Introduction
Breast cancer is the most common cancer in women.
Recurrence rate and mortality remains high in women with breast
cancer (1). Despite advances, the
clinical prognosis for breast cancer depends on the stage of the
tumor. Further studies investigating the molecular mechanisms
underlying the pathogenesis of breast cancer and identifying novel
therapeutic approaches are required.
DNA polymerase is required for DNA replication in
eukaryotic cells. DNA polymerase (Pol) δ belongs to the B family of
DNA polymerases. Pol δ is the most important replicase in
eukaryotic DNA replication and serves a primary role for the
synthesis of lagging strand, and has a 3′-5′exonuclease activity.
It also participates in DNA damage repair. Pol δ is involved in DNA
replication and repair of damaged DNA, and mutations in Pol δ may
be associated with the progression of cancer (2). DNA Pol δ consists of four subunits
(p125, p68, p50 and p12) (2). The
p125 subunit is a 125-kDa protein encoded by polymerase δ catalytic
subunit gene 1 (POLD1). The p125 catalytic subunit harbors
polymerase exonuclease catalytic domain, and serves an important
function in cellular growth and differentiation (3). Previous studies suggest that DNA Pol δ
may be involved in tumor progression. Sanefuji et al
(4) reported that p125 may serve an
important function in tumor invasion, thus leading to a poor
prognosis in hepatocellular carcinoma. They found that p125
expression in specimens was significantly correlated with cellular
differentiation and the degree of vascular invasion. It was also
significantly correlated with abnormal p53 expression, and in
vivo studies showed that p125 was upregulated in mutant
p53-transfected HepG2 cells. Venkatesan et al (5) mutations at the polymerase active site of
DNA Pol δ may promote genomic instability and accelerate
tumorigenesis. A previous study demonstrated that overexpression of
POLD1 is associated with an increased risk of breast cancer
(6). Therefore, DNA pol δ may a
crucial role in tumor progression. However, the expression and
clinical value of POLD1 in breast cancer remain unclear.
The aim of the present study was to detect the
expression and biologic function of POLD1 in breast cancer, and to
investigate its association with clinicopathological
characteristics and prognosis in patients with breast cancer.
Materials and methods
Patients and tissue specimens
A total of 84 female patients with breast cancer
were recruited between January 2011 and December 2013 in The
Affiliated Tumor Hospital of Guangxi University (Guangxi Zhuang
Autonomous Region, China). The mean age of patients was 49 years
(range, 34–65 years). The inclusion criteria were as follows: i)
Patients with invasive ductal breast carcinoma; ii) patients who
underwent mastectomy or wide local excision with axillary surgery;
iii) patients with paired fresh carcinoma and adjacent normal
tissues; and iv) patients with complete clinical and follow-up
information. Exclusion criteria were as follows: i) Patients who
received chemotherapy or endocrinotherapy prior to surgery; and ii)
lack of pathological data. All patients provided written informed
consent, and the study was approved by the Ethics Committee of The
Affiliated Tumor Hospital of Guangxi Medical University (Guangxi
Zhuang Autonomous Region, China). Fresh tissue specimens were
collected immediately after surgical resection. Tumor and adjacent
normal (2 cm distance from the edge of the tumor) tissues were
obtained.
Data collection and follow-up
period
Clinical data including age, pathological grade and
treatment of patients with breast cancer were collected. Follow-up
data were obtained by reviewing the medical records and from direct
communication with patients. Following surgery, patients were
followed-up until the date of mortality or censored at the date of
the last follow-up (March 2016). Disease relapse and metastasis
were determined by clinical examination and imaging evaluation,
including ultrasonography, X-ray, computed tomography (CT) or
magnetic resonance imaging (MRI). The primary endpoint was
disease-free survival (DFS), defined as the time interval from
surgery to the first evidence of recurrence and metastasis. The
follow-up period was 37 months (range, 15–62 months).
Immunohistochemistry
Expression of estrogen receptor (ER), progesterone
receptor (PR), human epidermal growth factor receptor 2 (HER-2),
p53 and Ki-67 was assessed using immunohistochemistry. A total of
84 samples were subjected to immunohistochemistry. All tissue
samples were fixed with 30% formalin at 28°C for 60 min and then
embedded with paraffin. The samples were sectioned to a thickness
of 5-mm. The sections were incubated at 60°C for 1 h, then placed
in xylene I and xylene II for 30 min in turn. All the slices were
dehydrated in a graded ethyl alcohol solution series (100, 95, 90
and 85%). Antigen retrieval was performed by placing the slices in
0.01 M potassium citrate solution (Fuzhou Maixin Biotech. Co.,
Ltd., Fuzhou, China) and microwaved at 90°C for 10 min prior to
cooling at room temperature. Slices were then rinsed in PBS three
times. Endogenous peroxidase activity was blocked with 3%
H2O2 and goat serum (Shanghai Haoran
Biological Technology Co., Ltd., Shanghai, China) at 37°C for 30
min, then immediately incubated with primary antibody at 4°C
overnight. The primary antibodies were: ER (dilution, 1:100;
catalog no. sc-420; Santa Cruz Biotechnology, Inc., Dallas, TX,
USA); PR (dilution, 1:1,000; catalog no. 8753; Cell Signaling
Technology, Inc., Danvers, MA, USA); HER-2 (dilution, 1:100;
catalog no. sc-33684; Santa Cruz Biotechnology, Inc.); p53
(dilution, 1:100; catalog no. sc-47698; Santa Cruz Biotechnology,
Inc.); and Ki-67 (dilution, 1:100; catalog no. sc56319; Santa Cruz
Biotechnology, Inc.). Subsequently, the slides were washed with PBS
three times prior to incubation with corresponding horseradish
peroxidase-conjugated mouse anti-goat secondary antibody (dilution,
1:100; catalog no. sc-516246; Santa-Cruz Biotechnology, Inc.) at
37°C for 1 h. The slides were then washed three more times in PBS,
then stained with DAB (Fuzhou Maixin Biotech. Co., Ltd.) at room
temperature for 3–5 min, rinsed 3 times with tap water and stained
with 0.5% hematoxylin (Fuzhou Maixin Biotech. Co., Ltd.), at room
temperature for 2 min, prior to sealing using mounting medium
(Fuzhou Maixin Biotech. Co., Ltd.). Images were captured using a
Leica DMLA light microscope (Leica Microsystems, Inc., Buffalo
Grove, IL, USA) with magnifications ×200 and ×400. Each section was
independently evaluated by 2 pathologists, who were blinded to the
data of the patients. The intensity was scored as follows: 0,
negative; 1, weak; 2, moderate; and 3, strong. The positive area
was defined as follows: 0, <5%; 1, 5%-25%; 2, 26%-50%; 3,
51%-75%; and 4, >75%. According to the St Gallen International
Expert Consensus (7), the cut-off
point for Ki-67 expression was determined as 14%, >14% was
considered high expression and ≤14% was considered low expression.
p53 expression referred to wild-type p53.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissues using TRIzol
reagent (Thermo Fisher Scientific, Inc., Waltham, MA, USA)
according to the manufacturer's protocol. RNA was
reverse-transcribed into cDNA using iScript cDNA Synthesis kit
(Bio-Rad Laboratories, Inc., Hercules, CA, USA). qPCR was performed
using SYBR Green PCR Master Mix (Qiagen GmbH, Hilden, Germany),
according to the manufacturer's protocol. The primer sequences for
POLD1 were as follows: 5′-GCTCCGCTCCTACACGCTCAA-3′ (forward) and
5′-GGTCTGGTCGTTCCCATTCTGC-3′ (reverse). The primer sequences for
GAPDH were as follows: 5′-AACGGATTTGGTCGTATTG-3′ (forward) and
5′-CTGGAAGATGGTGATGGG-3′ (reverse). The PCR thermocycling
conditions were as follows: 95°C for 30 sec, followed by 40 cycles
of 95°C for 5 sec, and 60°C for 30 sec. The results were analyzed
using the 2−ΔΔCq method (8) by Bio-Rad CFX Manger 3.0 (Bio-Rad
Laboratories, Inc.). Three individual experiments were
performed.
Western blot analysis
Paired tumor and adjacent normal tissues were
extracted using NP40 lysis buffer (50 mM Tris-Cl, pH 7.5, 150 mM
NaCl, 2% NP-40). Cellular proteins were extracted using a cell
lysis buffer (10 mM Tris-HCl, pH 7.4, 1% SDS, 1mM Na3VO4). Protein
concentration was quantified by bicinchoninic acid protein assay
kit. Equal amounts of protein (20 µg) were separated by SDS-PAGE
(5% gels) and transferred onto polyvinylidene fluoride membranes.
Following transfer, membranes were washed with TBS-T solution (1 ml
TBS solution + 20% Tween 20), and then blocked with 5% skimmed milk
(in PBS) and agitated at room temperature for 1 h. Next, membranes
were incubated at 4°C overnight with primary antibodies against
p125 (1:1,000; catalog no. 15646-1-AP; ProteinTech Group, Inc.,
Chicago, IL, USA) and β-actin (1:1,000; catalog no. sc-130300;
Santa Cruz Biotechnology, Inc.). Membranes were washed with 0.01%
Tween-20 (TBST) three times, for 5 min each time, prior to being
incubated with horseradish peroxidase-conjugated goat anti-rabbit
secondary antibody (1:10,000; catalog no. 7074; Cell Signaling
Technology, Inc.), agitated at room temperature for 1 h and washed
with 0.01% Tween-20 (TBST) three times, for 5 min each time. Immune
complexes were detected by incubation with enhanced
chemiluminescence western blotting substrate (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) and visualized using a LI-COR
Odyssey gel imaging scanner (LI-COR Biosciences, Lincoln, NE, USA).
Odyssey V3.0 software (LI-COR Biosciences) was used to analyze the
image. β-actin was used as an internal control. Patients with
breast cancer were divided into high and low expression group based
on the median expression level of p-125.
Cell culture
The human breast adenocarcinoma MCF-7 cell line was
obtained from American Type Culture Collection (ATCC; Manassas, VA,
USA). The cells were maintained in Dulbecco's modified Eagle's
medium (Thermo Fisher Scientific Inc.) supplemented with 10% fetal
bovine serum (Hangzhou Sijiqing Biological Engineering Materials
Co., Ltd., Hangzhou, China) at 37°C in a humidified atmosphere
containing 5% CO2.
Cell transfection
Short hairpin RNA (shRNA) targeting POLD1 (shPOLD1)
and the negative control shRNA (shControl) were purchased from
Shanghai GenePharma Co., Ltd. (Shanghai, China). shRNA was designed
using online shRNA tools (Invitrogen; Thermo Fisher Scientific,
Inc.), and the oligonucleotides encoding POLD1 shRNA were as
follows: 5′-CACCGGTCCACCTTCATCCGTATCACGAATGATACGGATGAAGGTGGACC-3′
(forward) and
5′-AAAAGGTCCACCTTCATCCGTATCATTCGTGATACGGATGAAGGTGGACC-3′ (reverse).
Synthetic interference POLD1 gene sequences were inserted into the
shRNA eukaryotic expression vector pLKO. 1-puro plasmid (GenePharma
Co., Ltd.). and identified by enzyme digestion and sequencing. For
cell transfection, MCF-7 cells (1×105 cells/well) were
plated in 6-well plates and then transfected with plasmid
(shControl and shPOLD1; 500 ng/µl) for 6 h using
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. Next, cells were
incubated in complete medium (Thermo Fisher Scientific, Inc.) for
48 h, with concentration of 5 µg/ml puromycin screening culture
medium for screening of stable transfection cell lines. After 24 h
of infection, the cells were used for subsequent
experimentation.
Cell proliferation assay
The proliferation of MCF-7 was assessed using a Cell
Counting Kit-8 detection kit (CCK-8; Beyotime Institute of
Biotechnology, Haimen, China), as previously described (9). MCF-7 cells were seeded into 96-well
plates at a density of 2×103 cells/well incubated in 10%
CCK-8 diluted in Dulbecco's modified Eagle's medium at 37°C.
Following transfection, cell proliferation rates were determined at
0, 24, 48, 72 and 96 h. A microplate reader (set at 450 nm) was
used to measure the absorbance of each well.
Cell cycle analysis
Flow cytometry was used to determine the cell cycle
distribution. Briefly, MCF-7 cells were cultured in 6-well plates
and were transfected with shControl or shPOLD1 plasmids. For cell
transfection, MCF-7 cells (1×105 cells/well) were plated
in 6-well plates and then transfected with plasmid (shControl and
shPOLD1; 500 ng/µl; Shanghai GenePharma Co., Ltd.) for 6 h using
Lipofectamine 2000 (Thermo Fisher Scientific, Inc.), as
aforementioned. After incubation for 96 h, cells were seeded in
6-well culture plates and cultured to 80% confluence. Cells were
harvested and single cell suspensions were prepared in 0.25% of
trypsin (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany).
Subsequently, the cells were washed with PBS twice and resuspended
at 2×106 cells/ml. Aliquots of 1 ml cells were placed in
a 15 ml polypropylene tube on ice and allowed to cool, followed by
the addition of 3 ml cold (−20°C) absolute ethanol. The cells were
fixed in 70% cold ethanol for at least 1 h at 4°C. Cells were
washed twice with PBS, and 1 ml PI (Sigma-Aldrich; Merck KGaA; 40
µg/ml PI in PBS) staining solution was added to the cell pellet and
mixed well. Stained samples were stored for up to a week at ٤°C in
the dark. Analysis of cell cycle was determined on a flow cytometer
(BD Biosciences, Franklin Lakes, NJ, USA) and using the CellQuest
Pro software (version 5.1; BD Biosciences), as previously described
(9).
Statistical analysis
Data were analyzed using SPSS software (version
16.0; SPSS, Inc., Chicago, IL, USA). All results are expressed as
the mean ± standard deviation from three independent replicate
experiments. Student's t test was used to analyze the expression of
POLD1 level in tumor and adjacent normal specimens. The association
between the expression of POLD1 and clinicopathological parameters
was assessed using a χ2 test. Survival analysis was
performed using Kaplan-Meier method and log-rank tests. Univariate
and multivariate Cox regression analyses were used to determine
independent prognostic factors. P<0.05 was considered to
indicate a statistically significant difference.
Results
Patient characteristics
A total of 84 females with invasive breast cancer
treated with radical mastectomy were retrospectively collected. The
diagnosis was confirmed by pathological examination and all
patients were diagnosed with duct carcinoma. None of the patients
received any therapy prior to surgery. Clinical data including age,
tumor size, histological grade, lymph node status and pathological
features are presented in Table I.
ER, PR, HER-2, p53 status and ki-67 index were routinely evaluated
by immunohistochemistry. According to the results, 37 samples were
negative for wild-type p53 index, whereas 47 samples demonstrated
p53-positive staining (Table I).
 | Table I.Clinicopathological characteristics of
patients with breast cancer. |
Table I.
Clinicopathological characteristics of
patients with breast cancer.
| Characteristic | Patients, n (%) |
|---|
| Age, years |
|
| ≤45 | 34 (40.5) |
|
>45 | 50 (59.5) |
| Tumor size, cm |
|
| ≤2.5 | 24 (28.6) |
|
>2.5 | 60 (71.4) |
| Histological
grade |
|
| Grade
1 | 18 (21.4) |
| Grade
2–3 | 66 (78.6) |
| Lymph node
status |
|
|
Negative | 31 (36.9) |
|
Positive | 53 (63.1) |
| ER status |
|
|
Negative | 28 (33.3) |
|
Positive | 56 (66.7) |
| PR status |
|
|
Negative | 21 (25.0) |
|
Positive | 63 (75.0) |
| HER-2 status |
|
|
Negative | 40 (47.6) |
|
Positive | 44 (52.4) |
| Ki-67 index |
|
| ≤14% | 16 (19.0) |
|
>14% | 68 (81.0) |
| p53 status |
|
|
Negative | 37 (44.0) |
|
Positive | 47 (56.0) |
POLD1 expression in breast cancer
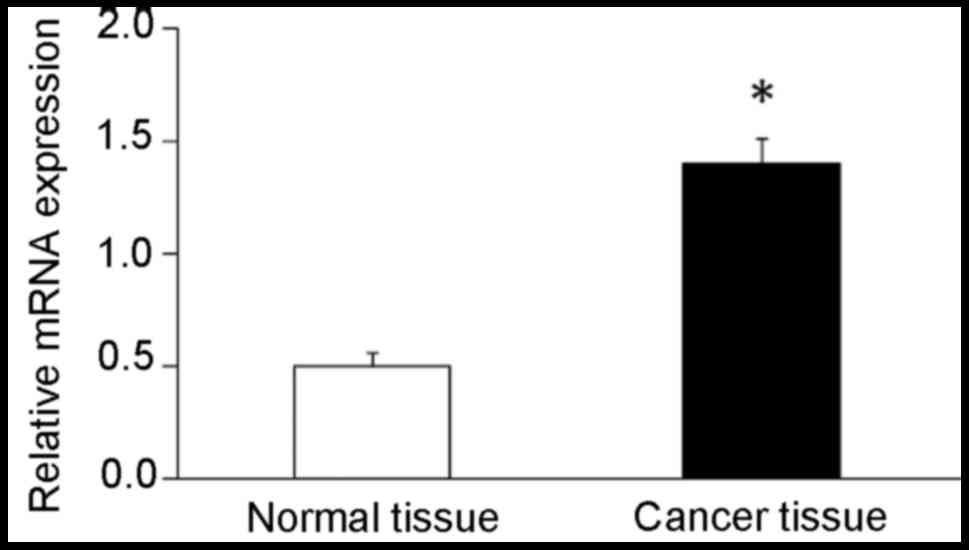
Gene expression level of POLD1 was assessed using
RT-qPCR. The results demonstrated that gene expression level of
POLD1 was significantly elevated (2.8-fold) in breast cancer
tissues compared with that in adjacent normal breast tissues
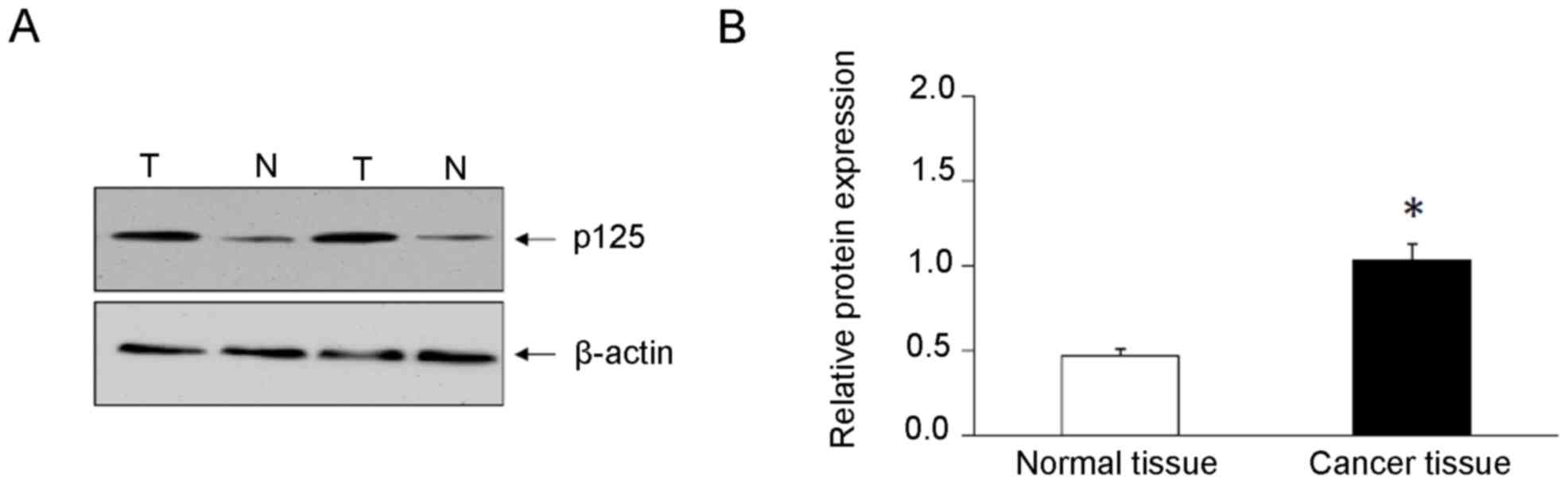
(Fig. 1). Western blot analysis
demonstrated that the expression levels of POLD1/p125 subunit were
significantly increased (2.2-fold) in cancer tissues compared with
adjacent normal breast tissues (Fig.
2).
Association between the expression of
POLD1 and clinicopathological characteristics
Median expression level of POLD1 was used as the
cut-off point (low score, <1.03; high score, ≥1.03) and patients
with breast cancer were divided into high and low expression
groups. The association between the expression of POLD1 and
clinicopathological variables in patients with breast cancer was
determined. The results demonstrated that increased expression of
POLD1 was associated with lymph node metastasis (P=0.028),
histological grade (P=0.025), p53 status (P<0.01) and ki-67
index (P=0.020; Table II). However,
the expression of POLD1 was not associated with the age, tumor
size, ER, PR and HER-2 status (P>0.05; Table II).
 | Table II.Association between POLD1 expression
and clinicopathological features. |
Table II.
Association between POLD1 expression
and clinicopathological features.
|
| POLD1 protein
expression |
|
|---|
|
|
|
|
|---|
| Variable | Low (n=41) | High (n=43) | P-value |
|---|
| Age, years |
|
| 0.478 |
|
≤45 | 15 | 19 |
|
|
>45 | 26 | 24 |
|
| Tumor size, cm |
|
| 0.073 |
|
≤2.5 | 8 | 16 |
|
|
>2.5 | 33 | 27 |
|
| Histological
grade |
|
| 0.025 |
| Grade
1 | 13 | 5 |
|
| Grade
2–3 | 28 | 38 |
|
| Lymph node
status |
|
| 0.028 |
|
Negative | 20 | 11 |
|
|
Positive | 21 | 32 |
|
| ER status |
|
| 0.123 |
|
Negative | 17 | 11 |
|
|
Positive | 24 | 32 |
|
| PR status |
|
| 0.059 |
|
Negative | 14 | 7 |
|
|
Positive | 27 | 36 |
|
| HER-2 status |
|
| 0.084 |
|
Negative | 17 | 23 |
|
|
Positive | 27 | 17 |
|
| Ki-67 index |
|
| 0.020 |
|
≤14% | 12 | 4 |
|
|
>14% | 29 | 39 |
|
| p53 status |
|
| <0.001 |
|
Negative | 9 | 28 |
|
|
Positive | 32 | 15 |
|
Prognostic value of POLD1 expression
in patients with breast cancer
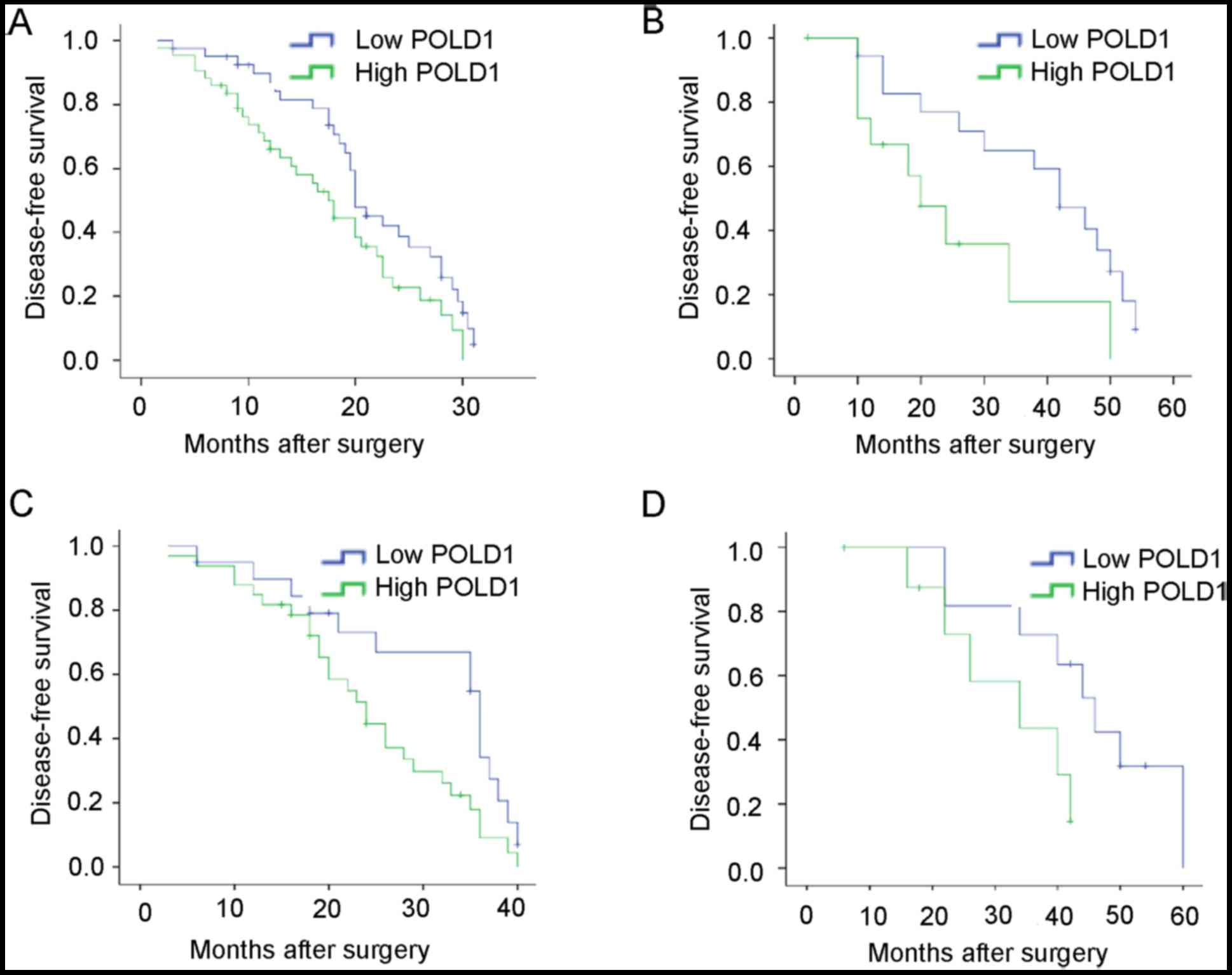
To further determine the importance of increased
expression of POLD1 in breast cancer, DFS was assessed in patients
with breast cancer patients using Kaplan-Meier method and log-rank
tests. The results indicated that patients with increased
expression of POLD1 exhibited shorter DFS compared with patients
with low expression of POLD1 (median, 31.5 vs. 38.6 months;
P=0.033; Fig. 3A). Additionally,
patients with increased expression of POLD1 exhibited shorter DFS
compared with those in the low expression group POLD1 at early
stage (P=0.037) or late stage (P=0.023) (Fig. 3B and C). Increased expression of POLD1
was associated with shorter DFS in patients with triple-negative
tumors (P=0.049; Fig. 3D).
Univariate analysis demonstrated that histological
grade (P=0.018), lymph node status (P=0.001), ki-67 index (P=0.019)
and POLD1 expression status (P<0.01) were associated with DFS.
These results suggest that POLD1 may be a valuable prognostic
factor in breast cancer. Multivariate analysis revealed that
expression of POLD1 [Hazard ratio (HR), 2.14; P=0.048],
histological grade (HR, 4.46; P=0.010), lymph node status (HR,
2.13; P=0.045) and ki-67 expression (HR, 4.61; P=0.021) were
independent factors associated with DFS (Table III).
 | Table III.Univariate and multivariate analysis
of clinicopathological factors for the DFS of 84 patients with
breast cancer. |
Table III.
Univariate and multivariate analysis
of clinicopathological factors for the DFS of 84 patients with
breast cancer.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Characteristic | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age | 0.81
(0.45–1.48) | 0.387 |
|
|
| Tumor size | 1.61
(0.88–2.93) | 0.119 |
|
|
| Histological
grade | 2.99
(1.20–7.45) | 0.018 | 4.46
(1.43–13.94) | 0.010 |
| Lymph node
status | 2.85
(1.50–5.38) | 0.001 | 2.13
(1.07–4.56) | 0.045 |
| ER status | 0.73
(0.36–1.47) | 0.376 |
|
|
| HER-2 status | 1.17
(0.59–2.32) | 0.654 |
|
|
| Ki-67 | 3.52
(1.23–10.12) | 0.019 | 4.61
(1.26–16.95) | 0.021 |
| POLD1 | 3.14
(1.67–5.91) | <0.001 | 2.14
(0.99–4.68) | 0.048 |
shPOLD1 inhibits the proliferation of
MCF7 cells
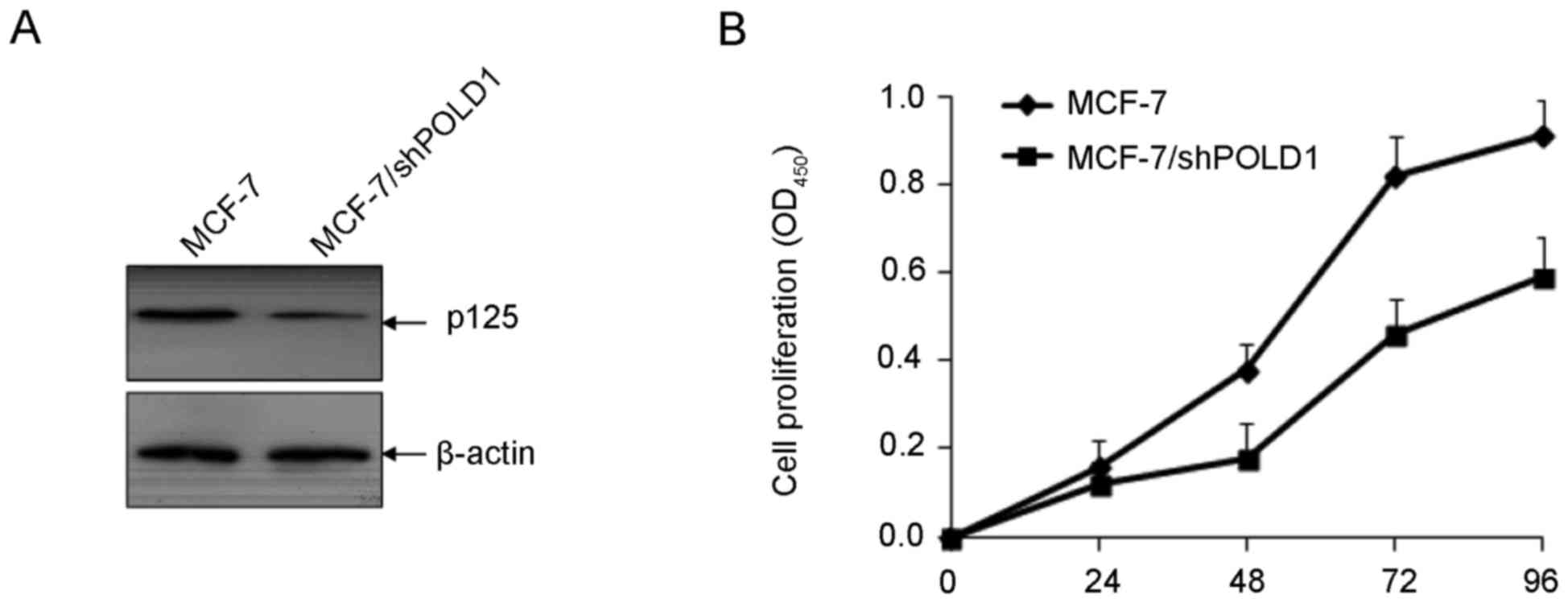
To investigate the function of POLD1 in cellular
proliferation in breast cancer, MCF-7 cells were transfected with
shPOLD1 and cell proliferation was assessed using a CCK-8 assay.
The expression of POLD1 was significantly downregulated following
transfection with shPOLD1 (Fig. 4A).
Downregulation of POLD1 significantly inhibited cell proliferation
compared with the control (P<0.01; Fig. 4B).
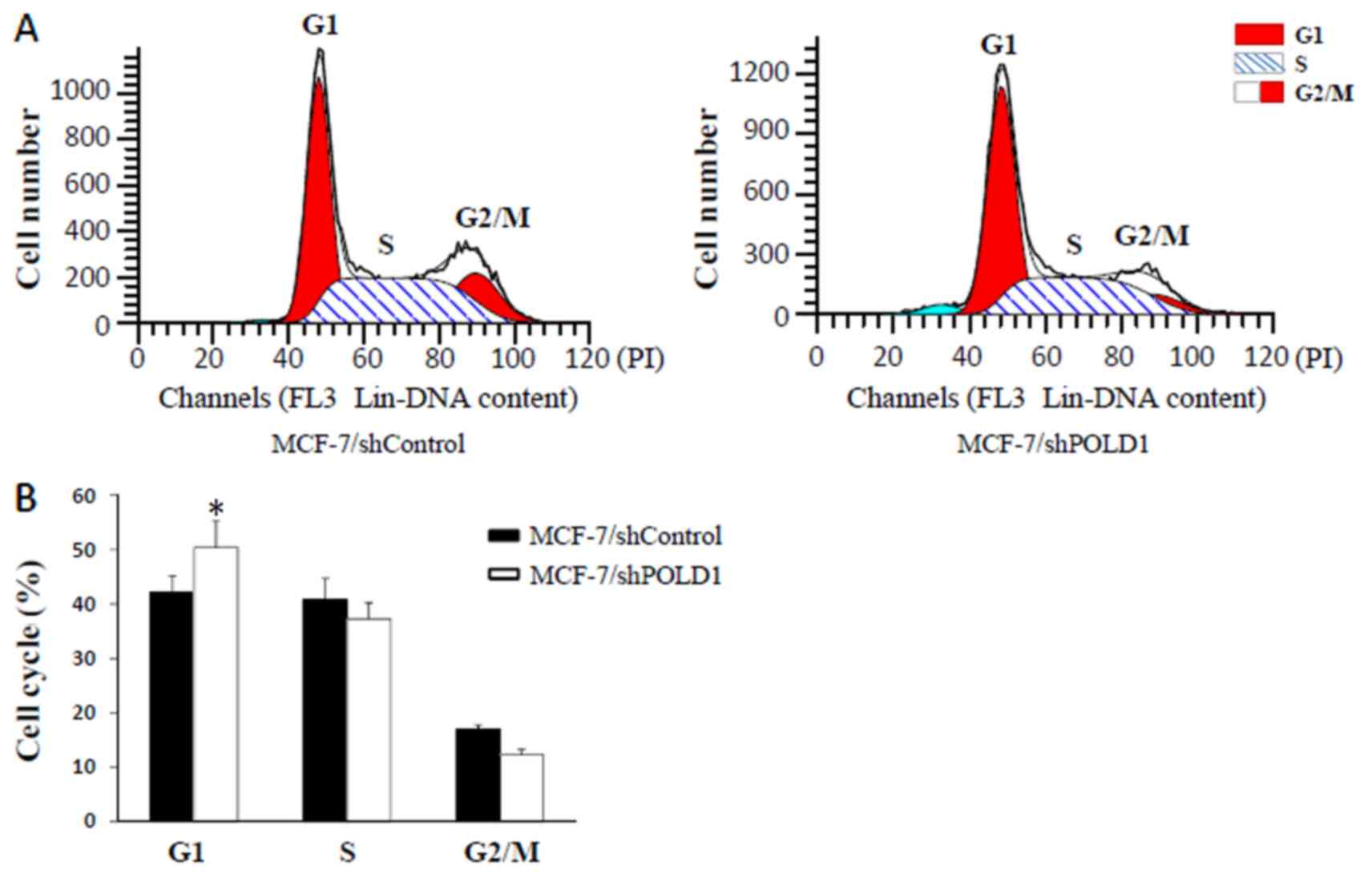
shPOLD suppresses cell cycle
progression
Following transient transfection of MCF-7 cells with
shPOLD1 or shControl, cell cycle distribution was analyzed using
flow cytometry. The results demonstrated that the percentage of
cells in G1 phase was increased following transfection with shPOLD1
compared with that in the control (P<0.05; Fig. 5), thereby suggesting that POLD1 shRNA
may inhibit the cell cycle at G1 phase in MCF-7 cells.
Discussion
POLD1 belongs to a family of human DNA polymerases
and exhibits polymerase and 3′ to 5′exonuclease activity. POLD1 is
responsible for the synthesis of the lagging strand during DNA
replication (10). It was recently
demonstrated that POLD1 may modulate cell cycle progression and
promote the proliferation of cancer cells (11). A previous study revealed that the
expression of POLD1 was increased in human breast cancer cells
(9). To the best of our knowledge,
the present study is the first to explore the association of the
expression of POLD1 with clinicopathological variables and survival
time in patients with breast cancer.
Breast cancer is a common cancer in females. In the
USA, >230,000 new cases of breast cancer were diagnosed and
>40,000 mortalities were reported in 2015 (12). Despite advances, the clinical
prognosis of breast cancer remains poor. Identification of novel
and reliable prognostic factors is required for the development of
therapeutic strategies in breast cancer.
Abnormal expression of POLD1 has been detected in
several types of solid tumors. However, the expression and clinical
value of POLD1 in breast cancer remains unclear (3). The results of the present study
demonstrated that gene and protein expression of POLD1 was elevated
in breast cancer tissues compared with adjacent normal tissues.
Additionally, increased expression of POLD1 was associated with
lymph node metastasis, differentiation grade, p53 status and ki-67
index, whereas there was no association with the remaining
clinicopathological variables. Additionally, the association
between POLD1 and breast cancer prognosis was assessed through
survival analysis. The results demonstrated that increased
expression of POLD1 was associated with shorter DFS in patients
with breast cancer. Multivariate analysis revealed that the
expression of POLD1, together with axillary lymph node status,
histological grade and ki-67 index may be considered as independent
prognostic factors for long-term outcomes in patients with breast
cancer. Therefore, POLD1 is a potential prognostic factor and
therapeutic target in patients with invasive breast carcinoma.
The results of the present study are consistent with
those of a previous study which reported that the expression of
POLD1 is associated with cellular differentiation and the degree of
vascular invasion in hepatocellular carcinoma (4), thus leading to poor prognosis. A
previous study identified that POLD1 may be used as a novel
biomarker for head and neck cancer (13). Narayan et al (14) employed cDNA array comparative genomic
hybridization to analyze 29 cervical cancer cases and detected
increased expression of POLD1 in the tissues. Previous studies have
demonstrated that genetic mutations in the POLD1 gene are
associated with epithelial (15),
endometrium (16) and colorectal
cancer (17). The hypothesis of the
present study was that POLD1 may be associated with tumorigenesis
and tumor progression, and may regulate proliferation and cell
cycle progression. A previous study reported that inhibition of
POLD1 promoted apoptosis of hepatocellular carcinoma cells
(18). Additionally, POLD1 may serve
an important function in cell cycle progression and repair of
damaged DNA (19). POLD1 may be
overexpressed in mesothelioma and its expression is associated with
pemetrexed/carboplatin resistance (20), thus suggesting that POLD1 may provide
protection against DNA damage through multiple molecular
mechanisms.
p53 may inhibit the expression of p125 by regulating
the methylation of POLD1 gene promoter (9). POLD1 is a transcriptional target of p53
(21), therefore the association
between the expression of wild-type p53 and POLD1 in breast cancer
was assessed in the present study. The results demonstrated that
the expression of POLD1was negatively associated with the
expression of wild-type p53. Previous studies demonstrated that
POLD1 may be upregulated in mutant p53-transfected cells (4). Further studies investigating the
association between the expression of mutant-type p53 and POLD1 in
clinical samples are required.
TNBC is a subtype of breast cancer, defined by lack
of expression of ER, PR and HER-2, and is associated with poor
prognosis (22). Further studies
identifying biomarkers in TNBC are required. The results of the
present study demonstrated that the expression of POLD1 was not
associated with ER, PR, HER-2, thus suggesting that there is no
association between the expression of POLD1 and TNBC. However,
POLD1 was associated with prognostic factors, including lymph node
metastasis and histological grade. Survival analysis indicated that
the expression of POLD1 was associated with shorter DFS in patients
with TNBC. The results suggested that the prognosis of breast
cancer is not associated with molecular subtypes and that a more
reliable prognostic factor is required. A previous study
demonstrated that POLD1 downregulation by shRNA suppressed cell
proliferation, cell cycle progression and DNA synthesis in HEK293
cells (19), suggested that POLD1
plays important role in the regulation of cell cycle progression;
therefore, the effects of POLD1 in regulating cell cycle and
proliferation of breast cancer cells was examined in MCF-7 cells
in vitro. The results revealed that downregulation of POLD1
suppressed cell cycle progression and cell proliferation in MCF-7
cells, thus suggesting that POLD1 may serve an important function
in tumor progression.
The present study has several limitations. In the
present study the proportion of HER-2 positive cells is high
compared with previous studies (23).
This may be due to the small sample size. Although protein
expression of POLD1 was detected using western blot analysis, its
expression should be also validated using immunohistochemistry.
Additionally, further studies are required to investigate the
molecular mechanisms underlying the effects of POLD1 in breast
cancer.
In conclusion, POLD1 may serve an important function
in breast cancer progression, and may be considered as a novel
therapeutic target in breast cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by National Natural
Science Foundation of China (grant no. 81360396) and Natural
Science Foundation of Guangxi (grant no. 2015GXNSFAA139204).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CW, QQT and QT conceived and designed the study,
performed patient collection and clinical data interpretation, and
wrote the manuscript. JL and WY helped with the tissue preparation
and conducted the in vitro experiments. QM and BL performed
the statistical analysis. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The Affiliated Tumor Hospital of Guangxi Medical
University (Guangxi Zhuang, China) and all participants provided
written informed consent.
Patient consent for publication
Written informed consent was obtained from all
participants for the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liu L, Mo J, Rodriguez-Belmonte EM and Lee
MY: Identification of a fourth subunit of mammalian DNA polymerase
delta. J Biol Chem. 275:18739–18744. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nicolas E, Golemis EA and Arora S: POLD1:
Central mediator of DNA replication and repair, and implication in
cancer and other pathologies. Gene. 590:128–141. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sanefuji K, Taketomi A, Iguchi T,
Sugimachi K, Ikegami T, Yamashita Y, Gion T, Soejima Y, Shirabe K
and Maehara Y: Significance of DNA polymerase delta catalytic
subunit p125 induced by mutant p53 in the invasive potential of
human hepatocellular carcinoma. Oncology. 79:229–237. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Venkatesan RN, Treuting PM, Fuller ED,
Goldsby RE, Norwood TH, Gooley TA, Ladiges WC, Preston BD and Loeb
LA: Mutation at the polymerase active site of mouse DNA polymerase
delta increases genomic instability and accelerates tumorigenesis.
Mol Cell Biol. 27:7669–7682. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sigurdson AJ, Hauptmann M, Chatterjee N,
Alexander BH, Doody MM, Rutter JL and Struewing JP: Kin-cohort
estimates for familial breast cancer risk in relation to variants
in DNA base excision repair, BRCA1 interacting and growth factor
genes. BMC Cancer. 4:92004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Goldhirsch A, Wood WC, Coates AS, Gelber
RD, Thurlimann B and Senn HJ: Panel members: Strategies for
subtypes-dealing with the diversity of breast cancer: Highlights of
the St. Gallen international expert consensus on the primary
therapy of early breast Cancer 2011. Ann Oncol. 22:1736–1747. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang L, Yang W, Zhu X and Wei C: p53
inhibits the expression of p125 and the methylation of POLD1 gene
promoter by downregulating the Sp1-induced DNMT1 activities in
breast cancer. Onco Targets Ther. 9:1351–1360. 2016.PubMed/NCBI
|
|
10
|
Lessel D, Hisama FM, Szakszon K, Saha B,
Sanjuanelo AB, Salbert BA, Steele PD, Baldwin J, Brown WT, Piussan
C, et al: POLD1 Germline mutations in patients initially diagnosed
with Werner syndrome. Hum Mutat. 36:1070–1079. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kashkin K, Chernov I, Stukacheva E,
Monastyrskaya G, Uspenskaya N, Kopantzev E and Sverdlov E: Cancer
specificity of promoters of the genes controlling cell
proliferation. J Cell Biochem. 116:299–309. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ceder R, Haig Y, Merne M, Hansson A, Zheng
X, Roberg K, Nees M, Iljin K, Bloor BK, Morgan PR, et al:
Differentiation-promoting culture of competent and noncompetent
keratinocytes identifies biomarkers for head and neck cancer. Am J
Pathol. 180:457–472. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Narayan G, Bourdon V, Chaganti S,
Arias-Pulido H, Nandula SV, Rao PH, Gissmann L, Dürst M, Schneider
A, Pothuri B, et al: Gene dosage alterations revealed by cDNA
microarray analysis in cervical cancer: Identification of candidate
amplified and overexpressed genes. Genes Chromosomes Cancer.
46:373–384. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Goldsby RE, Hays LE, Chen X, Olmsted EA,
Slayton WB, Spangrude GJ and Preston BD: High incidence of
epithelial cancers in mice deficient for DNA polymerase delta
proofreading. Proc Natl Acad Sci USA. 99:15560–15565. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wong A, Kuick CH, Wong WL, Tham JM, Mansor
S, Loh E, Jain S, Vikas NN, Tan SH, Chan SH, et al: Mutation
spectrum of POLE and POLD1 mutations in South East Asian women
presenting with grade 3 endometrioid endometrial carcinomas.
Gynecol Oncol. 141:113–120. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Palles C, Cazier JB, Howarth KM, Domingo
E, Jones AM, Broderick P, Kemp Z, Spain SL, Guarino E, Salguero I,
et al: Germline mutations affecting the proofreading domains of
POLE and POLD1 predispose to colorectal adenomas and carcinomas.
Nat Genet. 45:136–144. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cao B, Zhang Z, Zhang Y, Li J, Liang G and
Ling J: Effect of Smilax china L.-containing serum on the
expression of POLD1 mRNA in human hepatocarcinoma SMMC-7721 cells.
Exp Ther Med. 6:1070–1076. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Song J, Hong P, Liu C, Zhang Y, Wang J and
Wang P: Human POLD1 modulates cell cycle progression and DNA damage
repair. BMC Biochem. 16:142015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Roe OD, Szulkin A, Anderssen E, Flatberg
A, Sandeck H, Amundsen T, Erlandsen SE, Dobra K and Sundstrøm SH:
Molecular resistance fingerprint of pemetrexed and platinum in a
long-term survivor of mesothelioma. PLoS One. 7:e405212012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li B and Lee MY: Transcriptional
regulation of the human DNA polymerase delta catalytic subunit gene
POLD1 by p53 tumor suppressor and Sp1. J Biol Chem.
276:29729–29739. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lee A and Djamgoz MBA: Triple negative
breast cancer: Emerging therapeutic modalities and novel
combination therapies. Cancer Treat Rev. 62:110–122. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chang HR: Trastuzumab-based neoadjuvant
therapy in patients with HER2-positive breast cancer. Cancer.
116:2856–2867. 2010. View Article : Google Scholar : PubMed/NCBI
|