Introduction
Despite continued research to identify novel
antineoplastic agents, colon carcinoma remains the third most
common neoplastic malignancy globally and one of the leading causes
of cancer-associated mortality, due to its aggressiveness and
resistance to therapy (1). The
majority of patients respond to the initial treatment and achieve
clinical remission following chemotherapy, but a substantial
proportion of patients develop chemoresistance and relapse within 5
years, leading to a low survival rate. Chemoresistance and the
resultant low survival rate are thought to be associated with the
presence of cancer stem cells (CSCs), which represent a rare
population of undifferentiated cells that drive the growth of
tumors, differentiating into a variety of cell types corresponding
to the original tissue while maintaining the ability for
self-renewal (2).
Colon cancer stem-like cells were identified in 2007
(3). Another characteristic of CSCs
is their relative quiescence, which may endow CSCs with resistance
to well-defined chemotherapies that predominantly target
proliferating rather than quiescent cells. Resistance is also
exhibited through a wide spectrum of activities, including DNA
damage repair, altered cell cycle checkpoint control, and
malfunction of apoptosis, drug transporters and detoxifying enzymes
(4). Therefore, one strategy for the
treatment of chemo-resistant cancers is to develop an agent that
selectively targets quiescent and drug-resistant CSCs (5).
Retinoids are a class of natural and synthetic
derivatives of vitamin A that exert effects on critical biological
processes that include development, cell growth and
differentiation, metabolism and homeostasis. They demonstrate
anti-proliferative and cell differentiation activity in various
cancer cell lines in vitro and in vivo, and are
therefore promising candidates for the chemoprevention and
treatment of certain types of cancer (6,7). However,
their high degree of toxicity, which may reflect the broad
biological responses mediated by retinoid receptors, is the main
limitation for the clinical use of the retinoids available at
present. N-(4-hydroxyphenyl)-retinamide (fenretinide, also known as
4HPR), a synthetic derivative of all-trans-retinoic acid,
has emerged as a promising anticancer agent due to its distinct
advantages over other agents for treating cancer, as demonstrated
in numerous in vitro and in vivo studies, and
chemoprevention clinical trials (8–10). In
addition to its efficacy against a wide range of types of tumor,
fenretinide has minimal side effects and synergizes with other
anticancer agents, reinforcing their anticancer efficacy (11–13).
In the present study, sphere culture in serum-free
medium was used to isolate tumor spheres from two human colon cell
lines: HT29 and HCT116. The capacity for self-renewal,
chemoresistance, and tumor initiation was then assessed in the
tumor sphere cells. Fenretinide was demonstrated to preferentially
target colon sphere cells, which are believed to possess certain
stem-like characteristics. Transcriptome analysis of
fenretinide-treated HT29 sphere cells was then performed to
investigate the mechanisms involved, and a number of features
associated with cell cycle regulation and activation of reactive
oxygen species (ROS)-induced stress responses were identified.
These results are an important addition to the current knowledge
concerning of fenretinide, and provide a foundation for its
clinical application in the treatment of cancer.
Materials and methods
Cell lines, cell culture and
reagent
The human colon cancer cell lines HCT116 and HT29,
obtained from the Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences (Shanghai, China), were cultured in
RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) containing 10% fetal bovine serum (FBS; PAA Laboratories;
GE Healthcare Life Sciences, Chalfont, UK). The sphere cells were
obtained with similar protocol as illustrated in previous study
(14). Single HCT116 and HT29 cells
were plated in ultralow-attachment plates in serum-free RPMI-1640
medium at a density of 5,000 cells/ml. The sphere-forming medium
(SFM) was Dulbecco's modified Eagle's medium-F12 (Gibco; Thermo
Fisher Scientific, Inc.) supplemented with 2% B-27, 20 ng/ml
epidermal growth factor (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany), 5 µg/ml insulin (Sigma-Aldrich; Merck KGaA) and 0.4% FBS
(Ameresco, Inc., Framingham, MA, USA). Dissociated cells were
seeded in SFM with or without fenretinide treatment, and the
spheres were observed and photographed with an objective lens at
magnification, ×20 using an inverted microscope. All cells were
incubated at 37°C in a humidified atmosphere with 5%
CO2. Fenretinide was purchased from Sigma-Aldrich (Merck
KGaA) and dissolved in absolute ethanol.
Cell cycle and cell viability
assay
For the cell viability assay, HT29 and HCT116 cells
were incubated at 37°C in 48-well plates at a density of 10,000
cells/well overnight in RPMI-1640 medium containing 10 or 0.5% FBS
when comparing the sensitivity of colon cancer cells to fenretinide
in normal or low serum levels, respectively, and were then treated
with 6 µM fenretinide for 48 or 72 h. Fenretinide in absolute
ethanol was used as the negative control. For cell cycle analysis,
the trypsinized adherent cells were cultured for 48 h, then
collected and fixed with 75% ethanol (v/v) for 24 h at 4°C, stained
with propidium iodide at the final concentration of 50 µg/ml for 30
min at room temperature and analyzed by flow cytometry using the
FC500 flow cytometer (Beckman Coulter, Inc., Brea, CA, USA). HT29
and HCT116 cells were treated with the MTT solution (50 µl; 5 µg/ml
in PBS) to each well and the plate was incubated for 3 h at 37°C,
following which the medium was replaced by 200 µl dimethyl
sulfoxide. Cell viability was evaluated by measuring the absorbance
optical density at 595 nm on a DU 800 spectroscopy microplate
reader (Beckman Coulter, Inc., Brea, CA, USA). For sphere cell
viability, the cells were cultured in SFM for 7 days as described
in the cell culture methods. The sphere cells were then centrifuged
at 600 × g for 5 min at room temperature and collected in a new 50
ml centrifuge tube, then 0.05% trypsin (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) was added for enzymatic
dissociation. The sphere cells were incubated for 3 min at 37°C,
seeded in SFM in 48-well plates with 10,000 cells/well, and then
treated with 20 µM fluorouracil (5-FU; Sigma-Aldrich; Merck KGaA)
or 400 ng/ml epirubicin (EPB; Sigma-Aldrich; Merck KGaA) at 37°C
for 48 h. Concurrently, the parental HT29 cells were incubated in
48-well plates with 10,000 cells/well at 37°C overnight in
RPMI-1640 medium containing 10% FBS, and were then treated with 20
µM 5-FU or 400 ng/ml EPB for 48 h at 37°C. Sterile normal saline
was added into the plate as the negative control.
Cell apoptosis assay
For parental cell apoptosis assay, HT29 and HCT116
cells were seeded into 6-well plates (2×105/well) and
exposed to the fenretinide treatments with indicated concentration
for indicated time. Absolute ethanol was used as the negative
control. The treated cells were dissociated with 0.25% trypsin for
3 min at 37°C, collected in BD falcon centrifuge tubes, washed with
Annexin V binding buffer (BD Biosciences, San Jose, CA, USA) and
centrifuged at 350 × g for 5 min. Washing and centrifugation was
repeated twice. Then, the samples were incubated with 5 µl
fluorescein isothiocyanate (FITC)-stained Annexin V antibody and 5
µl propidium iodide stain for 15 min at room temperature according
to the protocol of the manufacturer of the Apoptosis Detection Kit
(BD Biosciences; cat. no. 556547), and detected and analyzed by
flow cytometry using the FC500 flow cytometer (Beckman Coulter,
Inc., Brea, CA, USA). For the sphere cell apoptosis assay, the
sphere cells from HCT116 and HT29 were cultured in SFM,
trypsinized, and seeded into 6-well plates with 2×105
cells/well. Following treatment with 3 µm fenretinide, sphere cells
were collected and centrifuged with 1,000 × g for 5 min at room
temperature. Cell apoptosis was detected using a Fluorescein
Isothiocyanate (FITC)-Annexin V Apoptosis Detection kit (BD
Biosciences) according to the protocol of the manufacturer, and
analyzed by flow cytometry. The total percentage of Annexin
V+/PI–/+ cells was quantified and CXP
Analysis software (version 1.0; Beckman Coulter, Inc., Brea, CA,
USA) was used to analyze the apoptosis data.
Cell surface marker analysis
HT29 parental cells at 80% confluence were rinsed
twice with PBS, released from the culture dish with 1 mmol/l EDTA
(Sigma-Aldrich; Merck KGaA) for 10 min followed by 0.05% trypsin
(Sigma-Aldrich; Merck KGaA) for 1 min, washed with RPMI-1640
containing 10% FBS, and harvested by centrifugation at 300 × g for
3 min at room temperature. Sphere cells from HT29 was cultured in
SFM and obtained as described above. All cells were resuspended at
1×106 cells/ml in cold PBS buffer with 1% FBS (Ameresco,
Inc.) for 5 min at room temperature and then stained with
FITC-conjugated cluster of differentiation CD44-specific antibodies
(BD Biosciences; cat. no. 560977) at a dilution of 1:400 for 30 min
in the dark at room temperature. For investigating the difference
prior and subsequent to fenretinide treatment for sphere-derived
HT29 cells, sphere cells derived from HT29 cells were seeded into
6-well at a density of 2×105 cells/well. Sphere cells
were treated with 3 µM fenretinide or absolute ethanol for 48 h.
Cells were harvested, resuspended, and stained with CD44-specific
antibody as described above. The cells were analyzed by flow
cytometry using FC500 flow cytometer (Beckman Coulter, Inc.). The
data were analyzed with CXP analysis software (version 1.0; Beckman
Coulter, Inc.).
Subcutaneous model of
tumorigenesis
The animal experiments were approved by the
Committee on Laboratory Animal Research of Shanghai Jiao Tong
University (Shanghai, China), and were conducted according to the
guidelines of the Laboratory Animal Center of Shanghai Jiao Tong
University School of Medicine (Shanghai, China). Female non-obese
diabetic/severe combined immunodeficiency (NOD/SCID) mice (6–8
weeks old, 25 g) were purchased from Shanghai SLAC Animal Center
(Shanghai, China) and fed ad libitum in a sterile
environment and a relative humidity of 40–70% at 25°C in a 12 h
light/dark cycle. A total of four mice were used to compare
tumorigenic ability between HT29 sphere cells and parental cells.
HT29 sphere cells (1×104 cells) were injected
subcutaneously into the left inguinal area of each mouse and the
same number of parental cells was injected into the right inguinal
area of the same mouse. Tumor growth was monitored every 5 days,
and the xenograft mice were sacrificed after 3 weeks when the tumor
diameter of xenograft mice was >15 mm.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Cellular RNA was isolated using TRIzol®
reagent (Molecular Research Center, Inc., Cincinnati, OH, USA)
according to the manufacturer's protocol. DNA was removed from the
samples using DNase treatment (DNA-free kit; Ambion; Thermo Fisher
Scientific, Inc.), and cDNA was synthesized according to the
Moloney Murine Leukemia Virus Reverse Transcription kit (Promega
Corporation, Madison, WI, USA). The sequence of the GAPDH primer
was as follows: forward, GCACCGTCAAGGCTGAGAAC; reverse,
TGGTGAAGACGCCAGTGGA, and these sets were used to produce a
normalization control. qPCR was performed in triplicate with
SYBR-Green PCR Master Mix (Applied Biosystems; Thermo Fisher
Scientific, Inc.) and the 7900HT Fast Real-Time PCR machine
(Applied Biosystems; Thermo Fisher Scientific, Inc.) with the
following thermocycling conditions: 95°C for 10 min, followed by 40
cycles of 95°C for 15 sec and 60°C for 1 min, and a final
elongation step of 72°C for 10 min, was used to perform the
reaction. The expression levels of certain cell-cycle and
stress-response associated genes, including cyclin E2
(CCNE2), cell division cycle 25A (CDC25A), E2F
transcription factor 8 (E2F8), BRCA2, DNA repair associated
(BRCA2), cyclin A2 (CCNA2), sestrin 2 (SESN2),
transglutaminase 2 (TGM2), homocysteine inducible ER protein
with ubiquitin like domain 1 (HERPUD1) and calmegin
(CLGN) were assessed. GAPDH was used as a reference,
and the 2−ΔΔCq method was used to quantify the relative
expression level of those genes (15). Table I
summarizes the forward primers and reverse primers of all other
genes.
 | Table I.Forward and reverse primers of genes
in reverse transcription quantitative polymerase chain reaction
analysis. |
Table I.
Forward and reverse primers of genes
in reverse transcription quantitative polymerase chain reaction
analysis.
| Gene name | Forward
primers | Reverse
primers |
|---|
| CDC25A |
GTTAGACGTCCTCCGTCCAT |
AGACCTTTCCTTCCCAGGTT |
| CCNE2 |
GCATTATGACACCACCGAAG |
ATTGGCTAGGGCAATCAATC |
| E2F8 |
CCGCAACAGAGATCAGAAAA |
AAGTTCCTCTGCCACTTCGT |
| BRCA2 |
AGTGACCTTCCAGGGACAAC |
GCCCATTGATGGCTAAAACT |
| CCNA2 |
GGAGCTGCCTTTCATTTAGC |
TTGACTGTTGTGCATGCTGT |
| SESN2 |
GAGGACTTCACTCGGAGAGG |
GCATGGCGATGGTATTGTAG |
| TGM2 |
CCAGAACAGCAACCTTCTCA |
TCGTACTTGGTGCTCAGGTC |
| HERPUD1 |
TGGATTGGACCTATTCAGCA |
CAGGAGGAGGACCATCATTT |
| CLGN |
AACCAATGGACCTGGAAGAG |
CTCCAGATCCTGTGCTCTCA |
Microarray hybridization and data
mining
Total RNA from HT29 parental and sphere cells was
amplified and labeled with biotin according to the standard
Affymetrix protocol as outlined by Lockhart et al (16). The fragmented, biotinylated cDNA was
then subjected to hybridization with the GeneChip Human Genome-U133
Plus 2.0 array (Affymetrix, Inc., Santa Clara, CA, USA) according
to manufacturer's protocol. Analysis of the underlying biological
mechanisms of the significantly differentially expressed genes was
performed using the Database for Annotation, Visualization and
Integrated Discovery (DAVID 6.7 version) Bioinformatics Resources
database (17), with functional
relevance assessed via Gene Ontology (GO) enrichment analysis
(18). Genes that varied by
>2-fold following fenretinide treatment in each sample were
selected as the target genes, and mapped as Venn diagrams using
VennyDiagram software (http://bioinfogp.cnb.csic.es/tools/venny/). The
transcriptome profiles of HT29 enriched sphere cells treated with 6
µm fenretinide for 48 and 72 h and the unenriched cells treated
with 3 µm fenretinide for 24 and 48 h were deposited in NCBI's Gene
Expression Omnibus (GEO) and the microarray data are available at
the GEO accession no. GSE66983 (19).
The similar concentration of fenretinide and treatment time are
described in our previous study (14). For data mining, a software package of
Component Plane Presentation, Self-Organizing Map was implemented
using Matlab 6.5 as described previously (20,21).
Statistical analysis
Data and photographs were analyzed and drawn with
GraphPad Prism v5 (GraphPad Software, Inc., La Jolla, CA, USA). All
data were expressed as the mean ± standard error of the mean.
Statistical analysis was performed using Student's unpaired
t-tests. P<0.05 was considered to indicate a statistically
significant difference.
Results
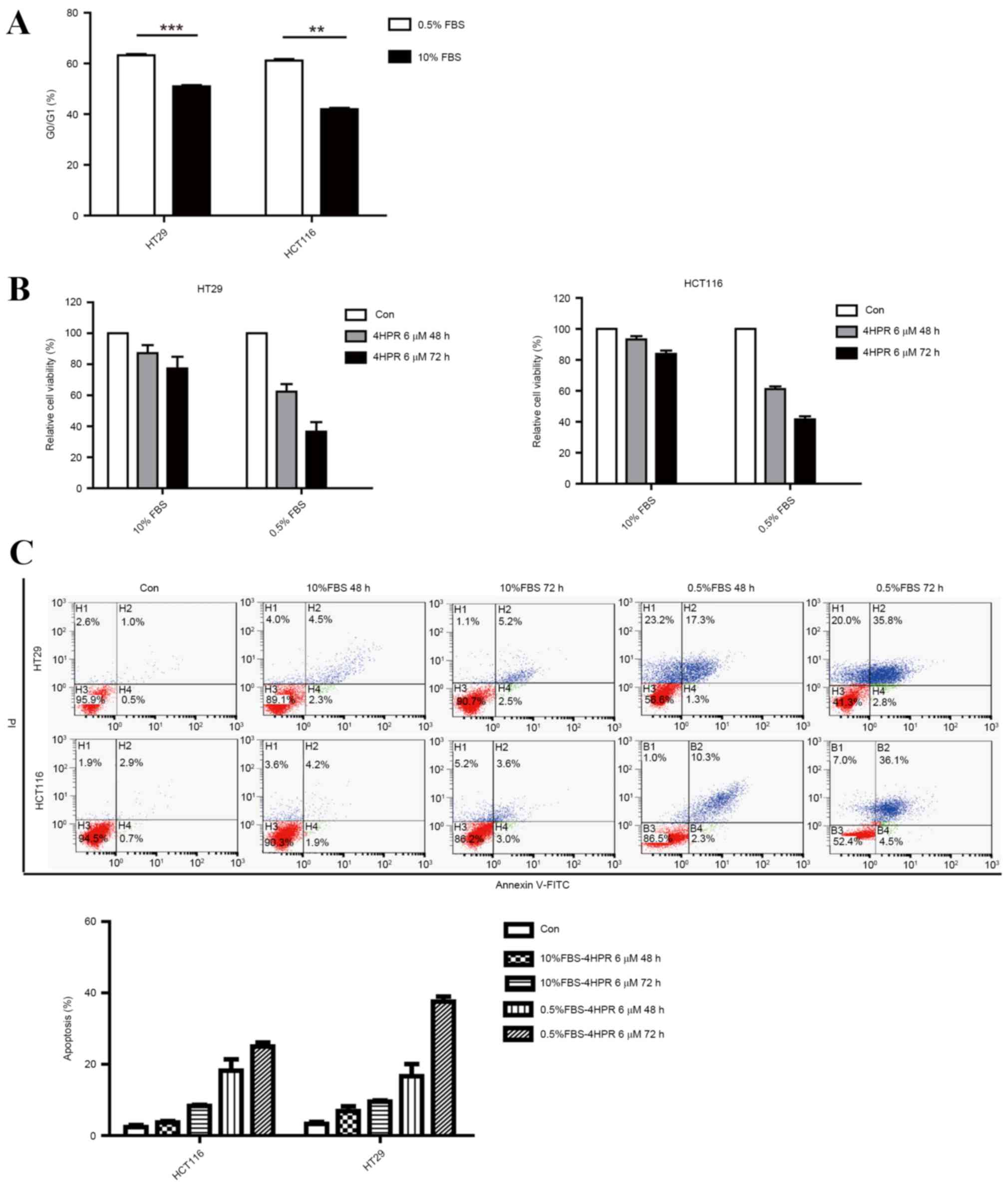
Cells cultured in low-serum medium
demonstrate greater sensitivity to fenretinide
The majority of CSC cells are in a quiescent state,
which is why the majority of current chemotherapy regimens fail to
achieve a cure. To evaluate the effects of fenretinide on quiescent
cells, HT29 and HCT116 cells were cultured in serum-starvation
conditions, which causes cell cycle arrest at the G0 phase and cell
quiescence (Fig. 1A). HT29 and HCT116
cells were incubated separately in low-serum medium and full-serum
medium, and their viability was assessed using the MTT cell
viability assay following treatment with 6 µm fenretinide for 48 or
72 h. Cells incubated in the low-serum culture medium were more
sensitive to fenretinide treatment compared with those incubated in
full-serum medium (Fig. 1B). In the
cell apoptosis assays, marked induction of apoptosis was observed
oonly when HCT116 and HT29 cells were treated with fenretinide in
low-serum medium (Fig. 1C). These
results indicated that the cells grown in low-serum medium
demonstrated greater sensitivity to fenretinide.
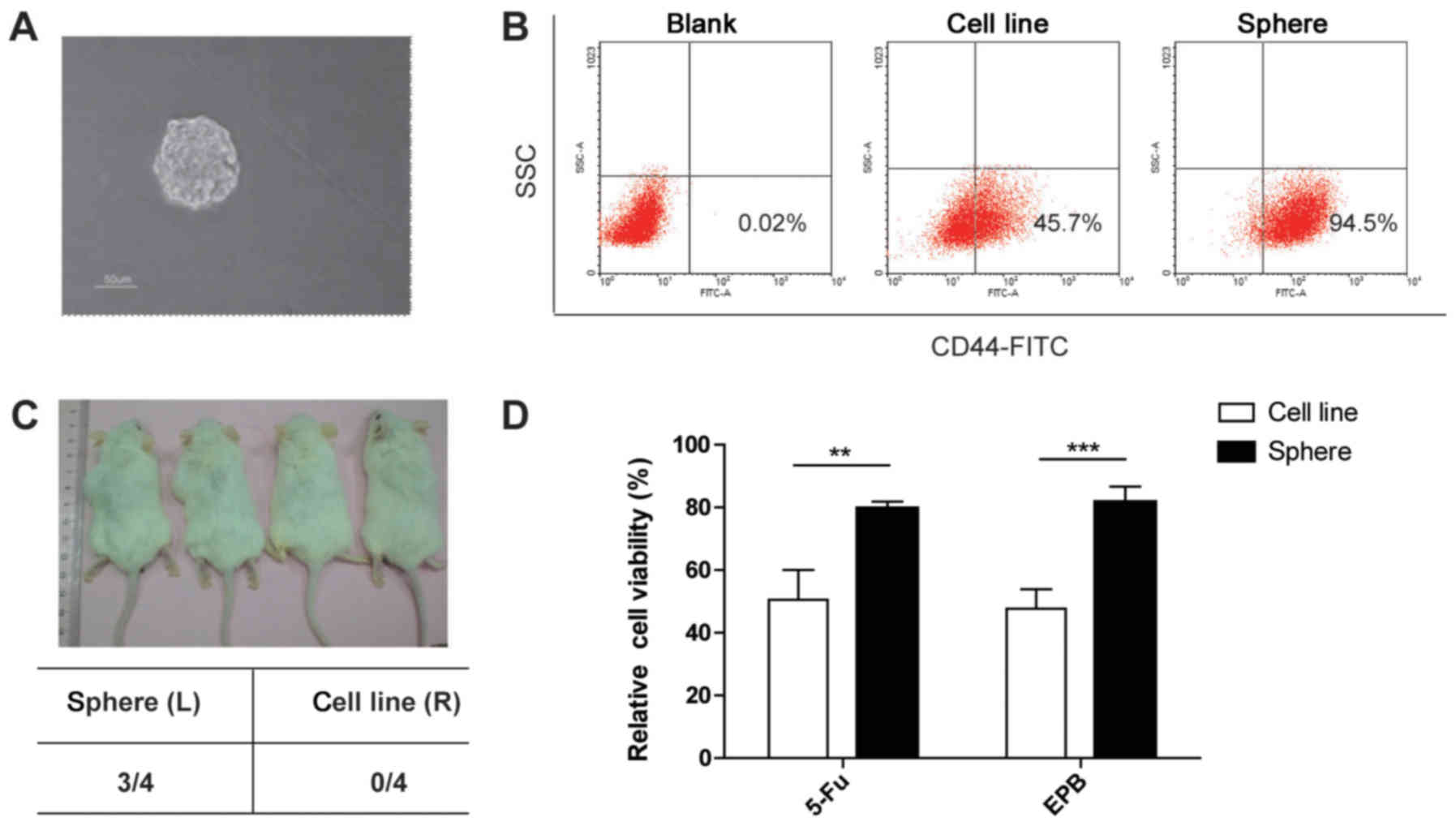
Colon spheres formed in the serum-free
medium demonstrate stem-like characteristics
Previous research has demonstrated that CSCs
generate three-dimensional spheres comprised of a small number of
CSCs that have the ability to self-renew and generate spheres on
serial passage, as well as progenitor cells capable of
multi-lineage differentiation (22).
Sphere-forming assays are used to enrich CSCs in vitro as an
operational surrogate for CSCs. Unsorted HCT116 and HT29 cells form
spheres in SFM following 5 days of culture. Spheres with a diameter
of 50–100 µm were observed (Fig.
2A).
CSCs are identifiable by their specific surface
epitopes. Colon CSCs express the surface markers CD44 and CD133
(23). CD44 has been demonstrated to
be a putative marker of colon CSCs in tumor specimens and cell
lines (24). Therefore, the
expression of CD44 was examined in HT29 parental and sphere cells.
The percentage of CD44+ cells was increased in sphere
cells compared with parental cells (Fig.
2B).
CSCs are considered to be drivers of tumor
initiation and progression. Accordingly, to determine the
tumorigenic ability of sphere cells, their ability to grow
subcutaneously in immune-deficient mice was analyzed. A total of
10,000 parental cells or sphere cells were injected into the
inguinal area of NOD/SCID mice. Significantly increased initiation
and growth of tumors were observed only in the inguinal areas
injected with the sphere cells (Fig.
2C). This result indicated the potent in vivo
self-renewal and tumor-initiating capacities of spheres, which is
consistent with the notion that CSCs drive tumor progression.
CSCs demonstrate resistance to a number of
conventional therapies, which may explain why it is difficult to
eradicate cancer cells completely and why recurrence is an
ever-present threat (25). To examine
whether sphere cells retained CSC chemo-resistance, the sensitivity
of sphere cells to fluorouracil (5-FU, Sigma-Aldrich; Merck KGaA)
and epirubicin (EPB, Sigma-Aldrich; Merck KGaA), two first-line
chemotherapeutic agents used to treat colon cancer, was assessed.
The cell viability of the parental HT29 cells and the sphere cells
treated with 5-FU and EPB was assessed. Marked inhibition of cell
growth was observed only in the parental HT29 cells treated with
fenretinide (Fig. 2D). These results
suggested that colon sphere cells mimic the status of colon CSC
cells in this setting.
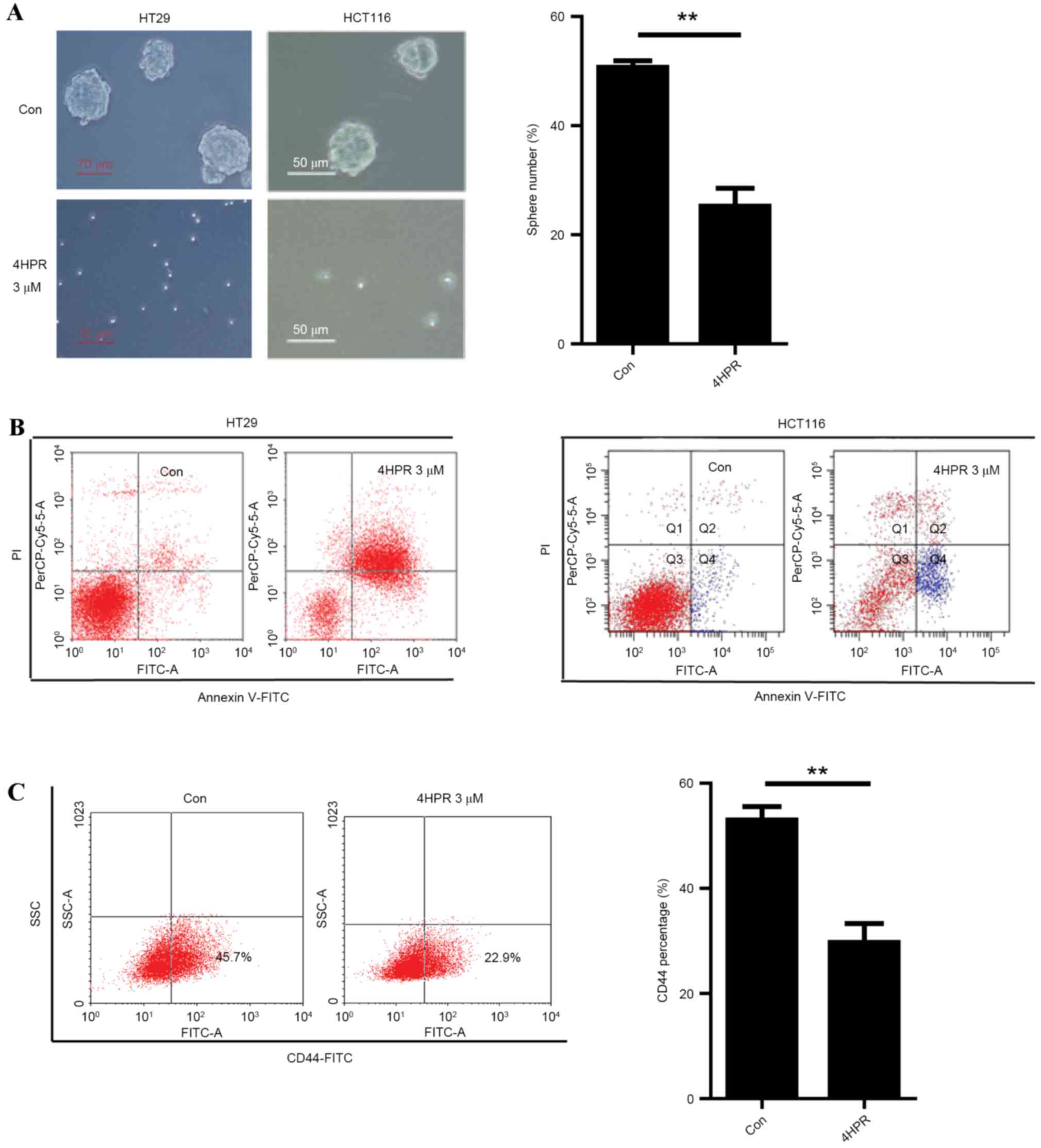
Fenretinide preferentially targets
sphere cells
Fenretinide has been demonstrated to preferentially
eradicate acute myeloid leukemia (AML) and chronic myeloid leukemia
(CML) stem cells, and CSCs in ovarian and breast cancer (9,10,14). Therefore, it is necessary to
investigate whether fenretinide can selectively target CSCs in
colon cancer and disturb their ability for self-renewal. HT29 and
HCT116 parental cells were insensitive to fenretinide (Fig. 1B). However, when cultured under
sphere-forming conditions, treatment with 3 µm fenretinide
inhibited the sphere formation of HT29 and HCT116 cells (Fig. 3A).
Marked induction of apoptosis was observed in HT29
and HCT116 sphere cells following treatment with 3 µm fenretinide
for 48 h (Fig. 3B). The percentage of
CD44+ cells was the measured in HT29 cells treated with
or without 3 µm fenretinide for 48 h. The percentage of
CD44+ cells was lower in the fenretinide-treated cells
compared with the corresponding control cells (Fig. 3C). These results suggested that
fenretinide is a potential candidate for eradicating colon CSCs, as
it reduced the CSC population below the control levels and because
the colon sphere cells were more sensitive to fenretinide compared
to the cell lines.
Microarray analysis of HT29 cells
treated with fenretinide
To identify the molecular mechanisms underlying
fenretinide-induced apoptosis, microarray gene expression profiling
of HT29 parental cells and sphere cells was performed. Parental
cells were treated with 6 µm fenretinide for 48 and 72 h, while
sphere cells were treated with 3 µm fenretinide for 24 or 48 h, as
sphere cells are not able to survive when treated with ≥6 µm
ferentinide or when treated with 3 µm for over 72 h. Total mRNA was
extracted and profiled using a whole-genome array. To recognize the
prominent features in the data, self-organizing map (SOM)-based
clustering analysis of the top 5,000 regulated genes was performed.
Genes were grouped into 13 clusters, and different changes in
expression were observed at each time (Fig. 4A).
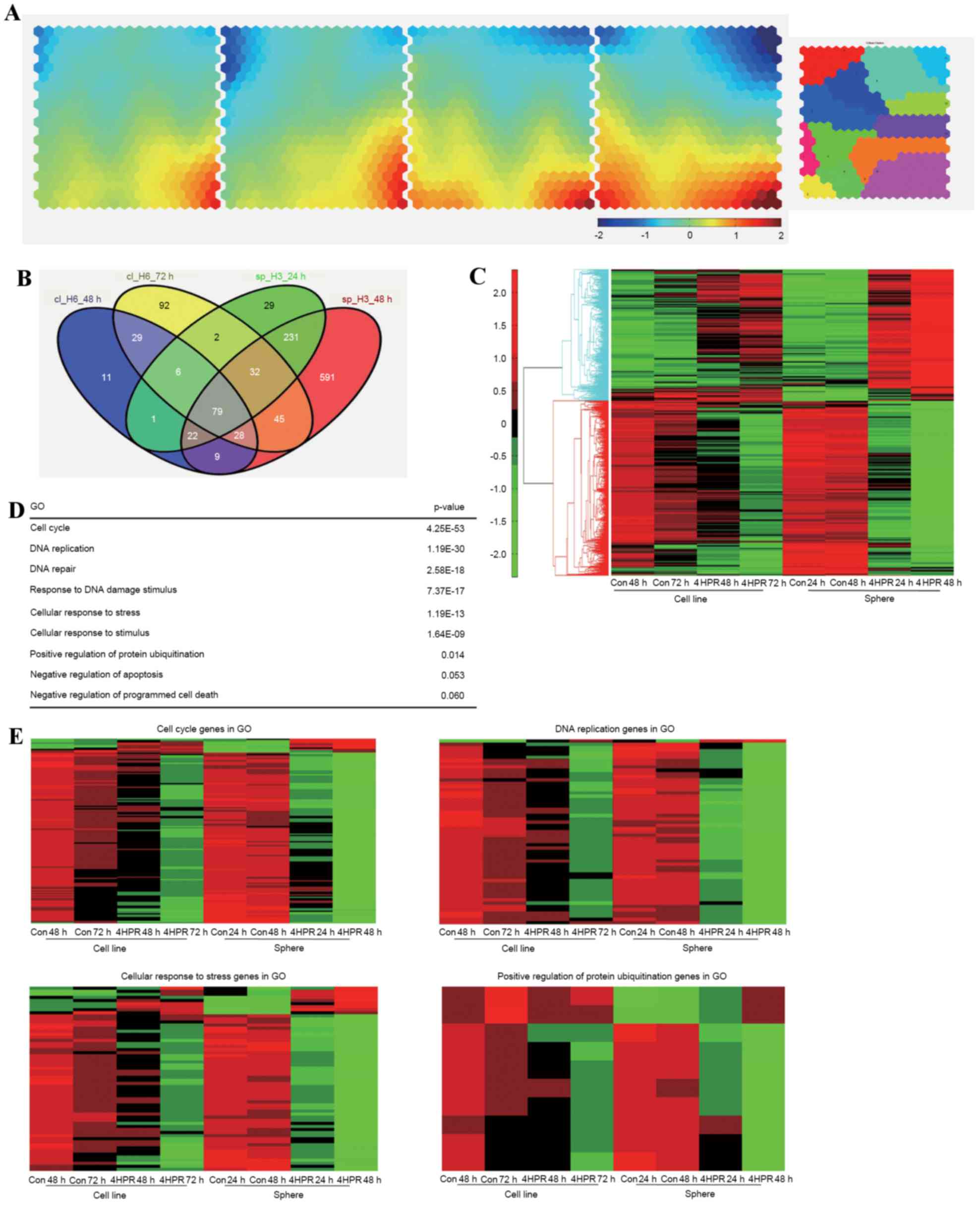 | Figure 4.Transcriptome profiles of HT29
enriched sphere cells and parental cells treated with 4HPR. (A)
Illustration of transcriptome changes using a Component Plane
Presentation, Self-Organizing Map. Each presentation illustrates a
sample-specific change, in which the upregulated genes (represented
in red), downregulated genes (represented in blue), and moderately
regulated genes (represented in yellow and green) are well
delineated. The color bar indicates the expression values (log
ratio in base 2), and brighter colors indicate higher values. (B)
The numbers of regulated genes that were selected based on a 2-fold
change threshold in each sample are depicted. (C) Heat map of the
microarray data revealing the expression levels of these selected
genes. (D) GO and Database for Annotation, Visualization and
Integrated Discovery analysis of the selected genes. (E) Heat maps
of significant GO clusters, revealing the specific changes in gene
regulation. 4HPR, fenretinide; GO, gene oncology; Con, control. |
The number of target genes in each sample is
depicted in Fig. 4B. A total of 851
genes from the sphere samples that did not overlap with the
parental cells were then selected for further analysis. These genes
exhibited more prominent changes following fenretinide treatment,
as depicted in Fig. 4C. To reveal the
functional relevance of these specifically regulated genes in
sphere cells, functional enrichment analysis was performed using
DAVID and GO (26). Significant
processes associated with specific regulation of sphere cells
treated with fenretinide were the cell cycle, DNA replication,
cellular response to stress or stimuli, cellular regulation of
protein ubiquitination and apoptosis (Fig. 4D). Heat maps of these gene clusters
are depicted in Fig. 4E.
Fenretinide induces genes that are
associated with cell cycle regulation and stress-responsive
activities
Cell cycle-associated genes were the most regulated
clusters in the sphere cells. This indicated that the effect of
fenretinide treatment on sphere cells was associated with cell
cycle regulation. Flow cytometry was used to analyze the cell cycle
of HCT116 and HT29 sphere cells. The percentage of sphere cells at
the G1 and G2 stages increased and the percentage in cells in S
phase decreased following treatment with 3 µm fenretinide for 24 h
(P<0.05; Fig. 5A). This result
indicated that sphere cell proliferation was inhibited by
fenretinide.
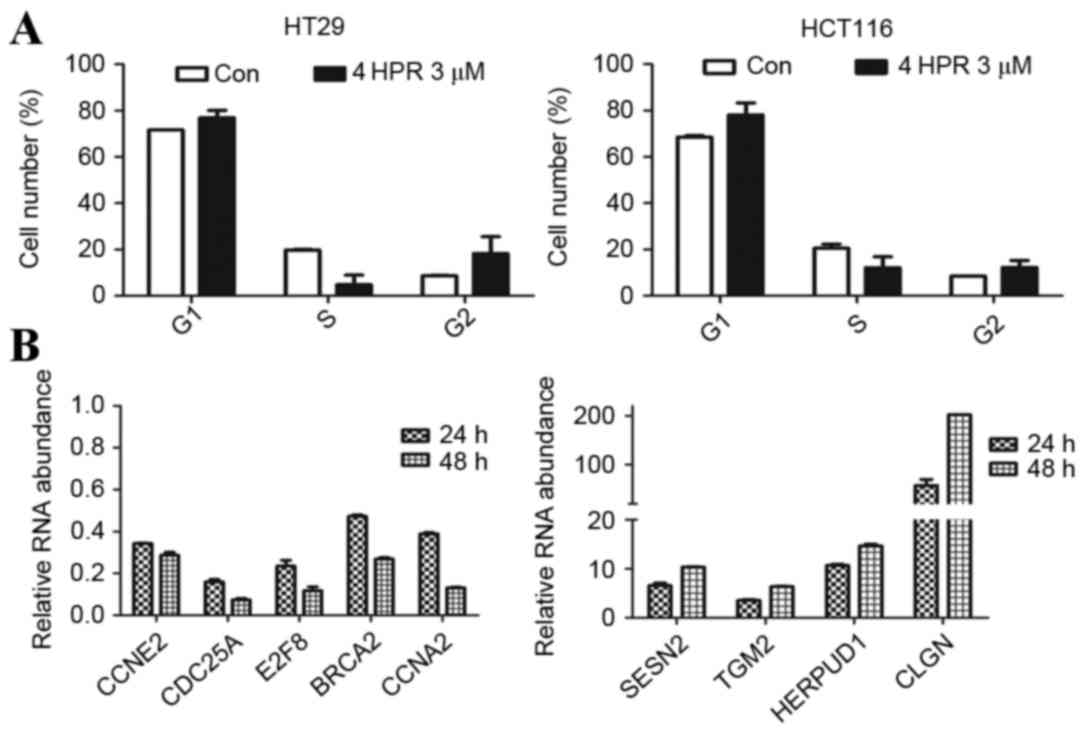 | Figure 5.Analysis of the cell cycle, cell
cycle-associated genes, and stress response-associated genes. (A)
Cell cycle analysis of HCT116 and HT29 sphere cells, assessed using
flow cytometry, following treatment with 3 µM 4HPR for 24 h. The
values are expressed as the mean ± standard deviation (n=3). (B)
Reverse transcription-quantitative polymerase chain reaction
analysis of the expression of cell cycle-associated genes, and
stress response-associated genes. 4HPR, fenretinide; Con, control;
CCNE2, cyclin E2; CDC25A, cell division cycle 25A; E2F8, E2F
transcription factor 8; BRCA2, BRCA2, DNA repair associated; CCNA2,
cyclin A2; SESN2, sestrin 2; TGM2, transglutaminase 2; HERPUD1,
homocysteine inducible ER protein with ubiquitin like domain 1;
CLGN, calmegin. |
Next, certain key genes involved in cell cycle
regulation for were selected for RT-qPCR analysis. Compared with
genes in the untreated cells, CCNE2, CDC25A, E2F8, BRCA2,
and CCNA2 were downregulated following fenretinide treatment
(Fig. 5B). CCNE2 encodes
cyclin E2, which belongs to the highly-conserved cyclin family.
This cyclin forms a complex with and functions as a regulatory
subunit of cyclin dependent kinase 2 (CDK2) and is involved in cell
cycle G1/S transition. A significantly increased expression level
of this gene is observed in tumor-derived cells. CCNA2
encodes cyclin A2, which binds to and activates CDK2 kinases, and
thus promotes G1/S and G2/M cell cycle transitions. CDC25A is a
phosphatase that is required for the progression from the G1 to the
S phase in the cell cycle. It activates the cyclin-dependent kinase
CDC2 by removing two phosphate groups. E2F8 regulates the
progression from G1 to S phase by ensuring the nucleus divides at
the proper time. BRCA2 is involved in the maintenance of genome
stability, and its mutation confers an increased lifetime risk of
developing cancer. These genes are essential for cell cycle
control. Fenretinide preferentially downregulated these genes in
HT29 sphere cells, which may inhibit cell proliferation and may
account for the fenretinide-induced apoptosis.
ROS balance is important for normal cell function,
and fenretinide is known to increase cellular ROS levels. Previous
research has demonstrated that fenretinide-induced apoptosis is
associated with the conversion of oxidative signaling into
downstream stress activities, including the redox response,
endoplasmic reticulum (ER) stress response/unfolded protein
response (UPR), and proteasome activation mediated by certain key
stress-responsive regulators (27).
The present study used RT-qPCR analysis to analyze certain key
genes involved in stress-response regulation. The accumulation of
unfolded proteins in the ER triggers the ER stress response. This
response includes the inhibition of translation to prevent further
accumulation of unfolded proteins, increased expression of proteins
involved in polypeptide folding, known as the UPR, and the
destruction of misfolded proteins by the ER-associated protein
degradation (ERAD) system (28).
SESN2 encodes sestrin 2, which may be involved in cellular
responses to different stress conditions. HERPUD1 may be involved
in the UPR and ERAD. HEPRUD1 expression is induced by the
UPR and has an ER stress response element in its promoter region,
whereas the encoded protein has an N-terminal ubiquitin-like
domain, which may interact with the ERAD system. This protein has
been demonstrated to interact with presenilin proteins and to
increase the level of amyloid-β protein when overexpressed
(29). TGM2 encodes
transglutaminase 2, which is a monomer that is induced by retinoic
acid, and may be involved in apoptosis. CLGN encodes a
specific ER chaperone protein, calmegin. The observed upregulation
of these genes suggested that significant antioxidative activity
occurred in sphere cells following fenretinide treatment, and may
partially explain why fenretinide preferentially targets sphere
cells.
Discussion
CSCs are widely accepted to be crucial in cancer
initiation, propagation, metastasis and relapse (30–32). The
residual CSC reservoir is correlated with the prognosis of patients
(33). Therefore, CSC-targeted
strategies are likely to be effective interventions to increase the
responsiveness to traditional therapeutic strategies, and to reduce
the risk of local recurrence and metastasis. At present,
substantial attention has been focused on the development of
strategies to selectively eliminate CSCs in the hope of preventing
the recurrence of cancer, and eventually curing the disease
(5,34,35).
Fenretinide has emerged as a promising candidate
with chemo-preventive properties (9,10,14). Clinical data have provided evidence
that fenretinide significantly reduces the risk of secondary breast
cancer in premenopausal women and may be able to eliminate cancer
cells in the early stages. In vitro studies suggest that the
anticancer activity of fenretinide may arise from its ability to
induce apoptosis in tumor cells. A number of previous
investigations have elucidated much about the apoptotic activity of
fenretinide. Diverse signaling molecules, including ROS, ceramide
and the ganglioside GD3, trigger the activation of cellular stress
response pathways and mediate the induction of apoptosis by
fenretinide in transformed, premalignant and malignant cells. In
the majority of cell types, the apoptotic activity of fenretinide
appears to be induced by mechanisms that are independent from
retinoic acid receptor activation and ultimately initiate the
intrinsic or mitochondria-mediated pathway of cell elimination.
Multiple proteins, including various transcription factors and
kinases, are responsive to stressful conditions in cells. Once
active, these proteins determine cell fate through the initiation
of apoptosis. Previous systematic detection of transcriptional
changes in response to oxidative signals generated in leukemia
cells following fenretinide treatment revealed the appearance of
multiple stress-responsive events during fenretinide-induced
apoptosis, including the redox response, the ER stress
response/UPR, translation repression and proteasome activation
(27).
In the present study, colon spheres were
demonstrated to form in serum-free medium with stem-like
characteristics. The unique anti-CSC effects of fenretinide were
stratified in a series of in vitro and in vivo
experiments, fenretinide was demonstrated to exert a selective
cytotoxic effect on HT29 sphere cells but not on their normal
counterparts. Mechanistic studies provided insight into the effects
of fenretinide in eradicating colon CSCs. Transcriptome analysis of
fenretinide-treated HT29 sphere cells identified several features
that highlighted the involvement of cell cycle regulation and
activation of ROS-induced stress responses.
ROS are involved in the regulation of CSCs and
confers resistance to radiotherapy. The ROS level in CSCs is low,
which makes it difficult to eliminate them by traditional
radiotherapy (36). However, the low
ROS levels in CSCs provide an opportunity to preferentially target
CSC cells using ROS inducers, including fenretinide.
Stress-responsive activities, including elevation of ROS levels in
cells treated with fenretinide, may explain the selective effect on
colon stem-like cells. However, further studies are required to
improve understanding of the specific underlying mechanisms.
Fenretinide is a promising agent that is able to
selectively target CSCs. Fenretinide does not induce point
mutations or chromosomal aberrations and causes few adverse side
effects, suggesting that fenretinide may be compatible for
long-term use as a chemo-preventive agent (37). Further investigations are warranted to
gain improved insight into its specific functions and the
underlying mechanisms.
To the best of our knowledge, the present study was
the first to investigate the effect of fenretinide on colon stem
cells. Fenretinide is a promising agent that is able to selectively
target CSCs. Fenretinide preferentially targeted colon sphere
cells, which are believed to possess certain stem-like
characteristics. These results are an important addition to current
knowledge about fenretinide, and provide a foundation for its
clinical application in the treatment of cancer.
Acknowledgements
The authors would like to thank Professor Zi-Jiang
Chen for her helpful suggestion and assistance.
Funding
The present study was supported in part by grants
from the National Natural Science Foundation (grant no. 81170503)
and Ministry of Science and Technology of China (grant no.
2013CB966802).
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author on reasonable
request. And the microarray data are available at the GEO accession
no. GSE66983.
Author's contributions
LL and YD designed the present study. JL and HW
performed the animal experiments. LL performed the experiments. LL,
JL, HZ and YD analyzed the data. LL and YD wrote the article. JL
and YD provided final approval of the version to be published.
Ethics approval and consent to
participate
The animal experiments were approved by the
Committee on Laboratory Animal Research of Shanghai Jiao Tong
University (Shanghai, China), and were conducted according to the
guidelines of the Laboratory Animal Center of Shanghai Jiao Tong
University School of Medicine (Shanghai, China).
Consent for publication
The present study did not use patient information
and related samples.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Scopelliti A, Cammareri P, Catalano V,
Saladino V, Todaro M and Stassi G: Therapeutic implications of
cancer initiating cells. Expert Opin Biol Ther. 9:1005–1016. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ricci-Vitiani L, Lombardi DG, Pilozzi E,
Biffoni M, Todaro M, Peschle C and De Maria R: Identification and
expansion of human colon-cancer-initiating cells. Nature.
445:111–115. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Todaro M, Francipane MG, Medema JP and
Stassi G: Colon cancer stem cells: Promise of targeted therapy.
Gastroenterology. 138:2151–2162. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Moon RC, McCormick DL, Becci PJ, Shealy
YF, Frickel F, Paust J and Sporn MB: Influence of 15 retinoic acid
amides on urinary bladder carcinogenesis in the mouse.
Carcinogenesis. 3:1469–1472. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ohshima M, Ward JM and Wenk ML: Preventive
and enhancing effects of retinoids on the development of naturally
occurring tumors of skin, prostate gland, and endocrine pancreas in
aged male ACI/segHapBR rats. J Natl Cancer Inst. 74:517–524.
1985.PubMed/NCBI
|
|
8
|
Malone W, Perloff M, Crowell J, Sigman C
and Higley H: Fenretinide: A prototype cancer prevention drug.
Expert Opin Investig Drugs. 12:1829–1842. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Du Y, Xia Y, Pan X, Chen Z, Wang A, Wang
K, Li J and Zhang J: Fenretinide targets chronic myeloid leukemia
stem/progenitor cells by regulation of redox signaling. Antioxid
Redox Signal. 20:1866–1880. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang H, Mi JQ, Fang H, Wang Z, Wang C, Wu
L, Zhang B, Minden M, Yang WT, Wang HW, et al: Preferential
eradication of acute myelogenous leukemia stem cells by
fenretinide. Proc Natl Acad Sci USA. 110:5606–5611. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lovat PE, Ranalli M, Bernassola F, Tilby
M, Malcolm AJ, Pearson AD, Piacentini M, Melino G and Redfern CP:
Distinct properties of fenretinide and CD437 lead to synergistic
responses with chemotherapeutic reagents. Med Pediatr Oncol.
35:663–668. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Maurer BJ, Melton L, Billups C, Cabot MC
and Reynolds CP: Synergistic cytotoxicity in solid tumor cell lines
between N-(4-hydroxyphenyl)retinamide and modulators of ceramide
metabolism. J Natl Cancer Inst. 92:1897–1909. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shan D, Gopal AK and Press OW: Synergistic
effects of the fenretinide (4-HPR) and anti-CD20 monoclonal
antibodies on apoptosis induction of malignant human B cells. Clin
Cancer Res. 7:2490–2495. 2001.PubMed/NCBI
|
|
14
|
Wang H, Zhang Y and Du Y: Ovarian and
breast cancer spheres are similar in transcriptomic features and
sensitive to fenretinide. Biomed Res Int. 2013:5109052013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lockhart DJ, Dong H, Byrne MC, Follettie
MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H
and Brown EL: Expression monitoring by hybridization to
high-density oligonucleotide arrays. Nat Biotechnol. 14:1675–1680.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization, and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gene Ontology Consortium, . Gene ontology
consortium: Going forward. Nucleic Acids Res. 43(Database Issue):
D1049–D1056. 2015.PubMed/NCBI
|
|
19
|
Edgar R, Domrachev M and Lash AE: Gene
expression omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Du Y, Wang K, Fang H, Li J, Xiao D, Zheng
P, Chen Y, Fan H, Pan X, Zhao C, et al: Coordination of intrinsic,
extrinsic, and endoplasmic reticulum-mediated apoptosis by imatinib
mesylate combined with arsenic trioxide in chronic myeloid
leukemia. Blood. 107:1582–1590. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xiao L, Wang K, Teng Y and Zhang J:
Component plane presentation integrated self-organizing map for
microarray data analysis. FEBS Lett. 538:117–124. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dontu G, Abdallah WM, Foley JM, Jackson
KW, Clarke MF, Kawamura MJ and Wicha MS: In vitro propagation and
transcriptional profiling of human mammary stem/progenitor cells.
Genes Dev. 17:1253–1270. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Haraguchi N, Ohkuma M, Sakashita H,
Matsuzaki S, Tanaka F, Mimori K, Kamohara Y, Inoue H and Mori M:
CD133+CD44+ population efficiently enriches colon cancer initiating
cells. Ann Surg Oncol. 15:2927–2933. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Du L, Wang H, He L, Zhang J, Ni B, Wang X,
Jin H, Cahuzac N, Mehrpour M, Lu Y and Chen Q: CD44 is of
functional importance for colorectal cancer stem cells. Clin Cancer
Res. 14:6751–6760. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Park CY, Tseng D and Weissman IL: Cancer
stem cell-directed therapies: Recent data from the laboratory and
clinic. Mol Ther. 17:219–230. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gene Ontology Consortium, . The gene
ontology project in 2008. Nucleic Acids Res. 36(Database Issue):
D440–D444. 2008.PubMed/NCBI
|
|
27
|
Wang K, Fang H, Xiao D, Zhu X, He M, Pan
X, Shi J, Zhang H, Jia X, Du Y and Zhang J: Converting redox
signaling to apoptotic activities by stress-responsive regulators
HSF1 and NRF2 in fenretinide treated cancer cells. PLoS One.
4:e75382009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Szegezdi E, Logue SE, Gorman AM and Samali
A: Mediators of endoplasmic reticulum stress-induced apoptosis.
EMBO Rep. 7:880–885. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li X, Uemura K, Hashimoto T, Nasser-Ghodsi
N, Arimon M, Lill CM, Palazzolo I, Krainc D, Hyman BT and
Berezovska O: Neuronal activity and secreted amyloid β lead to
altered amyloid β precursor protein and presenilin 1 interactions.
Neurobiol Dis. 50:127–134. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ward RJ and Dirks PB: Cancer stem cells:
At the headwaters of tumor development. Annu Rev Pathol. 2:175–189.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tang DG: Understanding cancer stem cell
heterogeneity and plasticity. Cell Res. 22:457–472. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kleffel S and Schatton T: Tumor dormancy
and cancer stem cells: Two sides of the same coin? Adv Exp Med
Biol. 734:145–179. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shien K, Toyooka S, Ichimura K, Soh J,
Furukawa M, Maki Y, Muraoka T, Tanaka N, Ueno T, Asano H, et al:
Prognostic impact of cancer stem cell-related markers in non-small
cell lung cancer patients treated with induction chemoradiotherapy.
Lung Cancer. 77:162–167. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dean M, Fojo T and Bates S: Tumour stem
cells and drug resistance. Nat Rev Cancer. 5:275–284. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jordan CT, Guzman ML and Noble M: Cancer
stem cells. N Engl J Med. 355:1253–1261. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Diehn M, Cho RW, Lobo NA, Kalisky T, Dorie
MJ, Kulp AN, Qian D, Lam JS, Ailles LE, Wong M, et al: Association
of reactive oxygen species levels and radioresistance in cancer
stem cells. Nature. 458:780–783. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hail N Jr, Kim HJ and Lotan R: Mechanisms
of fenretinide-induced apoptosis. Apoptosis. 11:1677–1694. 2006.
View Article : Google Scholar : PubMed/NCBI
|