCLC
compared with low-grade disease (17). Increased Hox transcript antisense RNA
and KLF4 levels were involved in cisplatin resistance in NSCLC
(18), but the underlying mechanism
remains unknown.
The present study aimed to explore whether and how
MALAT1 may affect the sensitivity of NSCLC towards cisplatin. It
was identified that MALAT1 and KLF4 levels were increased and
miR-145 levels were decreased in tumor tissues from patients with
cisplatin-refractory NSCLC compared with those from
cisplatin-sensitive NSCLC patients. Silencing of MALAT1 sensitized
A549rCDDP cells to cisplatin, while the overexpression of MALAT1
induced cisplatin resistance in A549 cells. Importantly, a direct
regulatory association between MALAT1 and miR-145 and the target
gene KLF4 was confirmed. Taken together, the present study
highlighted the role of MALAT1 in the development of
cisplatin-resistance in NSCLC.
Materials and methods
Patients
A total of 52 tumor tissue samples were collected
from 31 patients with NSCLC for whom cisplatin-based chemotherapy
was effective following surgery (patients who were
cisplatin-sensitive), and 21 patients with NSCLC for whom
cisplatin-based chemotherapy was ineffective following surgery
(patients who were cisplatin-resistant). Histopathological types
were assigned using WHO pathological staging criteria (19). All patients were administrated 100
mg/m2 cisplatin intravenously over 30 to 120 min on day
1 of the 28 day cycle. The total number of cycles administrated was
one. The samples were obtained from Shouguang People's Hospital
(Shouguang, China) from June 2012 to June 2015. The present study
was approved by the Ethics Committee of Shouguang People's
Hospital. All participants provided written informed consent prior
to surgery. Patients did not receive radiotherapy or chemotherapy
prior to surgery. The tissues were immediately frozen for protein
and RNA extraction.
Cell culture and agents
The human NSCLC A549 and H1299 cell lines were
purchased from American Type Culture Center (Manassas, VA, USA) and
the A549 cisplatin-resistant A549rCDDP subline was obtained from
the Cancer Hospital of Peking Union Medical College, Chinese
Academy of Medical Sciences (Beijing, China). All cell lines were
cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) containing 10% fetal bovine serum (Hyclone;
GE Healthcare Life Sciences, Logan, UT, USA). The medium for
A549rCDDP cells was additionally supplemented with 2 mg/l cisplatin
(Selleck Chemicals, Houston, TX, USA). All cell lines were
cultivated in a humid incubator at 37°C with 5% CO2. For
cisplatin treatment, 1, 2, 5 or 10 µM of cisplatin was added into
the culture medium of A549 cells or A549rCDDP cells for 48 h, and
then the cells were used for subsequent experimentation.
RNA extraction and reverse
transcription quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissues and A549, H1299
and A549rCDDP cells using the miRNeasy Mini kit (QIAGEN, Hilden,
Germany) according to the manufacturer's protocol. A Molony-Murine
Leukemia Virus kit (Life Technologies; Thermo Fisher Scientific,
Inc.) was used to synthesize cDNA. qPCR was performed with a CFX96
Touch™ system (Bio-Rad Laboratories, Inc., Hercules, CA,
USA) using SYBR Premix Ex Taq (Takara Bio, Inc., Otsu, Japan). The
thermocycling conditions for miR-145 and U6 were as follows: 95°C
for 10 min, 40 cycles of 95°C for 15 sec and 60°C for 1 min. The
thermocycling conditions for lncRNA, mRNA and GAPDH were as
follows: 95°C for 10 min, followed by 40 cycles at 95°C for 15 sec,
55°C for 30 sec and 72°C for 30 sec. GAPDH was used as an internal
control for lncRNA and mRNA, and U6 was applied as an internal
control for miRNA. The relative expression levels were calculated
using the 2−ΔΔCq method (20). The sequences for the primers were as
follows: MALAT1 forward, AGACCTTGAAATCCAT; MALAT1 reverse,
CTTCTGCTTCCTACTT; miR-145 forward, ACACTCCAGCTGGGGTCCAGTTTTCCCAGGA;
miR-145 reverse, TGGTGTCGTGGAGTCG; KLF4 forward,
GTCAGTTCATCTGAGCGGG; KLF4 reverse, AGAGTTCCCATCTCAAGGCA; GAPDH
forward, AGAAGGCTGGGGCTCATTTG; GAPDH reverse, AGGGGCCATCCACAGTCTTC;
U6 forward, CTCGCTTCGGCAGCACA; and U6 reverse,
TGGTGTCGTGGAGTCG.
Western blotting
Protein was measured with a bicinchoninic acid kit
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) according to
manufacturer's protocol. HRP-conjugated secondary antibodies for
mouse (SA00001-1) and rabbit (SA00001-2) were purchased from
Proteintech (Rosemont, IL). The antibody for GAPDH (catalog no.
G8795, 1:10,000) was purchased from Sigma Aldrich; Merck KGaA. The
KLF4 antibody (catalog no. 12173, 1:1,000) was purchased from Cell
Signaling Technology, Inc. (Danvers, MA, USA). A549 cells were
lysed in radioimmunoprecipitation assay lysis buffer (Beyotime
Institute of Biotechnology, Haimen, China) containing protease
inhibitor (Sigma Aldrich; Merck KGaA). 20 µg protein was separated
by 8% SDS-PAGE and then transferred to a polyvinylidene fluoride
membrane (EMD Millipore, Billerica, MA USA). Subsequent to blocking
at room temperature in 5% non-fat milk for 30 min, the membranes
were incubated with the indicated primary antibodies overnight at
4°C. The membranes were then incubated with a HRP-conjugated
secondary antibody for 1 h at room temperature; anti-mouse antibody
at 1:10,000 for GAPDH (catalog no. SA00001-1, Proteintech,
Rosemont, IL), and anti-rabbit at 1:10,000 for KLF4 (catalog no.
SA00001-2, Proteintech, Rosemont, IL). The membranes were developed
using Pierce ECL Plus substrate (Thermo Fisher Scientific, Inc.).
Images were captured with a densitometer (GS-700; Bio-Rad
Laboratories, Inc.), analysis of protein expression was achieved
using ImageJ software (Version 1.51k, Rawak Software, Inc.,
Germany). GAPDH served as an internal control.
Cell viability assay
Cell viability was assessed using a Cell Counting
Kit-8 (CCK-8; Dojindo Molecular Technologies, Kumamoto, Japan).
5,000 A549, H1299 or A549rCDDP cells were seeded in a 96-well plate
with DMSO (Beijing Solarbio Science & Technology Co., Ltd.,
Beijing, China) or 1, 2, 5 or 10 µM of cisplatin and cultured at
37°C for 72 h. Subsequently, 10 µl CCK8 solution was added into
each well and incubated at 37°C for 2 h. The absorbance of each
well at 450 nm was detected using a microplate reader (Bio-Rad
Laboratories, Inc.) to measure cell viability.
Cell apoptosis assay
A cell apoptotic assay was performed using an
Annexin-V/Dead Cell Apoptosis kit (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol.
Briefly, following siRNA transfection and DMSO or cisplatin
treatment (2 µM), A549rCDDP cells were collected by trypsinization
(Invitrogen; Thermo Fisher Scientific, Inc.) and suspended in
Annexin binding buffer (Invitrogen; Thermo Fisher Scientific,
Inc.). Next, propidium iodide and Annexin V provided by
Annexin-V/Dead Cell Apoptosis kit (Invitrogen; Thermo Fisher
Scientific, Inc.) were added into the cell suspension and incubated
at room temperature for 15 min, followed by analysis using a BD
FACSCalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ,
USA). The cell apoptosis rate was analyzed using FlowJo software
(Version 10.4.1, FlowJo LLC, Ashland, OR, USA). The cells positive
for Annexin-V staining were considered apoptotic cells.
Small interfering (si)RNA
transfection
A total of 2 MALAT1 siRNAs (siRNA1 and siRNA2) and
control siRNA were purchased from Invitrogen; Thermo Fisher
Scientific, Inc. The siRNA sequences were as follows: MALAT1
siRNA1, 5′-GGGCUUCUCUUAACAUUUAtt-3′; MALAT1 siRNA2,
5′-GGGCAAAUAUUGGCAAUUAtt-3′. For the siRNA transfection,
2×106 A549 or A549rCDDP cells were seeded on 6-well
plates. The following day, 50 pM siRNA were mixed with
Lipofectamine® RNAiMAX (Invitrogen; Thermo Fisher
Scientific, Inc.) in Opti-MEM (Thermo Fisher Scientific, Inc.) and
incubated at 37°C for 15 min. Subsequently, the siRNA1 or siRNA2
mixture was added into the culture medium of cells and cultured at
37°C for 24 h prior to cisplatin or vehicle treatment.
Construction of plasmid and
transfection
Full-length MALAT1 was amplified from cDNA of 293
cells and cloned into a pcDNA3 plasmid (Addgene, Inc., Cambridge,
MA, USA) using PrimeSTAR Max DNA Polymerase (Takara Bio, Inc.). The
PCR thermocycling conditions were as follows: Denaturation at 98°C
for 10 sec, annealing at 55°C for 10 sec and elongation at 72°C for
10 sec, repeated for 35 cycles. For the overexpression of MALAT1, 5
µg pcDNA3-MALTA1 plasmid was transfected into A549 cells using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), RT-PCR was used to confirm elevation of MALAT1
after 48 h according to the procedure above. The primer sequences
for MALAT1 amplification were as follows: MALAT1 forward:
GGCGGTACCATGAAACAATTTGGAGAAG; MALAT1 reverse:
GCGCTCGAGCTAAGTTTGTACATTTTGCC.
Luciferase reporter assay
The 3′ untranslated region (UTR) of KLF4 was
amplified from cDNA of 293 cells and inserted into pGL-3 (Promega
Corporation, Madison, WI, USA) using PrimeSTAR Max DNA Polymerase
(Takara Bio, Inc.). The PCR thermocycling conditions were as
follows: denaturation at 98°C for 10 sec, annealing at 55°C for 10
sec and elongation at 72°C for 10 sec, repeated for 35 cycles.
Then, 293 cells were co-transfected with pGL3-KLF4 3′UTR WT,
miR-145 mimics or miR-negative control mimics, in combination with
a pcDNA3-MALAT1 plasmid or pcDNA3 and internal control pRL-TK
plasmid using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). The sequence of miR-145 mimics was
5′-GUCCAGUUUUCCCAGGAAUCCCU-3′. The sequence of miR-negative control
mimics was 5′-UCACAACCUCCUAGAAAGAGUAGA-3′. After 24 h, luciferase
activity and Renilla activity were measured using a Dual
Luciferase Reporter Assay kit (Promega Corporation) according to
manufacturer's protocol. Renilla activity was used as
control.
Statistical analysis
All statistical analyses were performed using
GraphPad Prism 5.0 software (GraphPad Software, Inc., La Jolla, CA,
USA). The data were expressed as the mean ± standard deviation.
Differences between two groups were analyzed using an unpaired
t-test. Differences from multiple groups were firstly analyzed
using a one-way analysis of variance followed by
Student-Newman-Keuls post-hoc analysis. P<0.05 was considered to
indicate a statistically significant difference. All experiments
were repeated three times.
Results
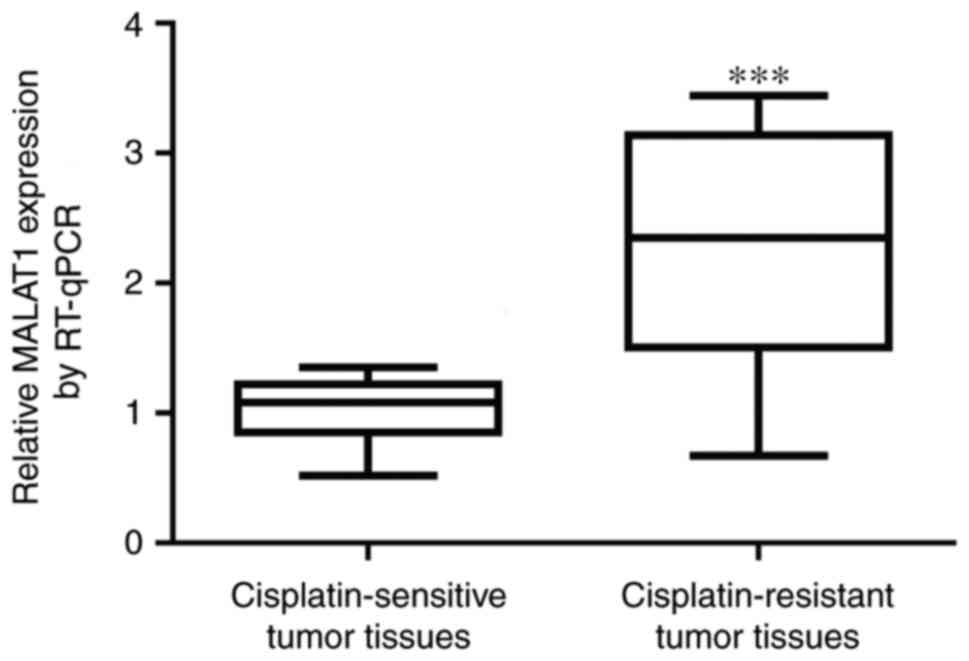
MALAT1 levels are increased in tumor
tissues from patients with cisplatin-resistant NSCLC compared with
those from patients with cisplatin-sensitive NSCLC
To explore whether MALAT1 was involved in cisplatin
resistance in NSCLC, MALAT1 levels from tumor tissues of 31
patients with cisplatin-sensitive NSCLC and 21 patients with
cisplatin-resistant NSCLC were measured. Significant elevation of
MALAT1 was detected in tumor tissues from patients with
cisplatin-resistant NSCLC (Fig. 1).
This suggested that the deregulation MALAT1 may contribute to
cisplatin resistance in patients with NSCLC.
MALAT1 levels are associated with
histological grades and metastasis
As summarized in Table
I, there were 21 male patients and 31 female patients in the
present study, with 32 patients aged <60 years old and 20
patients aged ≥60 years old, 20 patients with well to intermediate
differentiation and 32 patients with poor differentiation, and 41
patients with no metastasis and 11 patients with metastasis.
 | Table I.Expression of MALAT1 in tissues from
patients with non-small cell lung cancer. |
Table I.
Expression of MALAT1 in tissues from
patients with non-small cell lung cancer.
| Clinicopathological
factors | Cases, n | MALAT, mean ± SD | P-value |
|---|
| Sex |
|
| 0.70 |
| Male | 21 | 1.51±0.21 |
|
|
Female | 31 | 1.48±0.23 |
|
| Age, years |
|
| 0.35 |
|
<60 | 32 | 1.46±0.24 |
|
| ≥60 | 20 | 1.52±0.23 |
|
| Histological
grade |
|
| <0.01 |
| Well to
intermediate differentiation | 20 | 1.10±0.32 |
|
| Poor
differentiation | 32 | 1.93±0.58 |
|
| Metastasis |
|
| <0.01 |
| No | 41 | 1.16±0.38 |
|
|
Yes | 11 | 1.84±0.47 |
|
As for the difference of MALAT1 expression levels
between male (1.51±0.21) and female patients (1.48±0.23) or
patients aged <60 years old (1.46±0.24) and ≥60 years old
(1.52±0.23), there was no significant difference (P=0.70 and
P=0.35, respectively). There was significant difference of MALAT1
expression levels (P<0.01 and P<0.01, respectively) between
patients with well to intermediate differentiation (1.10±0.32) and
poor differentiation (1.93±0.58), and between patients with
metastasis (1.84±0.47) or no metastasis (1.16±0.38).
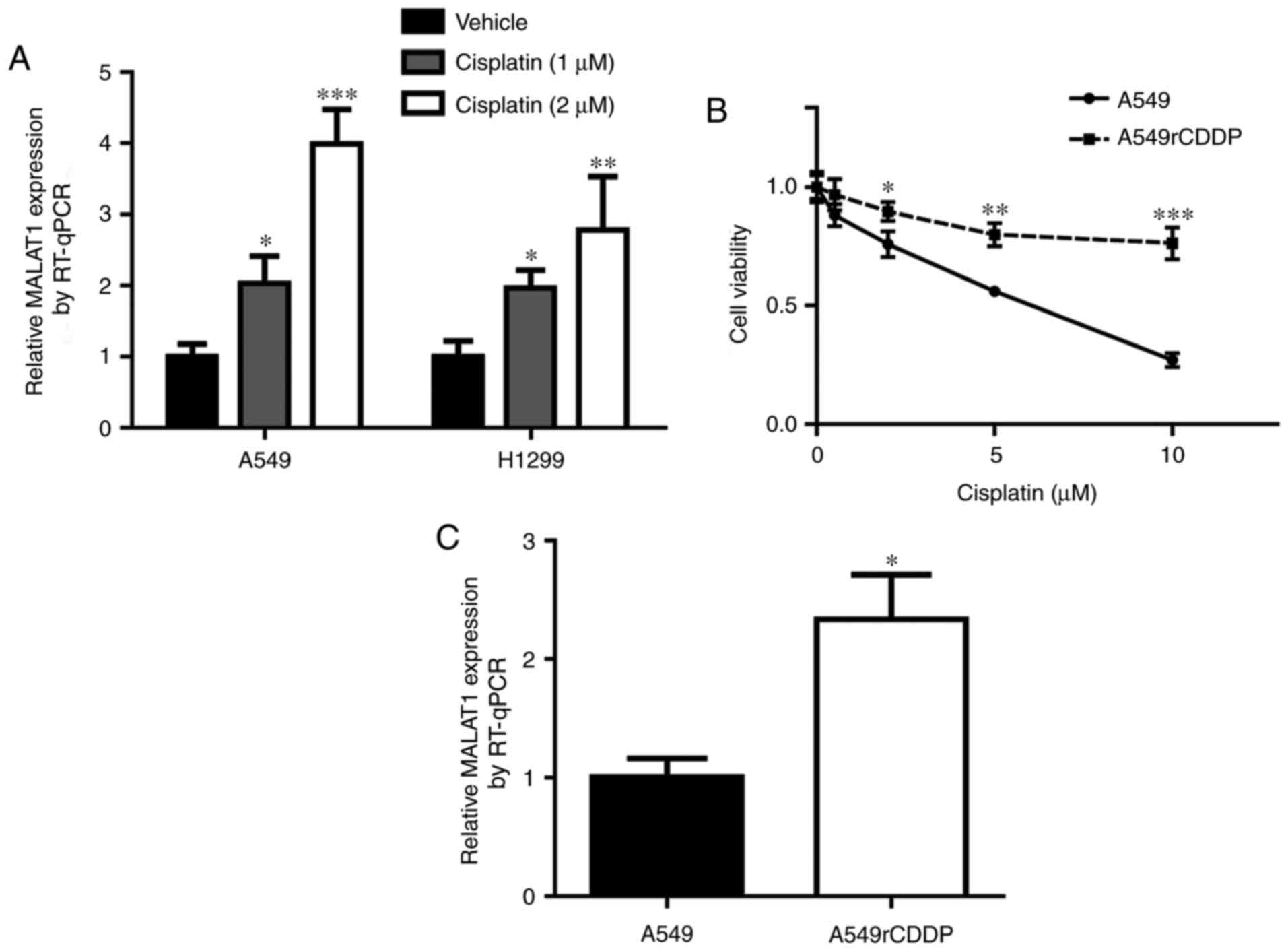
MALAT1 levels are increased in
cisplatin-resistant NSCLC cells and NSCLC cells treated with
cisplatin
To examine the role of MALAT1 during the development
of cisplatin resistance, 2 NSCLC cell lines, A549 cells and H1299,
were treated with (1 or 2 µM of cisplatin. Notably, there was an
elevation of MALAT1 level in response to cisplatin treatment in a
dose-dependent manner (Fig. 2A). In
addition, the present study aimed to analyze MALAT1 level in
cisplatin-resistant cells. The cell viability assay indicated that
A549rCDDP cells were relatively insensitive towards cisplatin
treatment compared with the parental A549 cells (Fig. 2B). Indeed, the MALAT1 level was
increased in A549rCDDP cells in comparison with A549 cells
(Fig. 2C). These data implied that
MALAT1 may contribute to cisplatin resistance in NSCLC cells.
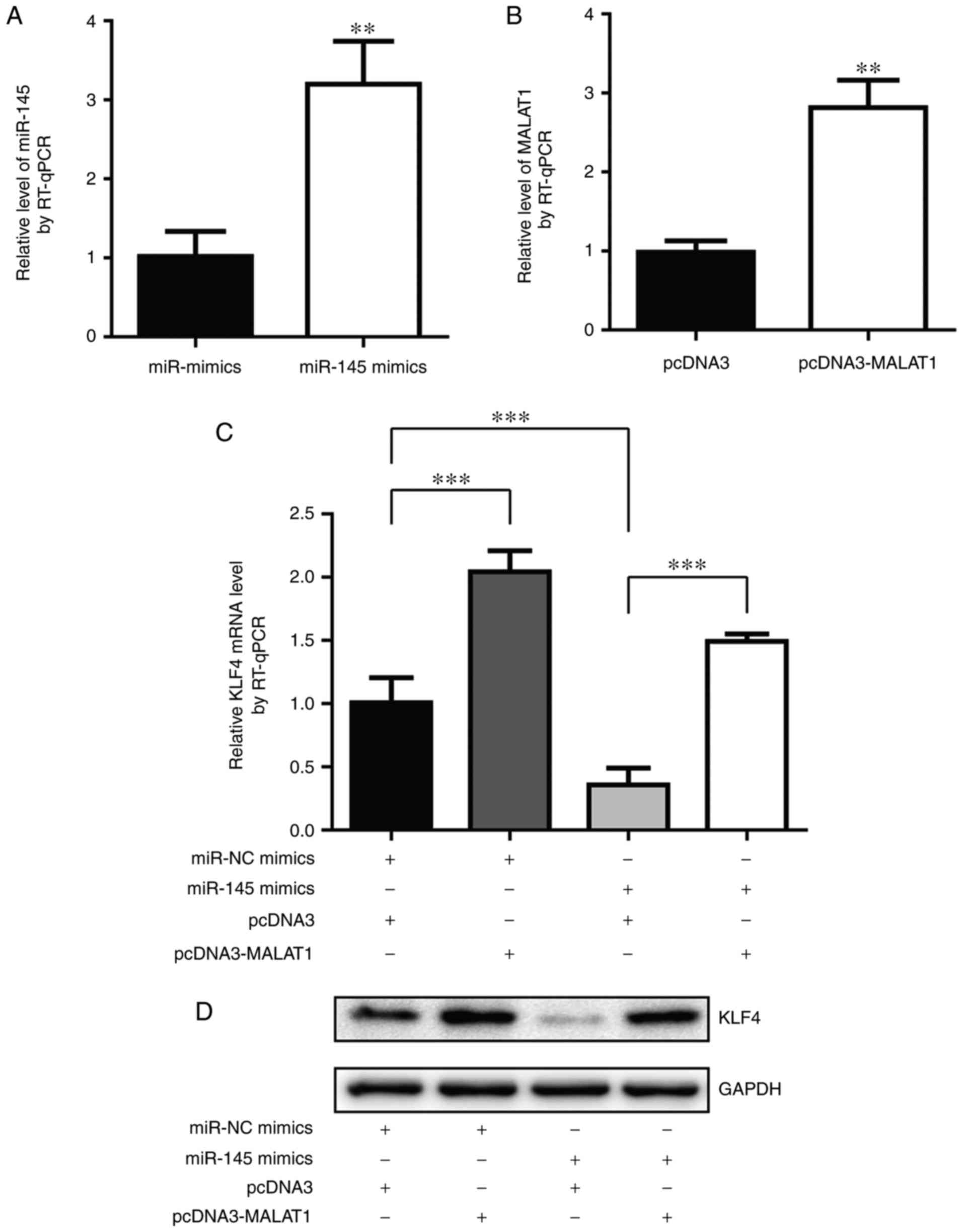
MALAT1 regulated KLF4, a chemotherapy
resistance associated oncogene, in A549 cells
KLF4 is a transcription factor that controls stem
cell reprogramming (17). Previously,
KLF4 was identified to be associated with cisplatin resistance in
several cancer types (16,21). A previous study indicated that miR-145
may target KLF4 in bladder cancer cells (22). Notably, MALAT1 was suggested to
repress miR-145 during endothelial to mesenchymal transition
(23). To investigate the role of
KLF4 and miR-145 in MALAT1-mediated cisplatin resistance, the
present study aimed to detect KLF4 level following MALAT1 or
miR-145 overexpression. As demonstrated in Fig. 3A and B, miR-145 mimics and
pcDNA-MALAT1 significantly increased miR-145 level and MALAT1 level
in A549 cells, respectively. As hypothesized, the overexpression of
MALAT1 markedly increased KLF4 mRNA level while miR-145 mimics
decreased KLF4 mRNA level, and a combination of MALAT1 and miR-145
overexpression rescued miR-145-induced KLF4 downregulation
(Fig. 3C). Consistently, MALAT1
overexpression increased KLF4 protein level compared with
transfection of pcDNA3 and miR-145 mimics triggered a decrease in
KLF4 protein level compared with cells transfected with miR-NC
mimics (Fig. 3D). These data
indicated that MALAT1 may function through the miR-145/KLF4 axis,
to confer cisplatin resistance in NSCLC cells.
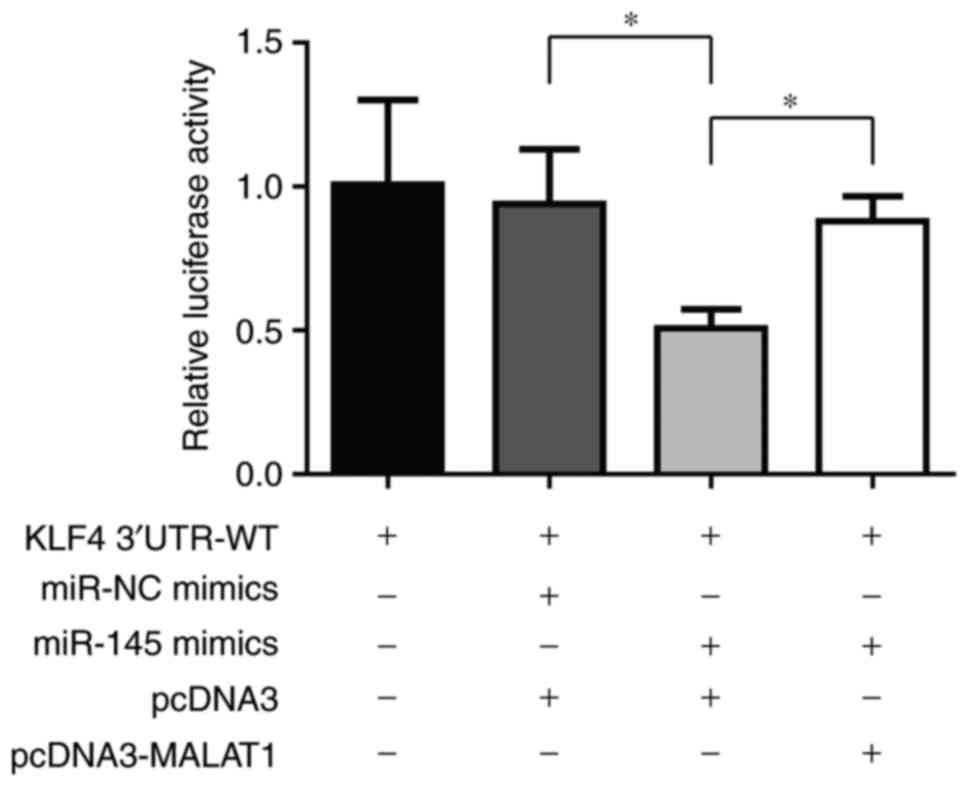
MALAT1 directly regulates miR-145 to
control KLF4 levels
To confirm whether MALAT1 increased KLF4 level via
targeting miR-145, a luciferase assay in 293 cells was performed.
In cells transfected with KLF4 3′UTR-WT, it was identified that
miR-145 mimics led to a decrease of luciferase activity and MALAT1
overexpression rescued this luciferase activity change (Fig. 4). This result indicated that KLF4 was
regulated by MALAT1 via miR-145.
MALAT1 silencing reverses cisplatin
resistance in A549rCDDP cells
The present study then aimed to confirm whether
MALAT1 contributed to cisplatin resistance in NSCLC cells by
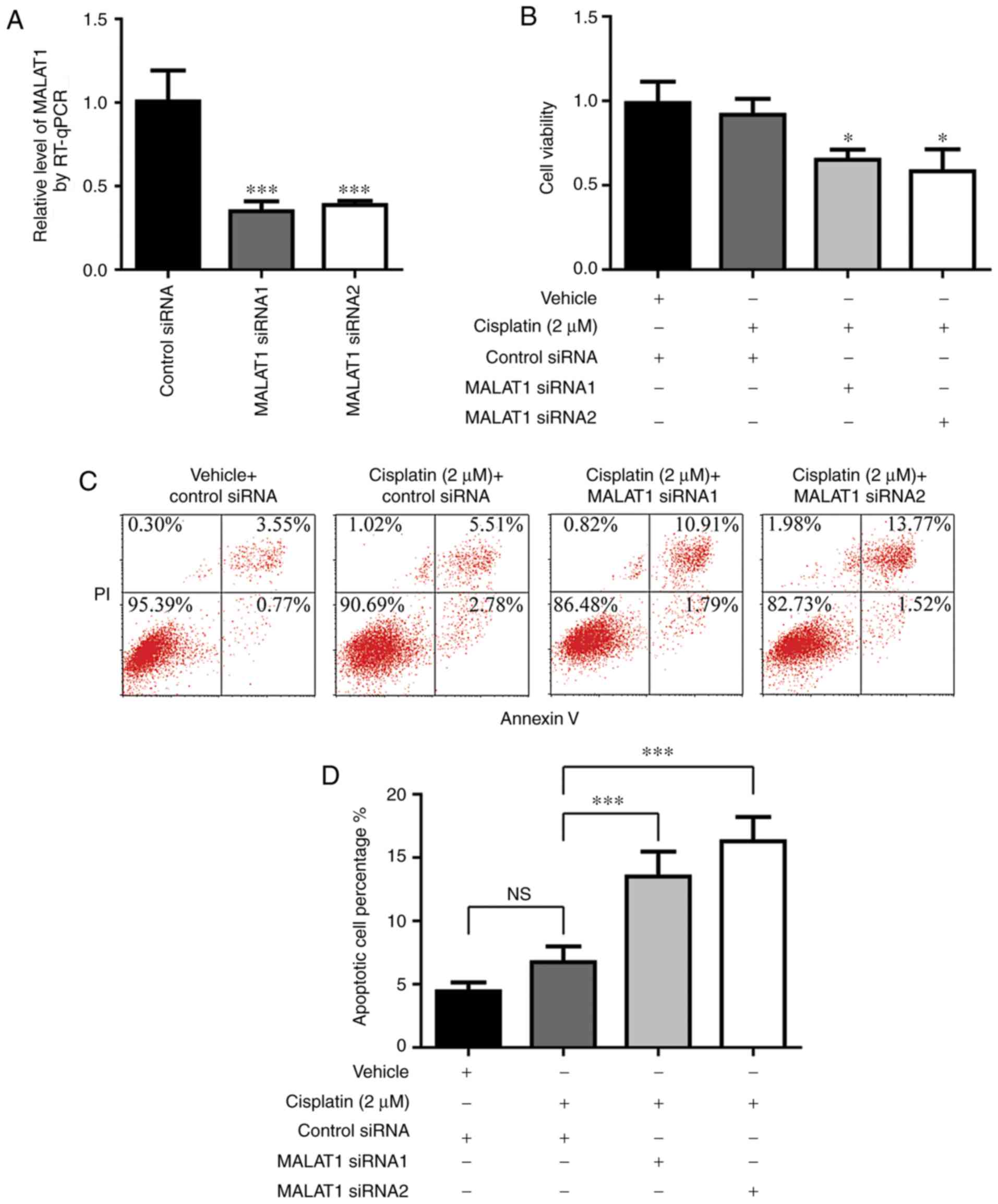
siRNA-mediated gene silencing. As demonstrated in Fig. 5A, MALAT1 siRNA1 and siRNA2
significantly knocked down MALAT1 expression in A549rCDDP cells.
Following MALAT1 silencing, cisplatin treatment induced a
significant decrease in cell viability in A549rCDDP cells (Fig. 5B). Furthermore, compared with
A549rCDDP cells treated with cisplatin, a significant increase in
the rate of cell apoptosis was observed in MALAT1-silenced
A549rCDDP cells following cisplatin treatment (Fig. 5C and D). These results directly
indicated that the increase of MALAT1 contributed to cisplatin
resistance in NSCLC.
Discussion
Cisplatin-based chemotherapy is a major therapeutic
approach for the treatment of patients with NSCLC. However,
although patients may respond to cisplatin at the initiation of the
treatment, cisplatin exposure would induce a series of adaptive
responses, leading to cisplatin resistance and tumor recurrence
(24). The present study identified a
MALAT1-miR-145-KLF4 axis was involved in driving cisplatin
resistance in NSCLC.
MALAT1 was a well-characterized oncogene involved in
the promotion of NSCLC cell proliferation and metastasis (25,26). In
the present study, it was also identified that MALAT1 levels were
positively associated with histological grade and occurrence of
metastasis. Recently, the upregulation of MALAT1 was demonstrated
to increase Histone-lysine N-methyltransferase EZH2 protein level
and contribute to resistance towards oxaliplatin-based chemotherapy
in patients with colorectal cancer (27). In the present study, cisplatin
treatment of A549 cells greatly increased MALAT1 level. In
addition, there was an elevation of MALAT1 level in A549rCDDP cells
compared with the parental A549 cells. Importantly, the silencing
of MALAT1 enhanced cisplatin-induced cell proliferation inhibition
and cell apoptosis in A549rCDDP cells. These results collectively
indicate that the abnormal expression of MALAT1 leads to cisplatin
resistance in NSCLC.
Cancer stem cells are only a small proportion of the
tumor cell population, and are crucial for tumor cell metastasis
and drug resistance (28,29). As a key transcription factor in
regulating cell reprogramming and an important gene in the
maintenance of cancer cell stemness (30,31), KLF4
has been suggested to be associated with chemotherapy resistance in
various types of cancer, including NSCLC (18,32). A
previous study identified that following benzo[a]pyrene treatment,
an increase in KLF4 and MALAT1 protein levels was detected during
the malignant transformation of the human bronchial epithelial
BEAS-2B cell line (33). However, to
the best of our knowledge, the regulatory association between KLF4
and MALTA1, and how their interplay contributed to cisplatin
resistance has not yet been studied. KLF4 is a direct target of
miR-145 (22). In the present study,
in the A549 cells, it was identified that KLF4 was negatively
regulated by miR-145 and positively regulated by MALAT1 at mRNA and
protein levels. Using a luciferase assay, it was confirmed that
MALAT1 indirectly regulated KLF4 at a transcription level via the
repression of miR-145. These data indicated that MALTA1 may
contribute to cisplatin resistance via regulating KLF4 level.
In conclusion, the data of the present study
demonstrated that the MALAT1-miR-145-KLF4 axis functions as an
important inducer of cisplatin resistance in NSCLC. Therefore,
MALAT1 may be a promising predictor of cisplatin response and
therapeutic target for patients with NSCLC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YC designed the study, analyzed the data and wrote
the manuscript. GL, XZ and FD collected the tissue samples,
performed the experiments, and analyzed the data. RZ performed the
experiments and analyzed the data.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Shouguang People's Hospital. Each patient provided
written consent to participate.
Patient consent for publication
Patients consented to the publication of their
data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Molina JR, Yang P, Cassivi SD, Schild SE
and Adjei AA: Non-small cell lun cancer: Epidemiology, risk
factors, treatment, and survivorship. Mayo Clin Proc. 83:584–594.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nishio K, Nakamura T, Koh Y, Suzuki T,
Fukumoto H and Saijo N: Drug resistance in lung cancer. Curr Opin
Oncol. 11:109–115. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Thum T: Noncoding RNAs and myocardial
fibrosis. Nat Rev Cardiol. 11:655–663. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim C, Kang D, Lee EK and Lee JS: Long
noncoding RNAs and RNA-binding proteins in oxidative stress,
cellular senescence, and age-related diseases. Oxid Med Cell
Longev. 2017:20623842017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lin R, Maeda S, Liu C, Karin M and
Edgington TS: A large noncoding RNA is a marker for murine
hepatocellular carcinomas and a spectrum of human carcinomas.
Oncogene. 26:851–858. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fűri I, Kalmár A, Wichmann B, Spisák S,
Schöller A, Barták B, Tulassay Z and Molnár B: Cell free DNA of
tumor origin induces a ‘Metastatic’ expression profile in HT-29
cancer cell line. PLoS One. 10:e01316992015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jin Y, Feng SJ, Qiu S, Shao N and Zheng
JH: LncRNA MALAT1 promotes proliferation and metastasis in
epithelial ovarian cancer via the PI3K-AKT pathway. Eur Rev Med
Pharmacol Sci. 21:3176–3184. 2017.PubMed/NCBI
|
|
8
|
Li H, Yuan X, Yan D, Li D1, Guan F, Dong
Y, Wang H, Liu X and Yang B: Long non-coding RNA MALAT1 decreases
the sensitivity of resistant glioblastoma cell lines to
temozolomide. Cell Physiol Biochem. 42:1192–1201. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Larne O, Hagman Z, Lilja H, Bjartell A,
Edsjö A and Ceder Y: miR-145 suppress the androgen receptor in
prostate cancer cells and correlates to prostate cancer prognosis.
Carcinogenesis. 36:858–866. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chang S, Gao L, Yang Y, Tong D, Guo B, Liu
L, Li Z, Song T and Huang C: miR-145 mediates the antiproliferative
and gene regulatory effects of vitamin D3 by directly targeting
E2F3 in gastric cancer cells. Oncotarget. 6:7675–7685.
2015.PubMed/NCBI
|
|
11
|
Ju BL, Chen YB, Zhang WY, Yu CH, Zhu DQ
and Jin J: miR-145 regulates chemoresistance in hepatocellular
carcinoma via epithelial mesenchymal transition. Cell Mol Biol
(Noisy-le-grand). 61:12–16. 2015.PubMed/NCBI
|
|
12
|
Ye Z, Shen N, Weng Y, Li K, Hu L, Liao H,
An J, Liu L, Lao S and Cai S: Low miR-145 silenced by DNA
methylation promotes NSCLC cell proliferation, migration and
invasion by targeting mucin 1. Cancer Biol Ther. 16:1071–1079.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lopez-Ayllon BD, Moncho-Amor V, Abarrategi
A, Ibañez de Cáceres I, Castro-Carpeño J, Belda-Iniesta C, Perona R
and Sastre L: Cancer stem cells and cisplatin-resistant cells
isolated from non-small-lung cancer cell lines constitute related
cell populations. Cancer Med. 3:1099–1111. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen YJ, Wu CY, Chang CC, Ma CJ, Li MC and
Chen CM: Nuclear Krüppel-like factor 4 expression is associated
with human skin squamous cell carcinoma progression and metastasis.
Cancer Biol Ther. 7:777–782. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shi M, Cui J, Du J, Wei D, Jia Z, Zhang J,
Zhu Z, Gao Y and Xie K: A novel KLF4/LDHA signaling pathway
regulates aerobic glycolysis in and progression of pancreatic
cancer. Clin Cancer Res. 20:4370–4380. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang J, Liu K, Song D, Ding M, Wang J,
Jin Q and Ni J: Krüppel-like factor 4 promotes high-mobility group
box 1-induced chemotherapy resistance in osteosarcoma cells. Cancer
Sci. 107:242–249. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fadous-Khalifé MC, Aloulou N, Jalbout M,
Hadchity J, Aftimos G, Paris F and Hadchity E: Krüppel-like factor
4: A new potential biomarker of lung cancer. Mol Clin Oncol.
5:35–40. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu MY, Li XQ, Gao TH, Cui Y, Ma N2, Zhou
Y2 and Zhang GJ: Elevated HOTAIR expression associated with
cisplatin resistance in non-small cell lung cancer patients. J
Thorac Dis. 8:3314–3322. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Petersen I and Warth A: Lung cancer:
Developments, concepts, and specific aspects of the new WHO
classification. J Cancer Res Clin Oncol. 142:895–904. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jia Y, Zhang W, Liu H, Peng L, Yang Z and
Lou J: Inhibition of glutathione synthesis reverses Krüppel-like
factor 4-mediated cisplatin resistance. Cancer Chemother Pharmacol.
69:377–385. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Minami K, Taniguchi K, Sugito N, Kuranaga
Y, Inamoto T, Takahara K, Takai T, Yoshikawa Y, Kiyama S, Akao Y
and Azuma H: MiR-145 negatively regulates Warburg effect by
silencing KLF4 and PTBP1 in bladder cancer cells. Oncotarget.
8:33064–33077. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xiang Y, Zhang Y, Tang Y and Li Q: MALAT1
modulates TGF-β1-induced endothelial-to-mesenchymal transition
through downregulation of miR-145. Cell Physiol Biochem.
42:357–372. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Galluzzi L, Vitale I, Michels J, Brenner
C, Szabadkai G, Harel-Bellan A, Castedo M and Kroemer G: Systems
biology of cisplatin resistance: Past, present and future. Cell
Death Dis. 5:e12572014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Guo F, Guo L, Li Y, Zhou Q and Li Z:
MALAT1 is an oncogenic long non-coding RNA associated with tumor
invasion in non-small cell lung cancer regulated by DNA
methylation. Int J Clin Exp Pathol. 8:15903–15910. 2015.PubMed/NCBI
|
|
26
|
Guo F, Jiao F, Song Z, Li S, Liu B, Yang
H, Zhou Q and Li Z: Regulation of MALAT1 expression by TDP43
controls the migration and invasion of non-small cell lung cancer
cells in vitro. Biochem Biophys Res Commun. 465:293–298. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li P, Zhang X, Wang H, Wang L, Liu T, Du
L, Yang Y and Wang C: MALAT1 is associated with poor response to
oxaliplatin-based chemotherapy in colorectal cancer patients and
promotes chemoresistance through EZH2. Mol Cancer Ther. 16:739–751.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Farnie G, Sotgia F and Lisanti MP: High
mitochondrial mass identifies a sub-population of stem-like cancer
cells that are chemo-resistant. Oncotarget. 6:30472–30486. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang Z, Wu T, Liu AY and Ouyang G:
Differentiation and transdifferentiation potentials of cancer stem
cells. Oncotarget. 6:39550–39563. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yu S, Cai X, Wu C, Wu L, Wang Y, Liu Y, Yu
Z, Qin S, Ma F, Thiery JP and Chen L: Adhesion glycoprotein CD44
functions as an upstream regulator of a network connecting ERK, AKT
and Hippo-YAP pathways in cancer progression. Oncotarget.
6:2951–2965. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li Y, Xian M, Yang B, Ying M and He Q:
Inhibition of KLF4 by statins reverses adriamycin-induced
metastasis and cancer stemness in osteosarcoma cells. Stem Cell
Reports. 8:1617–1629. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lund RJ, Huhtinen K, Salmi J, Rantala J,
Nguyen EV, Moulder R, Goodlett DR, Lahesmaa R and Carpén O: DNA
methylation and transcriptome changes associated with cisplatin
resistance in ovarian cancer. Sci Rep. 7:14692017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu Y, Lu R, Gu J, Chen Y, Zhang X, Zhang
L, Wu H, Hua W and Zeng J: Aldehyde dehydrogenase 1A1 up-regulates
stem cell markers in benzo[a]pyrene-induced malignant
transformation of BEAS-2B cells. Environ Toxicol Pharmacol.
45:241–250. 2016. View Article : Google Scholar : PubMed/NCBI
|