Introduction
Renal cell carcinoma (RCC) is the most prevalent
adult kidney malignancy, and its incidence has been increasing in
recent decades (1). RCC is associated
with high rates of mortality and resistance to chemotherapy and
radiotherapy (2–4). Patients with early-stage disease can be
treated with surgical resection, but ~20% of patients present with
metastatic disease at the initial diagnosis (5). Moreover, up to 20% of RCC patients
suffer from metastatic lesions even if nephrectomy is performed
(6). Therefore, the identification of
new sensitive, reliable biomarkers that predict RCC progression and
prognosis and the development of new targeted therapies that
improve RCC patient prognosis are necessary.
Proline-rich tyrosine kinase 2 (Pyk2), also known as
PTK2B, FAK2, RAFTK, and CAKB, regulates different signal
transduction cascades that control cell proliferation, migration
and invasion (7–10). Pyk2 has high homology with focal
adhesion kinase (FAK) at the structural level (11). Pyk2 is overexpressed in hepatocellular
carcinoma (HCC) cells, and its expression is associated with poor
prognosis (12). Pyk2 overexpression
promotes HCC cell migration and invasion via ERK pathway activation
(10,13). Pyk2 is critical for the malignant
phenotype in breast cancer (14) and
plays a significant role in facilitating epithelial-to-mesenchymal
transition in breast cancer (15).
Moreover, Pyk2 is a common downstream effector of ErbB and IL8
receptors, and it integrates these signaling pathways through a
positive feedback loop to potentiate breast cancer invasion
(16). Poor prognosis in lung cancer
has been proven to correlate with aberrant Pyk2 upregulation
(17). Pyk2 also plays an important
role in astrocytic tumor angiogenesis through VEGF regulation
(18) and is a key downstream
signaling molecule of chemokine receptor 7 in squamous cell
carcinoma of the head and neck, promoting tumorigenesis and
progression (19). However, the role
of Pyk2 in RCC remains less explored.
In our study, we demonstrated that Pyk2 is highly
expressed at the mRNA and protein levels in RCC tissues compared
with paired adjacent nontumor (NT) tissues. Moreover, we found that
Pyk2 upregulation is associated with poor clinical outcomes in RCC
patients. By using loss-of-function approaches, we found that Pyk2
knockdown reduced viability, invasive ability, and migratory
ability and increased apoptosis in RCC cell lines. In contrast,
Pyk2 overexpression promoted tumor cell proliferation, invasion and
migration and reduced apoptosis. Overall, our findings describe the
pro-oncogenic role of Pyk2 in RCC and thus provide molecular
evidence for a novel Pyk2-targeting therapeutic strategy in
RCC.
Patients and methods
Patients and clinical samples
All cases included in the study were clinically and
pathologically identified as RCC. This study included 60 RCC
tissues and paired adjacent NT tissues obtained from patients who
underwent surgery at Changzheng Hospital, Second Military Medical
University (Shanghai, China). No patients received anticancer
treatments before surgery in this study. The median fellow-up time
of these 60 RCC patients was 60 months. Written informed consent
was obtained from all patients. The Ethics Committee of Changzheng
Hospital, Second Military Medical University, approved the use of
these tissues in this study.
Cell lines and culture conditions
Human RCC cell lines (A498, ACHN, CAKI-2, OS-RC-2,
769P and 786-O) and the normal kidney cell line HK-2 were obtained
from the Shanghai Institute of Life Sciences Cell Resource Center
(Shanghai, China). OS-RC-2, 769P and 786-O cell lines were cultured
in RPMI modified medium (GE Healthcare Life Sciences, Logan, UT,
USA), and A498 and ACHN cell lines were cultured in minimum
essential medium (Eagle; Corning Inc., Corning, NY, USA). The
CAKI-2 cell line was cultured in McCoy's 5A medium (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA), and the HK-2 cell line
was cultured in DMEM (GE Healthcare Life Sciences). All media were
supplemented with 10% fetal bovine serum (FBS) and 1%
penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.),
according to the American Type Culture Collection. All cell
cultures were maintained at 37°C in a humidified atmosphere with 5%
CO2. According to the expression of Pyk2 in RCC cell
lines, we selected ACHN cells to generate Pyk2 knockdown cells and
A498 cells to generate stable Pyk2 overexpression cells.
Tissue microarray (TMA) construction
and immunohistochemical (IHC) detection
IHC was performed with the TMA using a two-step
immunoperoxidase technique. After heating the sections in 10 mmol/l
citrate buffer for antigen retrieval, the sections were incubated
with primary antibody against Pyk2 (Abcam, Cambridge, MA, USA;
dilution 1:60) at 4°C overnight and then with appropriate secondary
antibody for one h at room temperature.
RNA extraction, cDNA preparation and
qRT-PCR
Total RNA was extracted from cells and tissues using
TRIzol reagent (Takara Bio, Inc., Otsu, Japan), according to the
manufacturer's instructions. Total RNA quality was assessed using a
Nanodrop 2000 and agarose gel electrophoresis. First-strand cDNA
was generated from 2 µg of total RNA using M-MLV reverse
transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.) with
random primers. qRT-PCR was performed according to the SYBR Green
protocol in a Step One Plus System (Applied Biosystems, Foster
City, CA, USA), and β-actin served as the endogenous control.
Primer sequences were as follows: Pyk2 5′-GTGGGAGATCCTGAGCTTTG-3′
(forward) and 5′-TAAAGGACCGGTGGACAGAG-3′ (reverse); and β-actin
5′-CTGGTGCCTGGGGCG-3′ (forward) and 5′-AGCCTCGCCTTTGCCGA-3′
(reverse). Relative mRNA expression levels were calculated based on
the corresponding relative quantity (RQ) values and were normalized
to β-actin expression.
Western blot analysis
Total cell and tissue lysates were prepared in 1X
sodium dodecyl sulfate buffer. Identical quantities of protein were
separated by SDS gel electrophoresis and transferred onto
nitrocellulose filter membranes. After incubating with antibodies
specific for Pyk2 (ab32571; Abcam) and GAPDH (sc-25778; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA), the blots were incubated
with IRDye 800-conjugated goat anti-rabbit IgG, and bands were
detected using an Odyssey infrared scanner (Li-Cor Biosciences,
Lincoln, NE, USA). GAPDH was used as the loading control.
siRNA transfection
Pyk2 siRNA was synthesized by GenePharma (Shanghai,
China), with a sequence of 5′-GCTTCGAGAGCAACAGCTT-3′. A
non-silencing siRNA oligonucleotide that does not recognize any
known mammalian gene homolog (GenePharma) was used as a negative
control. We selected ACHN cells to generate Pyk2 knockdown cells,
which we named si-Pyk2 cells; control cells were named si-NC cells.
ACHN cells were transfected with Pyk2 siRNA (50 nmol/l) or control
siRNA (50 nmol/l) via Lipofectamine 2000 transfection reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions.
Lentiviral vectors and infection
The lentivirus encoding Pyk2 plasmids was packaged
and purified at Hanbio Biotechnology (Shanghai, China), and cells
were infected following the manufacturer's instructions. We
selected A498 cells to generate stable Pyk2 overexpression cells,
which we named A498-Pyk2 cells; control cells were named A498-Ctrl
cells.
Flow cytometric analysis
Cell apoptosis was quantified using flow cytometric
analysis (BD Biosciences, San Jose, CA, USA). For apoptosis
experiments, RCC cells were collected and washed twice with
ice-cold PBS and re-suspended in 200 µl of binding buffer.
FITC-conjugated Annexin V was added at a final concentration of 0.5
µg/ml and incubated for 20 min at room temperature in the dark;
then, 1 µg/ml propidium iodide (PI) was added. Samples were
immediately analyzed by flow cytometry.
Wound-healing migration assay
RCC cells were seeded at 5×105 cells/well
in 6-well plates and cultured until the cells were confluent. The
cell monolayer was scraped in a straight line using a 10-µl pipette
tip and was washed with PBS twice, and the medium was replaced with
serum-free medium. To evaluate cell migration, images were captured
at 0, 12, 24 and 48 h following the initial scratch.
Transwell assays
Polycarbonate membranes with a pore size of 8 µm and
24-well culture insert plates (EMD Millipore, Billerica, MA, USA)
were used for transwell assays. First, the insert plates were
equilibrated with 0.5 ml of serum-free culture medium for 1 h at
37°C in 5% CO2. Then, the medium in the lower chambers
was replaced with 0.5 ml of culture medium supplemented with 10%
FBS. Serum pre-starved RCC cells (5×104) in 400 µl of
serum-free medium were seeded into the upper chambers. After a 48-h
incubation period, the inserts were rinsed with PBS, and cells on
the upper surface of the membrane were scraped off. Cells on the
bottom side of the membrane were stained with crystal violet stain
and counted by a microscope. Cells were counted from 8 randomly
chosen fields (magnification, ×200).
Cell counting kit 8 (CCK8) assay
RCC cells were cultured for 12, 24, 36, 48 and 60 h.
Wells with only culture medium added served as blanks. At different
time points, the supernatant was removed, and 100 µl of culture
medium containing 10 µl of CCK8 reagent (Dojindo Molecular
Technologies, Inc., Kumamoto, Japan) was added to each well for
another 2-h incubation at 37°C. Absorbance was recorded at 450 nm
using a microplate reader (Varioskan Flash; Thermo Fisher
Scientific, Inc.). Viability (%) was calculated based on optical
density (OD) values as follows: (OD of time sample-blank)/(OD of
control sample-blank) ×100. All experiments were independently
repeated in triplicate on separate occasions.
Statistical analysis
Data are expressed as means ± sd. of three
independent experiments. All statistical analyses were performed
using SPSS version 17.0 software (Abbott Laboratories, Chicago, IL,
USA). For comparisons, Student's t-test (two-tailed), Dunnet-t
test, Analysis of Variance (ANOVA), Rank-sum test, Fisher's exact
test, Pearson correlation analysis were performed as appropriate.
Kaplan-Meier survival analysis was utilized to compare RCC patient
survival based on dichotomized Pyk2 expression by log-rank test. A
P<0.05 was considered to indicate a statistically significant
difference.
Results
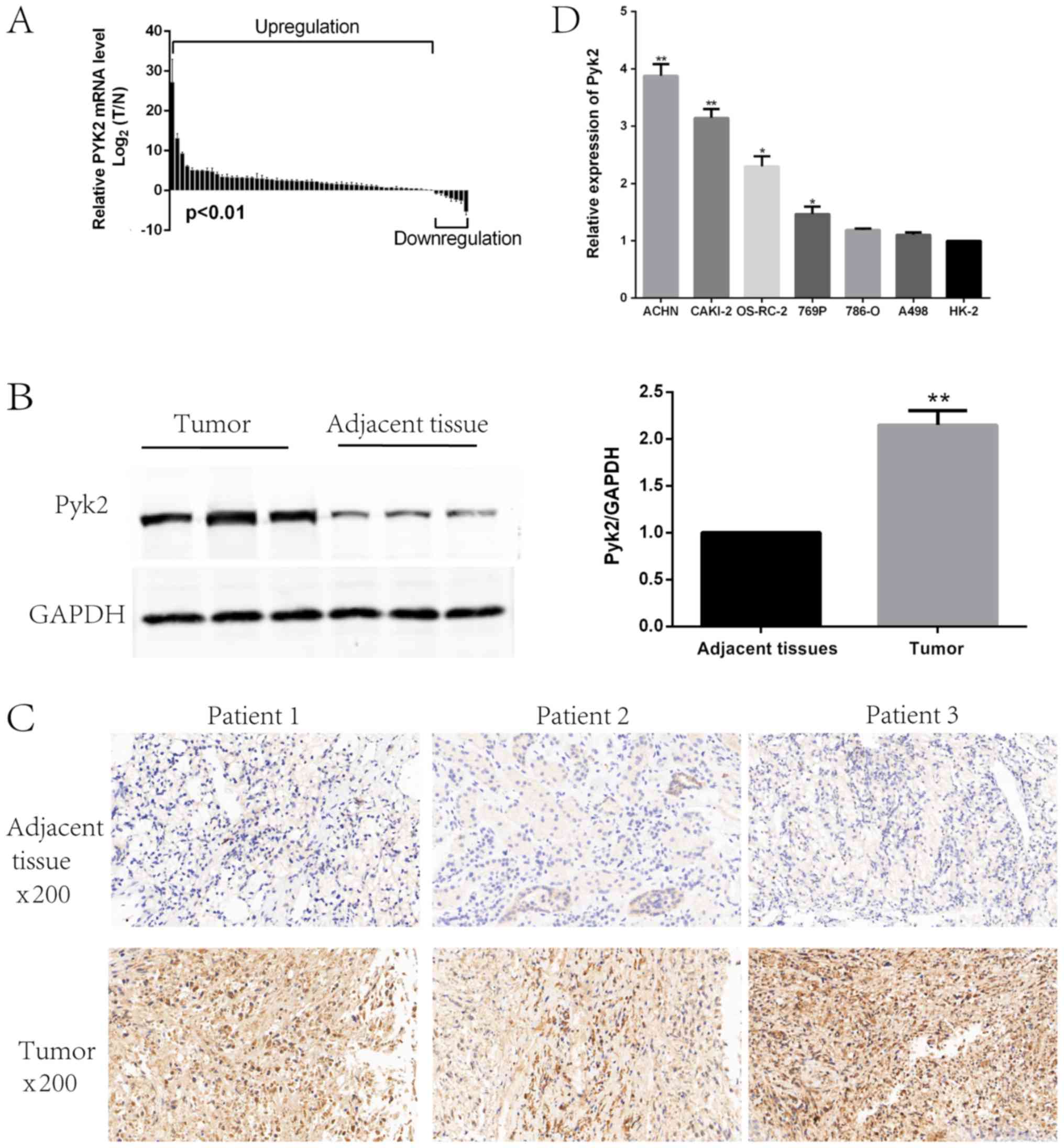
Pyk2 was highly expressed in RCC
We examined Pyk2 mRNA and protein expression levels
in RCC tissues and paired adjacent NT tissues. Pyk2 expression was
significantly upregulated in RCC tissues compared with paired
adjacent NT tissues (Fig. 1A and B).
Using IHC analysis of RCC tissues and paired adjacent NT tissues,
we confirmed Pyk2 upregulation at the protein level in RCC patients
(Fig. 1C). To explore the biological
functions of Pyk2 in RCC in vitro, we detected Pyk2
expression levels in several RCC cell lines (Fig. 1D).
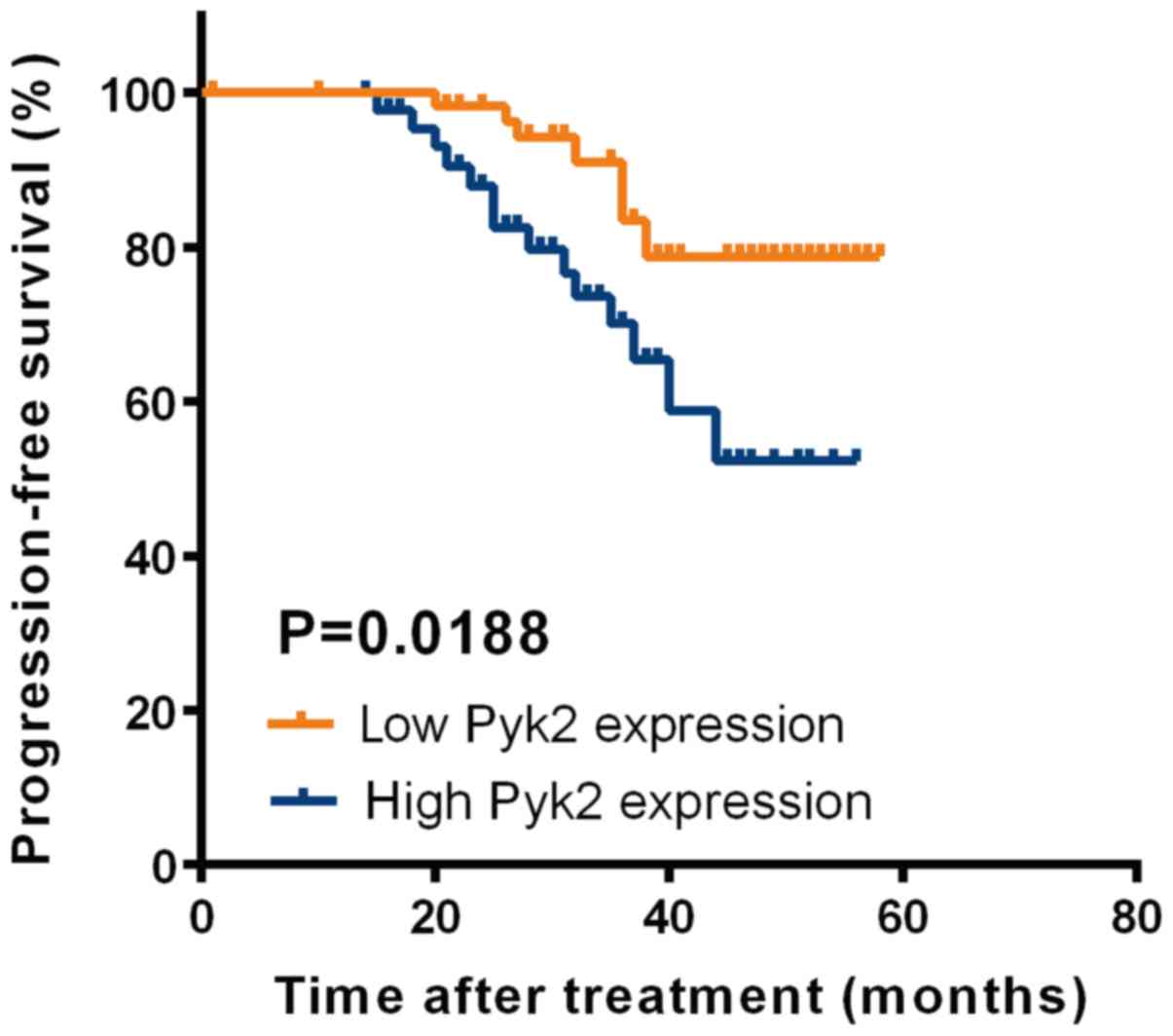
Pyk2 upregulation served as a
prognostic factor for patients with RCC
To determine the prognostic value of Pyk2 in RCC, we
generated Kaplan-Meier survival curves and performed log-rank tests
in qRT-PCR cohorts. The median expression level was used as the
cutoff. Remarkably, we found that progression-free survival (PFS)
was significantly shorter in patients with increased Pyk2 mRNA
levels than in those with reduced Pyk2 mRNA levels (Fig. 2). Collectively, our findings indicated
that Pyk2 can be used as a prognosis predictor in RCC patients.
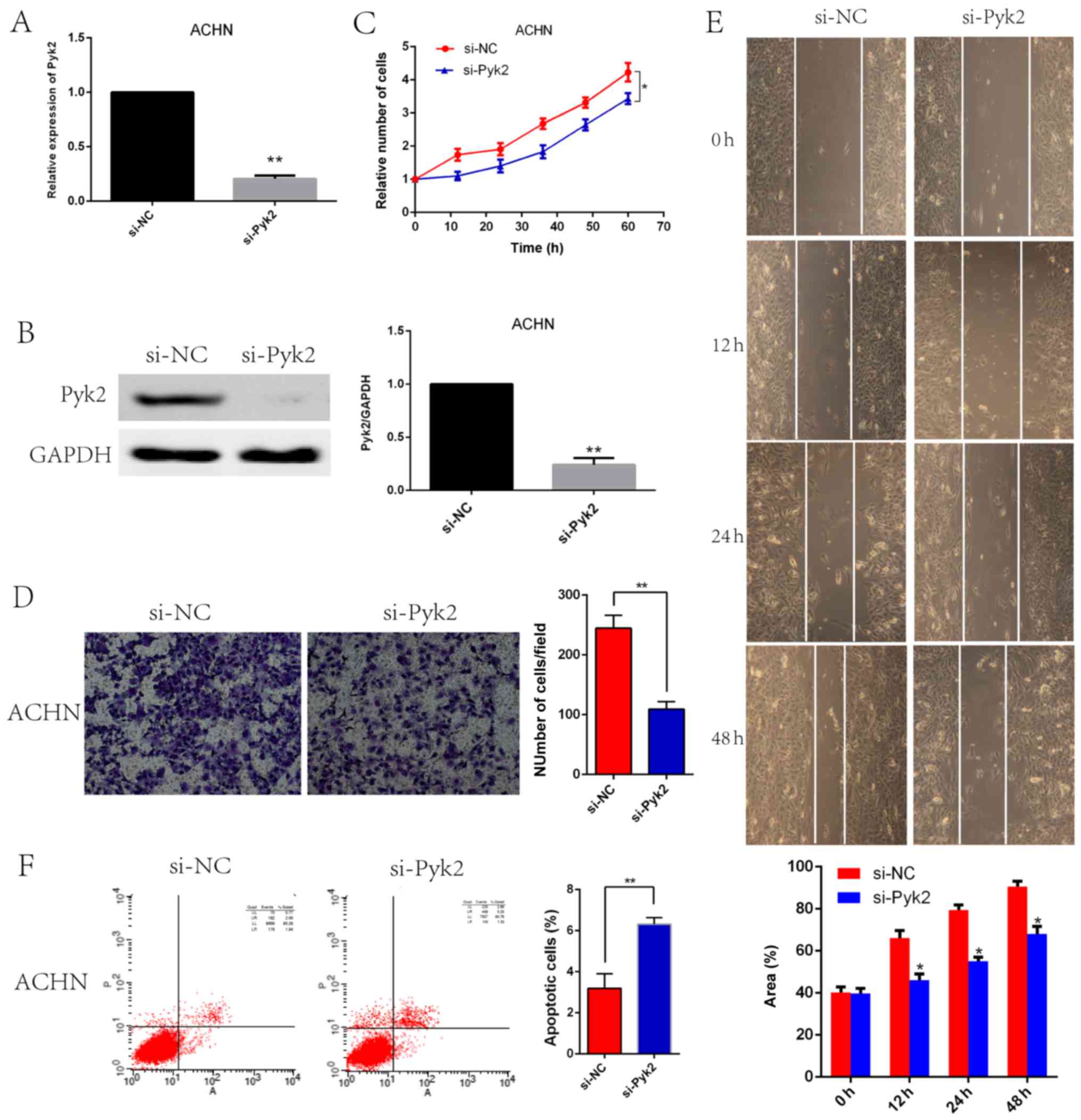
Pyk2 knockdown led to reduced
viability, invasive ability, and migratory ability and increased
apoptosis in vitro
Because Pyk2 is highly expressed in the ACHN cell
line, we selected this cell line to generate Pyk2 knockdown cells.
Pyk2 knockdown efficiency was confirmed by qRT-PCR (Fig. 3A) and western blotting (Fig. 3B). CCK8 assays revealed that si-Pyk2
cells presented with a significant reduction in cell proliferation
compared with si-NC cells (Fig. 3C).
Transwell assays also revealed that compared with si-NC cells,
si-Pyk2 cells displayed decreased migration and invasion (Fig. 3D). In wound-healing migration assays,
microscopic examination at 0, 12, 24 and 48 h revealed that
compared with si-NC cell migration, si-Pyk2 cell migration was
significantly reduced (Fig. 3E). Flow
cytometric analysis revealed that apoptosis is increased in si-Pyk2
cells compared with si-NC cells (Fig.
3F).
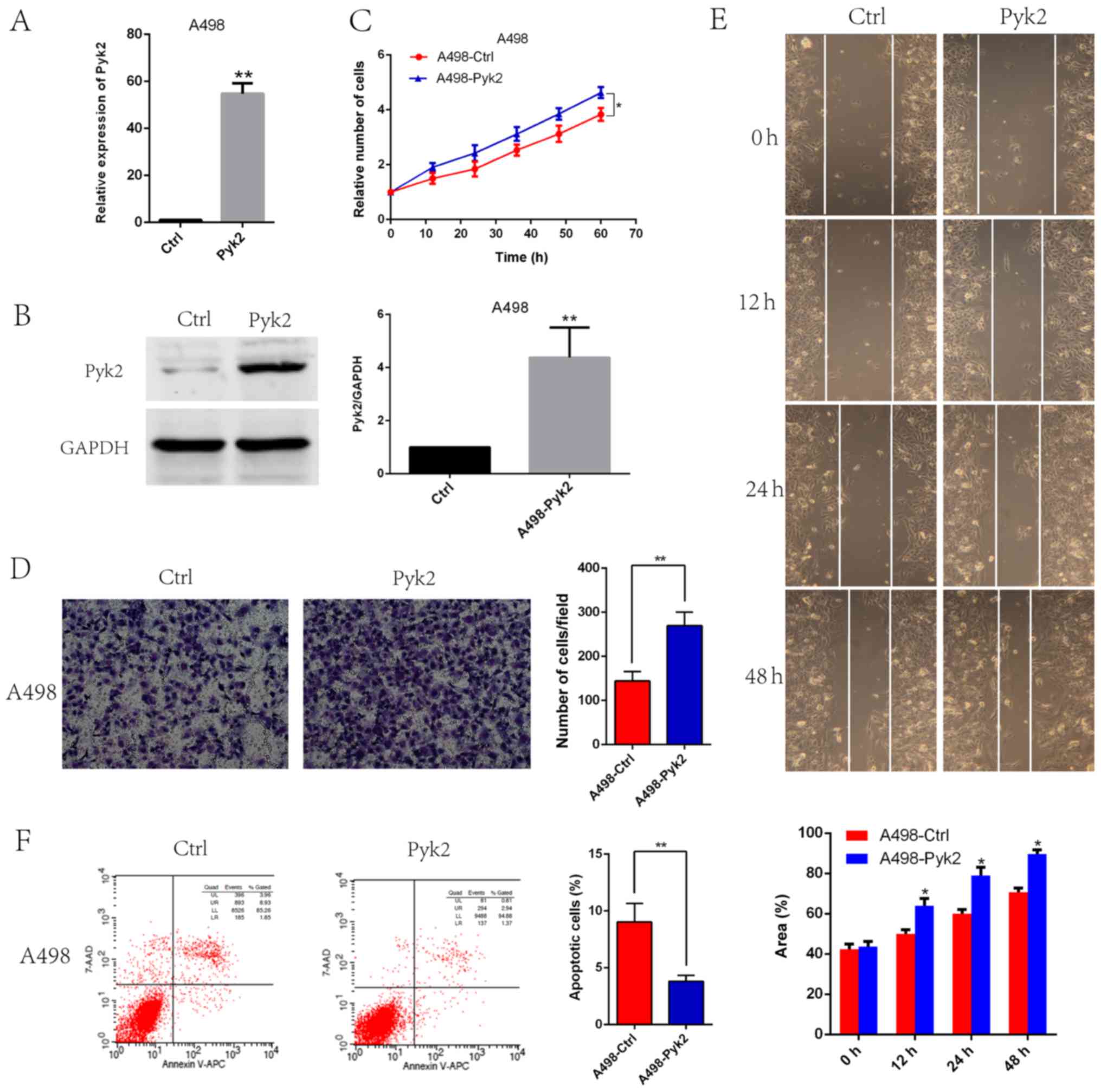
Pyk2 overexpression promoted tumor
cell proliferation, invasion and migration and reduced
apoptosis
Because of the low expression of Pyk2 in the A498
cell line, we selected this cell line to generate stable Pyk2
overexpression cells. Pyk2 overexpression efficiency was confirmed
by qRT-PCR (Fig. 4A) and western
blotting (Fig. 4B). CCK8 assays
revealed that A498-Pyk2 cells exhibited increased viability
(Fig. 4C). We also found that Pyk2
overexpression enhanced A498-Pyk2 cell invasion (Fig. 4D) and migration (Fig. 4E). Flow cytometry data revealed that
apoptosis was reduced in A498-Pyk2 cells compared with A498-Ctrl
cells (Fig. 4F).
Discussion
Partial nephrectomy is the recommended standard
treatment for localized RCC (20);
however, cancer metastasis is a serious problem in clinical
treatment, warranting a significant change in therapeutic
strategies and predicting poor outcomes in RCC (3,21). Once a
tumor has metastasized, the mortality burden faced by RCC patients
is significant (22). Thus, the
ability to determine which patients at high risk of developing
metastasis may benefit from radical nephrectomy and adjuvant
treatment is urgently needed. Targeted therapy is the main
treatment for metastatic RCC patients. Unfortunately, because of
acquired resistance and other drawbacks, both therapies for
metastatic RCC have limited efficacy and remain unsatisfactory in
respect to patient outcomes (23–25).
Therefore, gaining a better understanding of the molecular
mechanisms that underlie RCC metastasis may enable researchers to
identify reliable biomarkers to clinically diagnose affected
patients, predict the prognosis of these patients and target
therapies to treat these patients.
Many reports indicate that the expression of Pyk2, a
non-receptor kinase of the FAK family, is associated with the
prognosis of many tumors, such as colon cancer and gastric cancer.
We sought to determine whether Pyk2 is linked with RCC and detected
the expression of Pyk2 in tumor and NT tissues from RCC patients.
We discovered that the expression of Pyk2 was upregulated in tumor
tissues. We also found that the high expression of Pyk2 was
associated with poor prognosis in RCC. These results illustrated
that Pyk2 may play a role in RCC.
We detected Pyk2 expression level in several RCC
cell lines and HK2 cells and found the highest level in ACHN cells
and the lowest level in A498 cells.
We subsequently found that knocking down Pyk2 in
ACHN cells by siRNA can reduce cell invasion and metastatic
abilities, whereas Pyk2 overexpression in A498 cells can
significantly increase RCC invasion and metastatic abilities in
vitro. These results suggest that Pyk2 may be a potential
therapeutic target in RCC metastasis.
In summary, our study demonstrated that Pyk2 plays a
critical role in promoting cell proliferation and metastasis in RCC
and may serve as an independent predictor of clinical outcomes in
RCC patients. Based on these findings, targeting Pyk2 may represent
a potential therapeutic strategy to curb RCC progression. In the
future, we will explore the molecular mechanism by which Pyk2
promotes RCC progression and metastasis.
Acknowledgements
The authors would like to thank Professor Fu Yang
(Second Military Medical University) for providing valuable
suggestions on the writing of the original manuscript.
Funding
The present study was supported by the National
Nature Science Foundation of China (grant no. 81572521).
Availability of data and materials
The datasets generated/analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TZ, YB and XL performed the experiments, analyzed
the data and wrote the initial draft. TZ, YB and YH designed the
study and revised the manuscript. TZ, XG, JW and BL performed the
follow-up work of patients and created the figures. LW designed
this experiment, provided clinical specimens and clinical data of
renal cell carcinoma, participated in the writing and revision of
the paper and made great contributions to the article. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Changzheng Hospital, Second Military Medical
University and written informed consent was obtained from all
participants.
Patient consent for publication
The study participants provided consent for the data
and any associated images to be published.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Busch J, Ralla B, Jung M, Wotschofsky Z,
Trujillo-Arribas E, Schwabe P, Kilic E, Fendler A and Jung K:
Piwi-interacting RNAs as novel prognostic markers in clear cell
renal cell carcinomas. J Exp Clin Cancer Res. 34:612015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rini BI, Campbell SC and Escudier B: Renal
cell carcinoma. Lancet. 373:1119–1132. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ljungberga B, Cowan NC, Hanbury DC, Hora
M, Kuczyk MA, Merseburger AS, Patard JJ, Mulders PF and Sinescu IC:
European Association of Urology Guideline Group: EAU guidelines on
renal cell carcinoma: The 2010 update. Eur Urol. 58:398–406. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sekar RR, De La Calle CM, Patil D, Holzman
SA, Baum Y, Sheikh U, Huang JH, Osunkoya AO, Pollack BP, Kissick
HT, et al: Major histocompatibility complex I upregulation in clear
cell renal cell carcinoma is associated with increased survival.
Asian J Urol. 3:75–81. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gong J, Maia MC, Dizman N, Govindarajan A
and Pal SK: Metastasis in renal cell carcinoma: Biology and
implications for therapy. Asian J Urol. 3:286–292. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zisman A, Pantuck AJ, Wieder J, Chao DH,
Dorey F, Said JW, deKernion JB, Figlin RA and Belldegrun AS: Risk
group assessment and clinical outcome algorithm to predict the
natural history of patients with surgically resected renal cell
carcinoma. J Clin Oncol. 20:4559–4566. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang B, Yin C, Li H, Shi L, Liu N, Sun Y,
Lu S, Liu Y, Sun L, Li X, et al: Nir1 promotes invasion of breast
cancer cells by binding to chemokine (C-C motif) ligand 18 through
the PI3K/Akt/GSK3β/Snail signalling pathway. Eur J Cancer.
49:3900–3913. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang Q, Tang Y, Yu H, Yin Q, Li M, Shi L,
Zhang W, Li D and Li L: CCL18 from tumor-cells promotes epithelial
ovarian cancer metastasis via mTOR signaling pathway. Mol Carcinog.
55:1688–1699. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Okigaki M, Davis C, Falasca M, Harroch S,
Felsenfeld DP, Sheetz MP and Schlessinger J: Pyk2 regulates
multiple signaling events crucial for macrophage morphology and
migration. Proc Natl Acad Sci USA. 100:10740–10745. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sun CK, Man K, Ng KT, Ho JW, Lim ZX, Cheng
Q, Lo CM, Poon RT and Fan ST: Proline-rich tyrosine kinase 2 (Pyk2)
promotes proliferation and invasiveness of hepatocellular carcinoma
cells through c-Src/ERK activation. Carcinogenesis. 29:2096–2105.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang Y, Moschetta M, Huynh D, Tai YT,
Zhang Y, Zhang W, Mishima Y, Ring JE, Tam WF, Xu Q, et al: Pyk2
promotes tumor progression in multiple myeloma. Blood.
124:2675–2686. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lev S, Hernandez J, Martinez R, Chen A,
Plowman G and Schlessinger J: Identification of a novel family of
targets of PYK2 related to Drosophila retinal degeneration B (rdgB)
protein. Mol Cell Biol. 19:2278–2288. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun CK, Ng KT, Lim ZX, Cheng Q, Lo CM,
Poon RT, Man K, Wong N and Fan ST: Proline-rich tyrosine kinase 2
(Pyk2) promotes cell motility of hepatocellular carcinoma through
induction of epithelial to mesenchymal transition. PLoS One.
6:e188782011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Naylor TL, Greshock J, Wang Y, Colligon T,
Yu QC, Clemmer V, Zaks TZ and Weber BL: High resolution genomic
analysis of sporadic breast cancer using array-based comparative
genomic hybridization. Breast Cancer Res. 7:R1186–R1198. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wendt MK, Schiemann BJ, Parvani JG, Lee
YH, Kang Y and Schiemann WP: TGF-β stimulates Pyk2 expression as
part of an epithelial-mesenchymal transition program required for
metastatic outgrowth of breast cancer. Oncogene. 32:2005–2015.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Selitrennik M and Lev S: PYK2 integrates
growth factor and cytokine receptors signaling and potentiates
breast cancer invasion via a positive feedback loop. Oncotarget.
6:22214–22226. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kuang BH, Zhang MQ, Xu LH, Hu LJ, Wang HB,
Zhao WF, Du Y and Zhang X: Proline-rich tyrosine kinase 2 and its
phosphorylated form pY881 are novel prognostic markers for
non-small-cell lung cancer progression and patients' overall
survival. Br J Cancer. 109:1252–1263. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li Z, Yuan X, Wu Z, Guo Z, Jiang P and Wen
Z: Expressions of FAK and Pyk2 in human astrocytic tumors and their
relationship with angiogenesis. Chin-Ger J Clin Oncol. 7:658–660.
2008. View Article : Google Scholar
|
|
19
|
Yue Y, Li ZN, Fang QG, Zhang X, Yang LL,
Sun CF and Liu FY: The role of Pyk2 in the CCR7-mediated regulation
of metastasis and viability in squamous cell carcinoma of the head
and neck cells in vivo and in vitro. Oncol Rep. 34:3280–3287. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Campbell SC, Novick AC, Belldegrun A,
Blute ML, Chow GK, Derweesh IH, Faraday MM, Kaouk JH, Leveillee RJ,
Matin SF, et al: Guideline for management of the clinical T1 renal
mass. J Urol. 182:1271–1279. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cohen HT and McGovern FJ: Renal-cell
carcinoma. N Engl J Med. 353:2477–2490. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Taneja SS: Re: Validation of the 2009 TNM
version in a large multi-institutional cohort of patients treated
for renal cell carcinoma: Are further improvements needed? J Urol.
185:12232011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gupta K, Miller JD, Li JZ, Russell MW and
Charbonneau C: Epidemiologic and socioeconomic burden of metastatic
renal cell carcinoma (mRCC): A literature review. Cancer Treat Rev.
34:193–205. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Motzer RJ, Bacik J, Schwartz LH, Reuter V,
Russo P, Marion S and Mazumdar M: Prognostic factors for survival
in previously treated patients with metastatic renal cell
carcinoma. J Clin Oncol. 22:454–463. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Weinstock M and McDermott D: Targeting
PD-1/PD-L1 in the treatment of metastatic renal cell carcinoma.
Ther Adv Urol. 7:365–377. 2015. View Article : Google Scholar : PubMed/NCBI
|