Introduction
Jumonji C domain-containing 1A (JMJD1A), also called
lysine demethylase 3A (KDM3A), is an enzyme that converts
dimethylated lysine 9 on histone H3 progressively into its mono-
and unmethylated form (1). Depending
on the degree of this demethylation process and where throughout a
gene body it occurs, this affects gene expression in different ways
(2). Accordingly, in different
contexts, JMJD1A was shown to activate or repress gene
transcription (1,3–7). Further,
JMJD1A may act independently of its enzymatic activity through
binding to nucleosome remodeling complexes, thereby modulating
long-range chromatin interactions (8).
Knockout of JMJD1A in mice has revealed multiple
physiological functions of this histone demethylase. Complete
JMJD1A knockout caused male-to-female sex reversal, most likely due
to deficient transcription of the sex-determining region Y gene
that is required to trigger the differentiation of the bipotential
gonads into testes (9). Moreover, in
a hypomorphic JMJD1A knockout mouse model, males were infertile and
displayed defective spermatogenesis, indicating that JMJD1A plays
an important role in adult testes, too (10). Unrelated to its reproductive role,
JMJD1A is also critical for normal homeostasis because JMJD1A
knockout mice became obese and developed metabolic syndrome
(11,12). In addition, JMJD1A is required for the
adaptation to cold stress by promoting thermogenesis (8,13).
Interestingly, the stem cell factor OCT4 appears to
upregulate JMJD1A gene transcription and depletion of JMJD1A can
lead to the differentiation of embryonic stem cells (14), suggesting that JMJD1A exerts more,
yet-to-be-discovered tasks in development, wound repair or tissue
regeneration that are all dependent on stem cell function. In
addition, JMJD1A promoted stemness in breast and ovarian cancer
cells that could enhance chemoresistance (15,16). In
HCT116 colon cancer cells, JMJD1A was needed for efficient tumor
growth in a xenograft model (17,18),
possibly because of its ability to stimulate stem cells (19). Despite these findings, the role of
JMJD1A in colorectal cancer that is the second leading cause of
cancer death in the Western hemisphere (20) has not been fully elucidated. In the
present study, we set out to analyze JMJD1A expression in
colorectal tumors, gain mechanistic insights by determining genes
that are regulated by JMJD1A, and explore the role of one of these
genes, α-thalassemia/mental retardation syndrome X-linked (ATRX),
in colon cancer.
Materials and methods
Analysis of databases
Microarray experiments were analyzed with Oncomine
(21) and respective data downloaded
from www.oncomine.org. The Human Protein
Atlas (22) served as a source for
survival data (www.proteinatlas.org) with the corresponding RNA
sequencing data originating from ‘The Cancer Genome Atlas’ (TCGA).
To assess coexpression of ATRX and JMJD1A in colorectal
adenocarcinomas, provisional TCGA RNA sequencing data were analyzed
with cbioportal (www.bioportal.org).
Cloning of shRNA
The retroviral vector pSIREN-RetroQ (Clontech, Palo
Alto, CA, USA) was linearized with the restriction enzymes
BamHI and EcoRI and ligated with double-stranded
oligonucleotides encoding shRNAs (23). Correct cloning was verified by DNA
sequencing. The control shRNA targets the sequence
5′-CAACAAGATGAAGAGCACCAA-3′, which displays at least four
mismatches to any known human gene. The human JMJD1A shRNAs target
the sequences 5′-GCAGGTGTCAATAGTGATA-3′ (#1 shRNA) and
5′-GTAGACCTAGTTAATTGTA-3′ (#2 shRNA), while the human ATRX shRNAs
target the sequences 5′-GGTGTTATGATCATAGGCTAT-3′ (#1 shRNA) and
5′-GGATTCAACCTCTTGAGGATA-3′ (#3 shRNA).
Cell culture and analyses
Human colorectal carcinoma cells HCT116 (CCL-247),
SW480 (CCL-228), DLD-1 (CCL-221) and HT-29 (HTB-38) as well as
human embryonic kidney 293T cells (CRL-3216; all American Type
Culture Collection, Manassas, VA, USA) were grown in a humidified,
5% CO2-containing atmosphere in Dulbecco's modified
Eagle's medium (10-013-CV; Mediatech; Corning Inc., Corning, NY,
USA) that was supplemented with 10% fetal bovine serum (S11150;
Atlanta Biologicals, Flowery Branch, GA, USA) as previously
described (24,25). Transfection of 293T cells was done by
the calcium phosphate coprecipitation method (26,27), the
precipitate washed off with phosphate-buffered saline (28), retrovirus collected from the
supernatant over the next 48 h (29)
and in some cases concentrated by precipitation with poly (ethylene
glycol)-8000 (30). HCT116 cells were
infected with retrovirus three times (31) and then selected with 1.5 µg/ml
puromycin for 3–4 days (32). To
measure growth, 2,000 or 2,500 cells were seeded in 96-wells and
growth determined essentially as described (33,34). For
clonogenic assays, 1,000 or 3,750 cells were seeded into 6-wells
and colony formation assayed as described (30).
Analysis of protein expression
To generate whole cell protein extracts, cells were
lysed in Laemmli sample buffer and boiled for ~10 min (35). For biochemical fractionation of cells,
the NE-PER nuclear and cytoplasmic extraction kit (78833; Pierce
Biotechnology, Rockford, IL, USA) was employed as described
(36). Proteins were then
electrophoretically separated on SDS polyacrylamide gels (37), transferred to polyvinylidene
difluoride membrane (38) and
incubated with primary antibodies as described (39,40).
Signals on blots were revealed utilizing appropriate secondary
antibodies (41) followed by
detection with chemiluminescence (42). Staining of a human colon cancer tissue
microarray (AccuMax A303 I, slide #40; Isu Abxis, Seongnam, South
Korea) was performed employing 20 min of antigen retrieval with
method 2 and a 1:100 dilution of anti-JMJD1A antibody as described
(43). The stained slide was
digitized and the digital image extracted with Aperio ImageScope
software (Leica, Wetzlar, Germany). Diaminobenzidine staining was
obtained by color deconvolution from this image, intensity of light
transmission was measured with Fiji version of ImageJ software
(http://fiji.sc) and the strength of JMJD1A staining
was defined as 100× (log maximum intensity-log intensity). The
following rabbit polyclonal antibodies were utilized: Anti-Actin
(A2066; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany); anti-ATRX
(NBP1-83077); anti-JMJD1A (NB100-77282; both Novus Biologicals,
Littleton, CO, USA); and anti-H3K27me1 (07–448; Upstate
Biotechnology, Lake Placid, NY, USA). Also used was a goat
polyclonal anti-Lamin B antibody (sc-6216; Santa Cruz
Biotechnology, Santa Cruz, CA, USA).
RNA sequencing
Total RNA from HCT116 cells was isolated employing
TRIzol (44) following standard
procedures (45). RNA was further
purified with the RNeasy Mini kit (74104; Qiagen, Hilden, Germany)
and sequenced in the Targeted DNA Methylation and Mitochondrial
Heteroplasmy Core at the Oklahoma Nathan Shock Center of Excellence
in the Biology of Aging (Oklahoma City, OK, USA). Reads were
aligned in Strand NGS (Agilent Technologies, Inc., Santa Clara, CA,
USA) against the hg38 human genome build (December 2013) with
Ensemble gene annotations (v87, January 2017). Three bases were
trimmed from 3′ and 5′ ends of reads and read quality <Q20 was
discarded prior to alignment. Alignment was to the full genome
build to detect novel genes and splice variants and required 90
percent identity, minimum read length of 25, and reads with
multiple matches were eliminated. A screening database for Illumina
adapter sequences was used to remove adapter sequences. Reads
counts were normalized by DESeq and a z-test was used to determine
differential expression between samples. Ingenuity Pathway Analysis
(Qiagen) was performed on genes whose mRNA levels were at least
1.5-fold different upon JMJD1A downregulation compared to the
control shRNA treated cells.
Luciferase assays
The human ATRX promoter spanning from −600 to +100
(the ATRX transcription start site was based on the human
transcript variant 1: NCBI reference sequence NM_000489.4) was
cloned into pGL2-Basic (Promega Corp., Madison, WI, USA). Human
293T and HCT116 cells, which were grown in 12-wells, were
transiently transfected with 100 ng of this luciferase reporter
construct, 900 ng pBluescript KS+, and 60 ng of
Flag-JMJD1A expression plasmid or empty vector pEV3S utilizing 2 µg
polyethylenimine (43). Approximately
42 h after transfection, cells were lysed as described (46) and luciferase activities determined in
a luminometer (47).
Chromatin immunoprecipitation
Human embryonic kidney 293T cells were grown in 10
cm dishes and transfected by the calcium phosphate coprecipitation
method with 3 µg ATRX luciferase reporter plasmid, 21 µg
pBluescript KS+, and 6 µg Flag-JMJD1A expression plasmid
or empty vector pEV3S. Cells were treated with formaldehyde and
processed for chromatin preparation and immunoprecipitation as
described (48,49). The following antibodies were used:
Normal mouse IgG (sc-2025; Santa Cruz Biotechnology);
anti-H3K9me2 mouse monoclonal antibody (ab1220); and
anti- H3K36me2 rabbit polyclonal antibody (ab9049; both
Abcam, Cambridge, MA, USA). Resultant DNA was then amplified by
PCR, which was performed with the GoTaq DNA polymerase kit (M3008;
Promega Corp.) and the following temperature program: 97°C for 2
min; 8 cycles of 97°C for 25 sec, 65°C (−1°C per cycle) for 25 sec,
72°C for 35 sec; 26 cycles of 97°C for 25 sec, 57°C for 25 sec,
72°C for 35 sec (+1 sec per cycle); 72°C for 4 min followed by
cooling down to 4°C. Primers used were ATRX-for2
(5′-GTAGGTTTGTCTACCTCAGAGAGTG-3′; spanning the ATRX promoter from
−332 to −308) and ATRX-rev2 (5′-ACAGCTCAAAGGCCGCTACCACTGC-3′;
spanning the ATRX promoter from +117 to +141). The PCR products
were electrophoresed in agarose gels and revealed by ethidium
bromide staining (50).
Statistics
Statistical significance was assessed with one- or
two-way analysis of variance (ANOVA) with post hoc Dunnett's or
Tukey's multiple comparisons test, an unpaired Student's t-test, or
a log-rank test. P<0.05 was deemed to show a statistically
significant difference.
Results
JMJD1A expression in colorectal
cancer
To examine the expression of JMJD1A mRNA in
colorectal tumors, we first interrogated publicly available
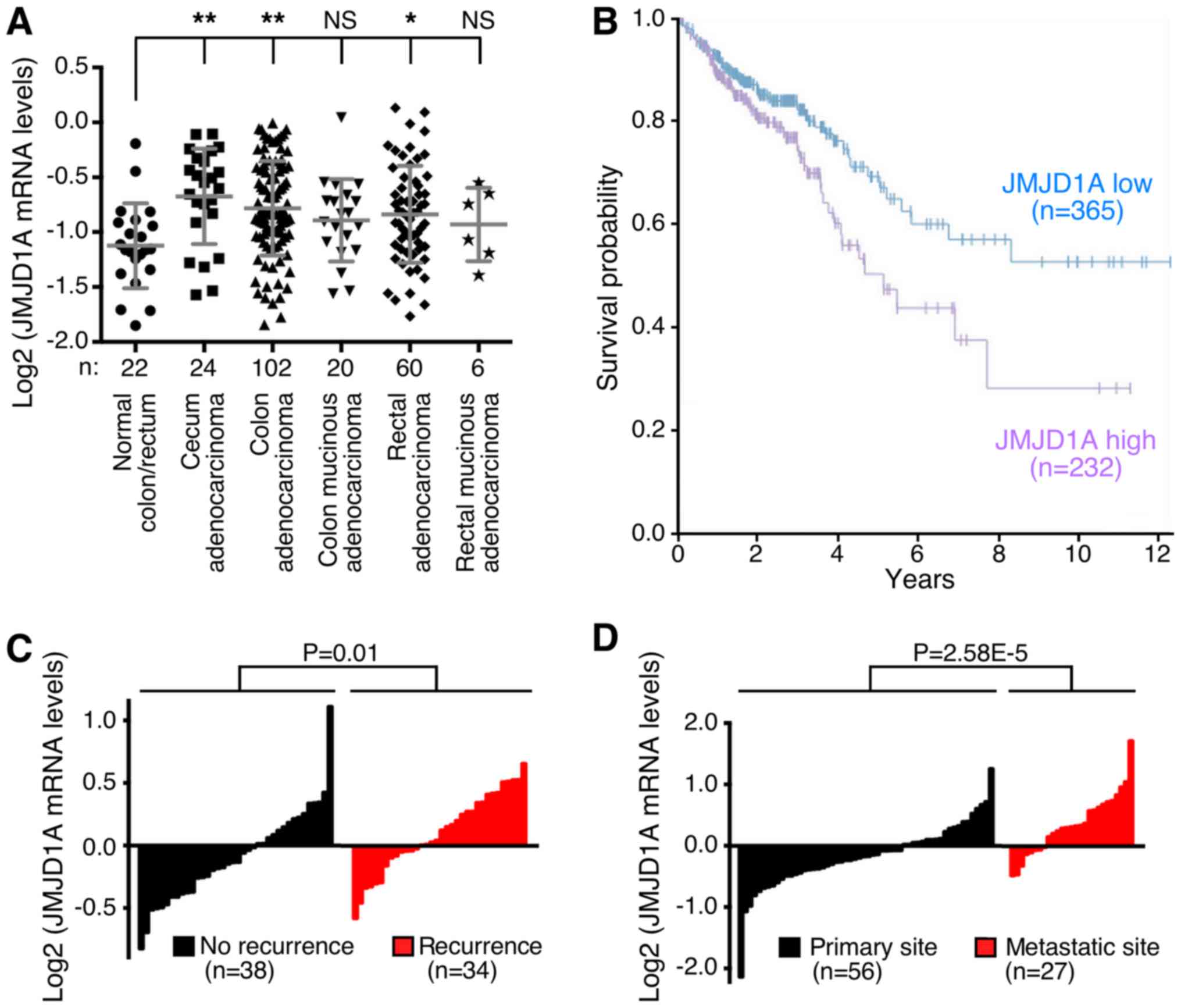
databases. In a study from TCGA (51), we found significant overexpression of
JMJD1A in cecum, colon and rectal adenocarcinomas and a trend
towards overexpression in mucinous tumors of the colon and rectum
(Fig. 1A). Importantly, high JMJD1A
mRNA levels were significantly correlated with reduced survival
(Fig. 1B). Further, we discovered
that JMJD1A was more highly expressed in patients with recurrent
disease [Fig. 1C; microarray data
retrieved from reference (52)] and
at metastatic sites compared to the primary colorectal tumors
[Fig. 1D; microarray data retrieved
from reference (53)]. Altogether,
these data indicate that JMJD1A mRNA is overexpressed in many
colorectal tumors and associated with a more aggressive
phenotype.
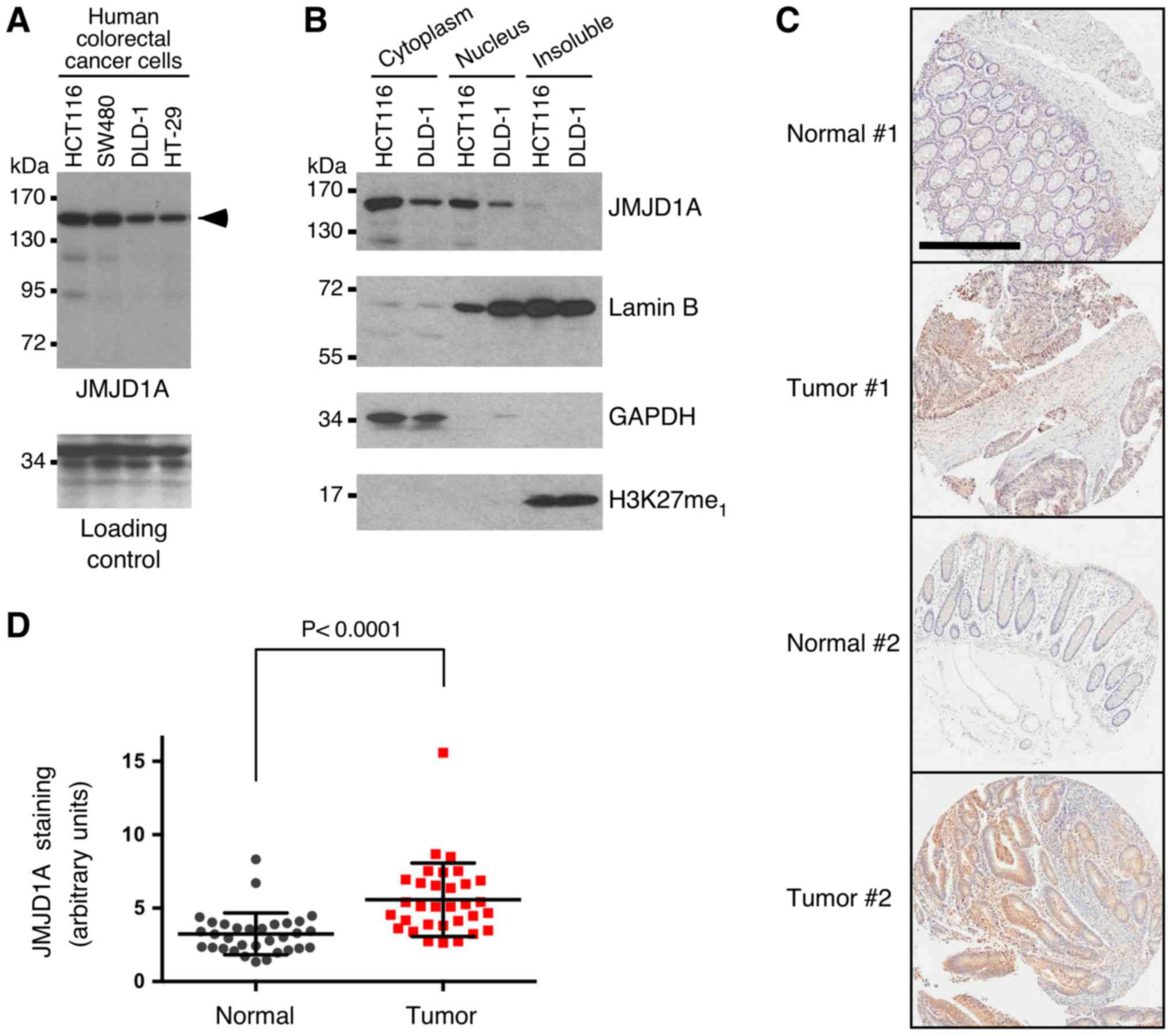
We then wanted to assess JMJD1A protein expression
in colorectal cancer. Expression of JMJD1A protein was confirmed in
all of the four tested human colorectal cancer cell lines by
western blotting (Fig. 2A). Further,
we biochemically fractionated HCT116 and DLD-1 colorectal cancer
cells and found that JMJD1A was present in both the cytoplasm and
nucleus, while very little JMJD1A was detectable in the insoluble
fraction that primarily consists of the nuclear matrix and
heterochromatin (Fig. 2B). Then, we
stained a human tissue microarray consisting of 32 matching normal
and cancerous colorectal tissues. Consistent with our biochemical
fractionation experiments, JMJD1A staining was present in both the
cytoplasm and nucleus. Importantly, a significant overexpression of
JMJD1A was observed in the tumors (Fig.
2C and D), further implicating that JMJD1A overexpression may
contribute to the development of colorectal cancer.
Impact of JMJD1A on HCT116 cells
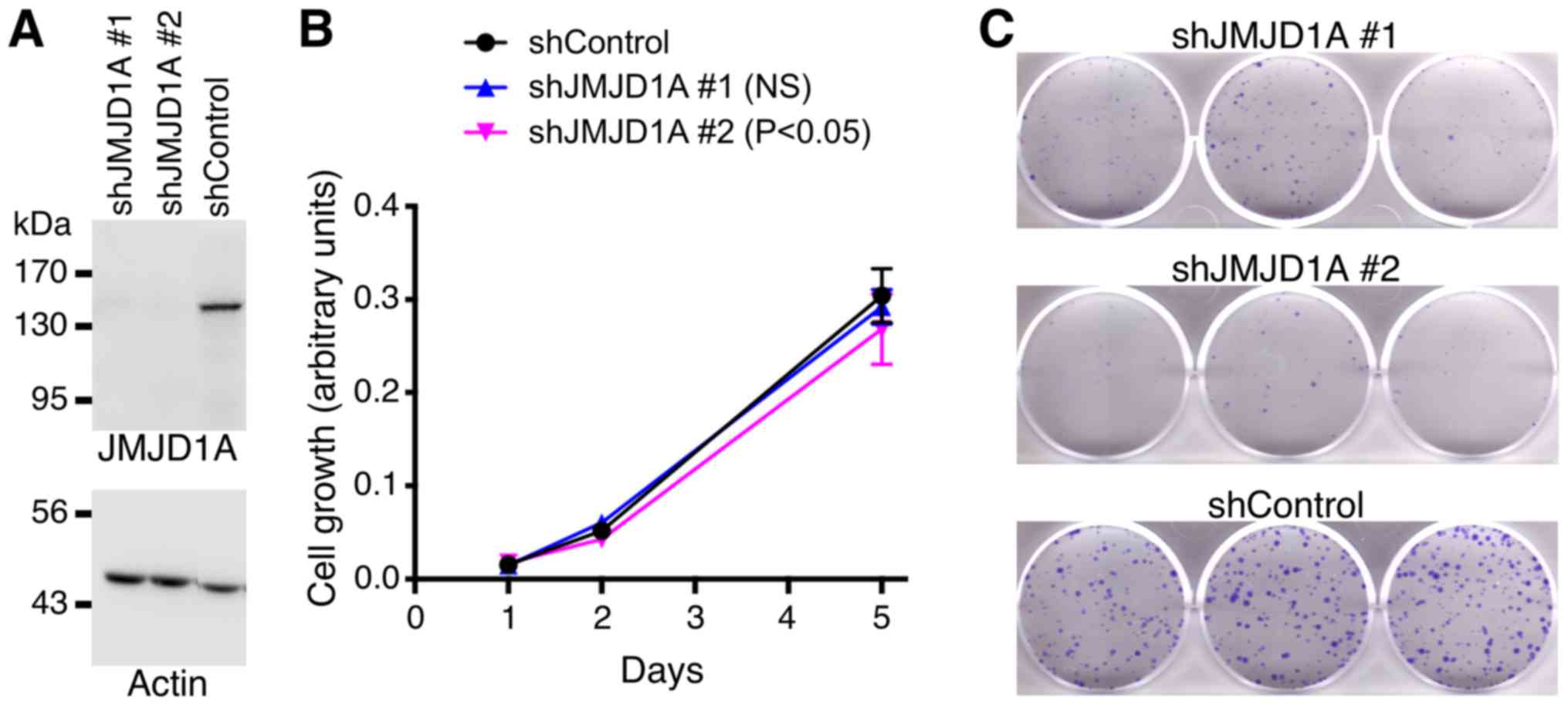
To assess a potential physiological role of JMJD1A,
we downregulated JMJD1A with two different shRNAs in HCT116
colorectal cancer cells. Both shRNAs induced a large reduction of
JMJD1A protein levels (Fig. 3A).
While JMJD1A shRNA#1 did not cause any change in HCT116 cell
growth, shRNA#2 displayed a significant, yet very small reduction
in cell growth (Fig. 3B). This
indicates that JMJD1A downregulation has only negligible effects on
HCT116 cell growth. In contrast, clonogenic activity of HCT116
colorectal cancer cells was robustly reduced upon JMJD1A
downregulation with either of the two shRNAs (Fig. 3C). The latter data indicate that
JMJD1A can influence the physiology of HCT116 cells in a manner
that is predicted to be tumor promoting.
Transcriptome analysis
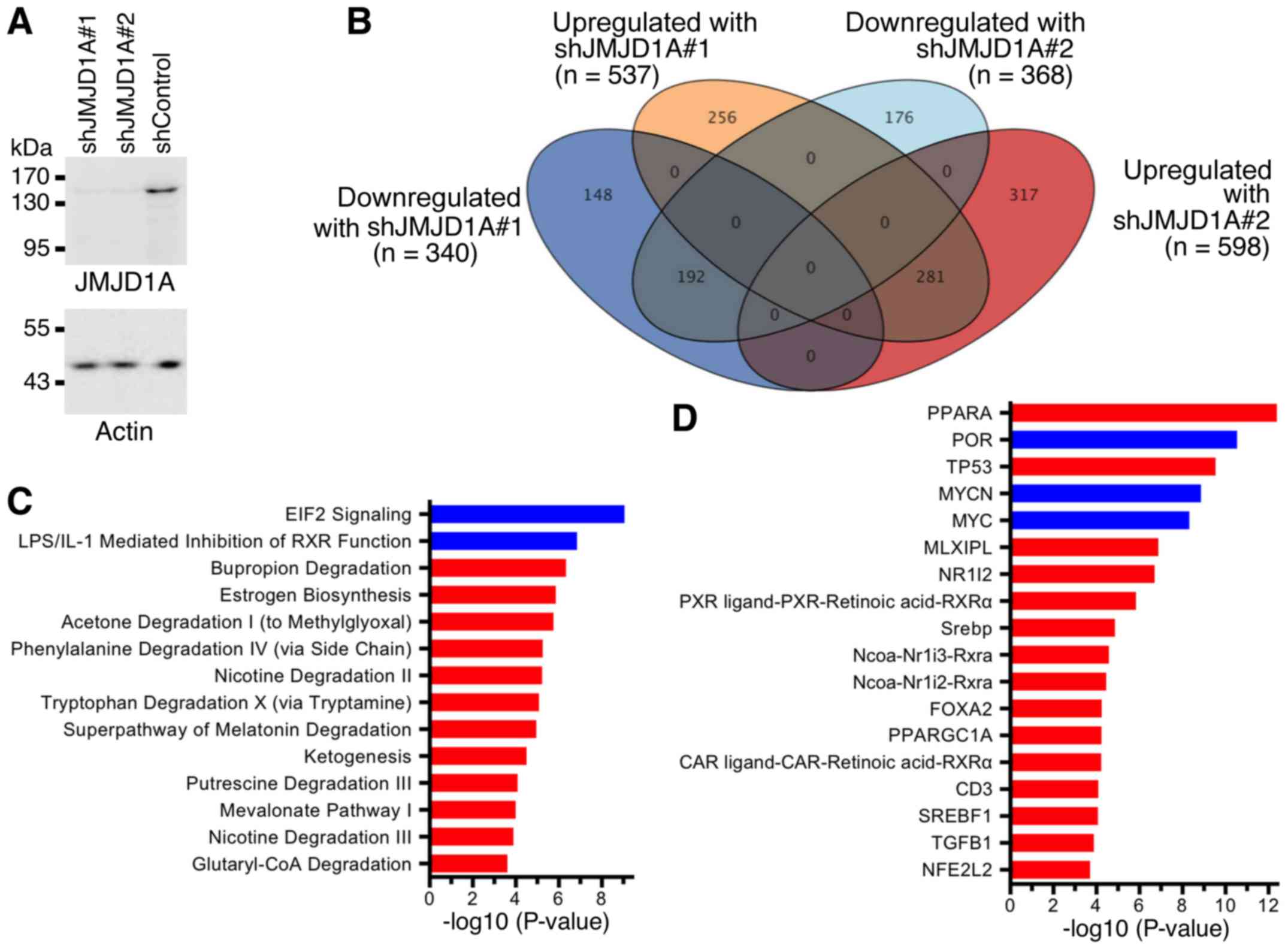
Next, we assessed how JMJD1A affects the
transcriptome of HCT116 cells. To this end, we again downregulated
JMJD1A with two different shRNAs (Fig.
4A) and performed RNA sequencing. Compared to control shRNA, we
found that 281 genes were >1.5-fold upregulated with both JMJD1A
shRNAs and 192 genes were >1.5-fold downregulated (Fig. 4B). Ingenuity Pathway Analysis revealed
that multiple metabolic (Fig. 4C) and
upstream regulatory pathways (Fig.
4D) were affected by JMJD1A downregulation. This indicates that
JMJD1A pleiotropically affects the gene expression program of
HCT116 colorectal cancer cells and may thereby be a determinant of
their oncogenic potential.
Identification of ATRX as a potential
JMJD1A target gene
Since JMJD1A as a histone demethylase modulates
chromatin structure, we were especially interested in other
proteins involved in chromatin regulation whose genes were found to
be affected by JMJD1A in our transcriptome analysis. One such gene
was ATRX, which encodes for a chromatin remodeler (54). Our RNA sequencing data showed that
JMJD1A downregulation by shRNA#1 or shRNA#2 led to a 2.6-fold or
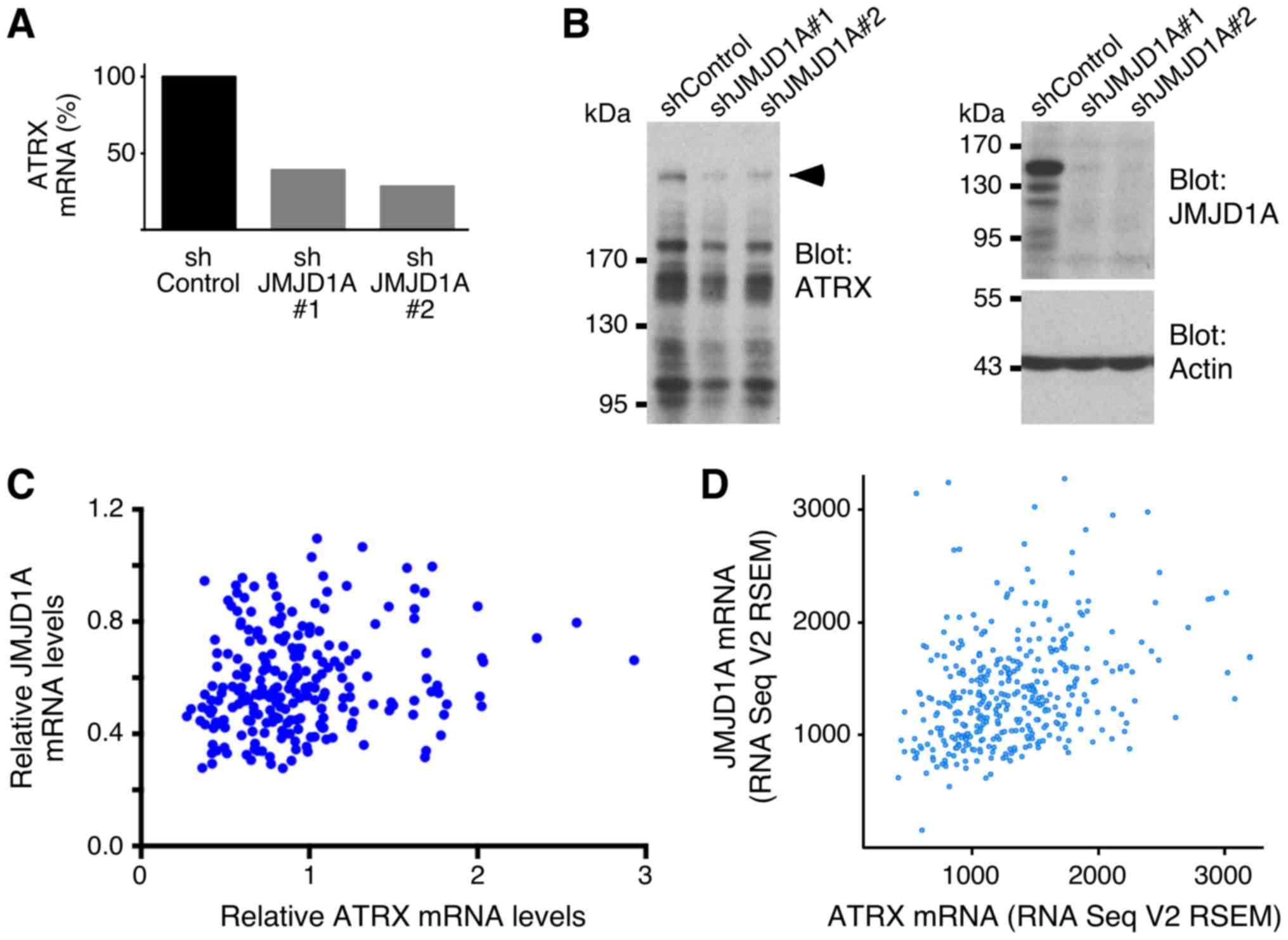
3.5-fold decrease in ATRX mRNA levels, respectively (Fig. 5A). Accordingly, ATRX protein levels
were also reduced in the presence of JMJD1A shRNAs (Fig. 5B). Please note that ATRX has a
calculated molecular weight of 282.6 kDa and that the ATRX gene is
composed of 35 exons, giving rise to multiple splice variants. This
complex structure, and possibly protein degradation, accounted for
the fact that multiple bands were detected with the anti-ATRX
antibody in our western blot analyses. In total, these data suggest
that JMJD1A can stimulate ATRX gene transcription. This notion is
strongly supported by the fact that JMJD1A and ATRX mRNA levels
were positively correlated across normal and malignant colorectal
tissue specimens (Fig. 5C) or across
adenocarcinomas in another data set (Fig.
5D).
To provide further evidence for a JMJD1A-ATRX axis,
we cloned the human ATRX gene promoter in front of a luciferase
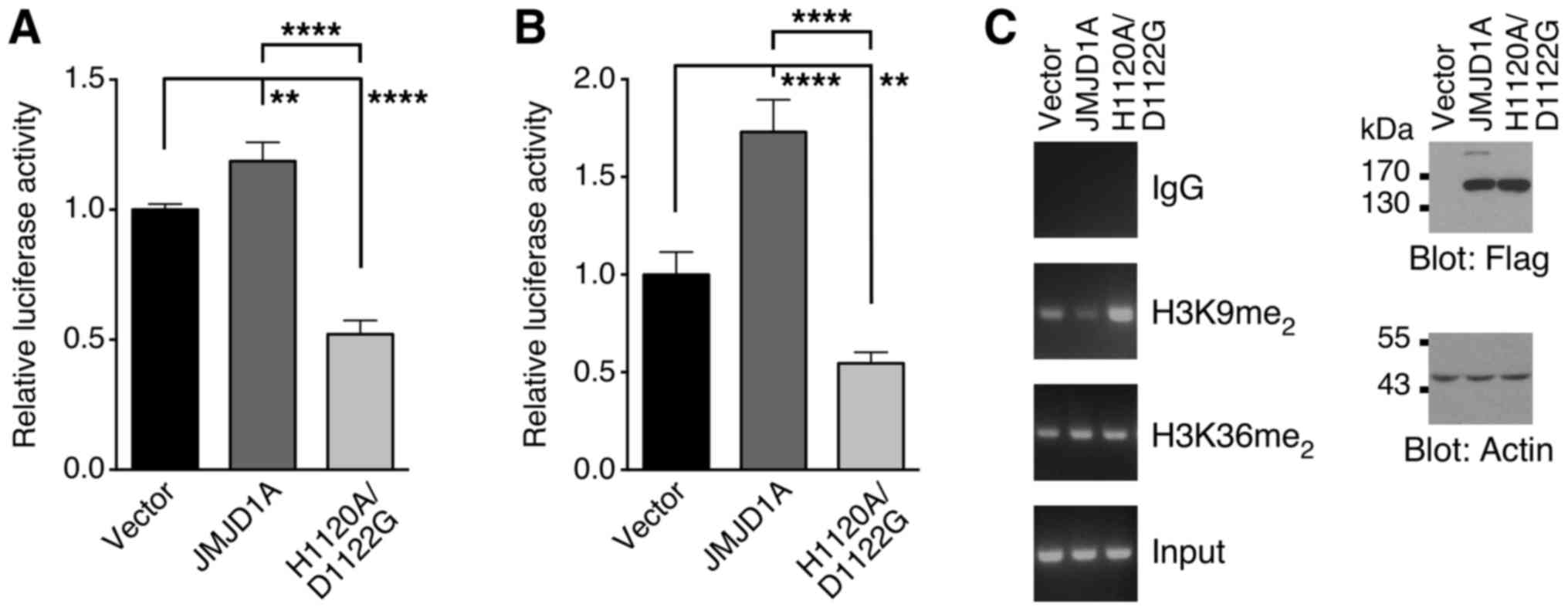
reporter gene. As shown in Fig. 6A,
overexpression of JMJD1A in 293T cells slightly stimulated the ATRX
promoter. In addition, we also overexpressed JMJD1A-H1120A/D1122G,
in which two amino acids within the JMJD1A catalytic center have
been mutated rendering it inactive (1). This mutant repressed the ATRX luciferase
reporter gene, presumably since it prevents endogenous JMJD1A from
interacting with and thereby activating the ATRX promoter. Similar
results were obtained with wild-type JMJD1A and its H1120A/D1122G
mutant in HCT116 colorectal cancer cells (Fig. 6B). Moreover, we found that JMJD1A
overexpression expectedly reduced dimethylation of histone H3 on
lysine 9, but not dimethylation on lysine 36 (Fig. 6C). On the other hand, the
H1120A/D1122G mutant predictably elevated levels of dimethylation
on lysine 9 of histone H3. Collectively, these data suggest that
JMJD1A interacts with the ATRX promoter and induces it by a
mechanism that involves reduction of H3K9me2 levels.
ATRX in colorectal cancer
The previous findings led to the question of whether
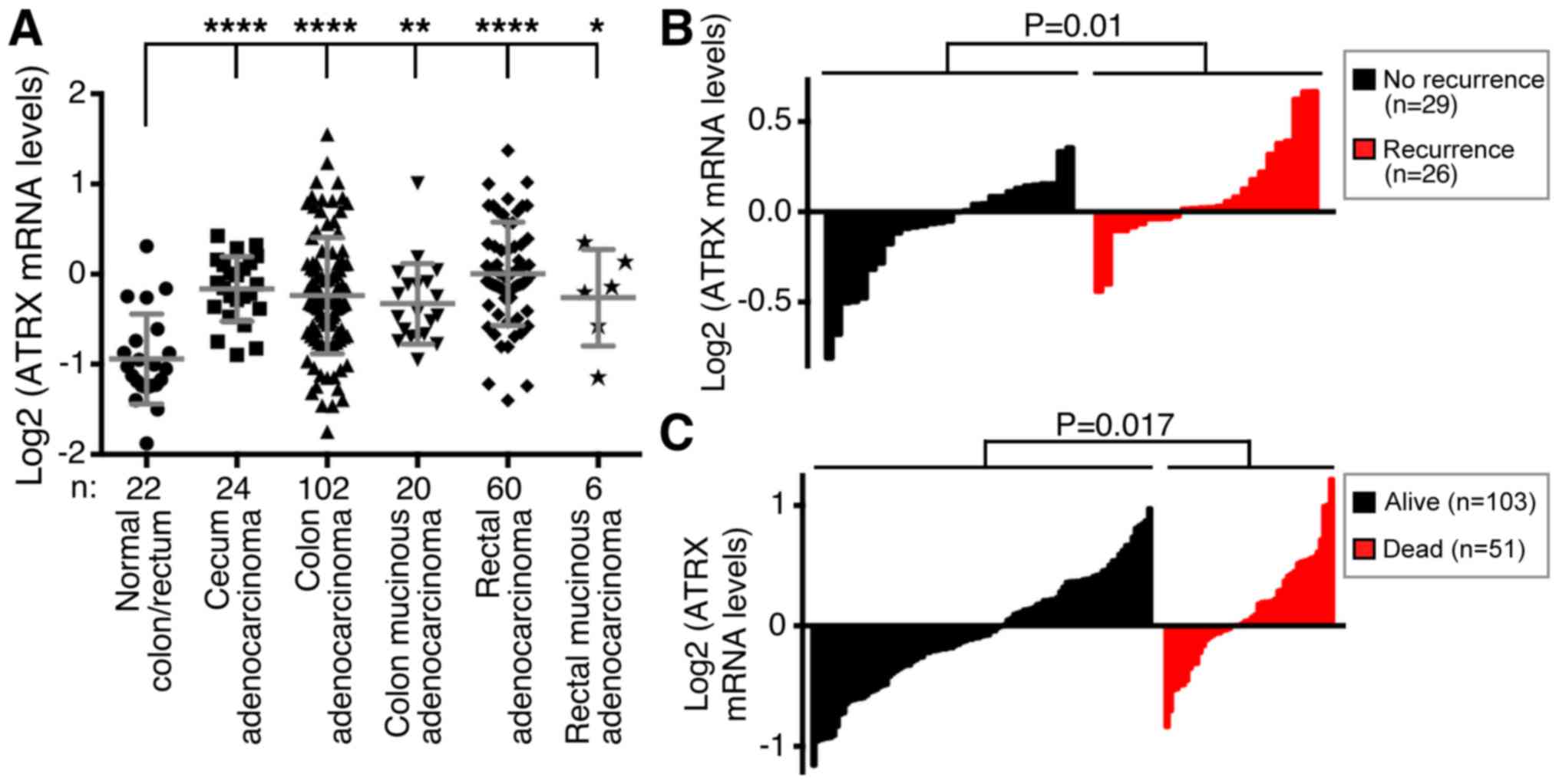
ATRX is overexpressed in colorectal cancer. Indeed, similar to
JMJD1A, ATRX mRNA was significantly upregulated in cecum, colon and
rectal adenocarcinomas, and even mucinous adenocarcinomas of the
colon and rectum displayed significant ATRX upregulation [Fig. 7A; microarray data from reference
(51)]. Excitingly, high ATRX
expression was positively correlated with increased disease
recurrence [Fig. 7B; microarray data
from reference (55)] and lethality
[Fig. 7C; microarray data from
reference (52)]. These data suggest
that ATRX might promote colorectal cancer.
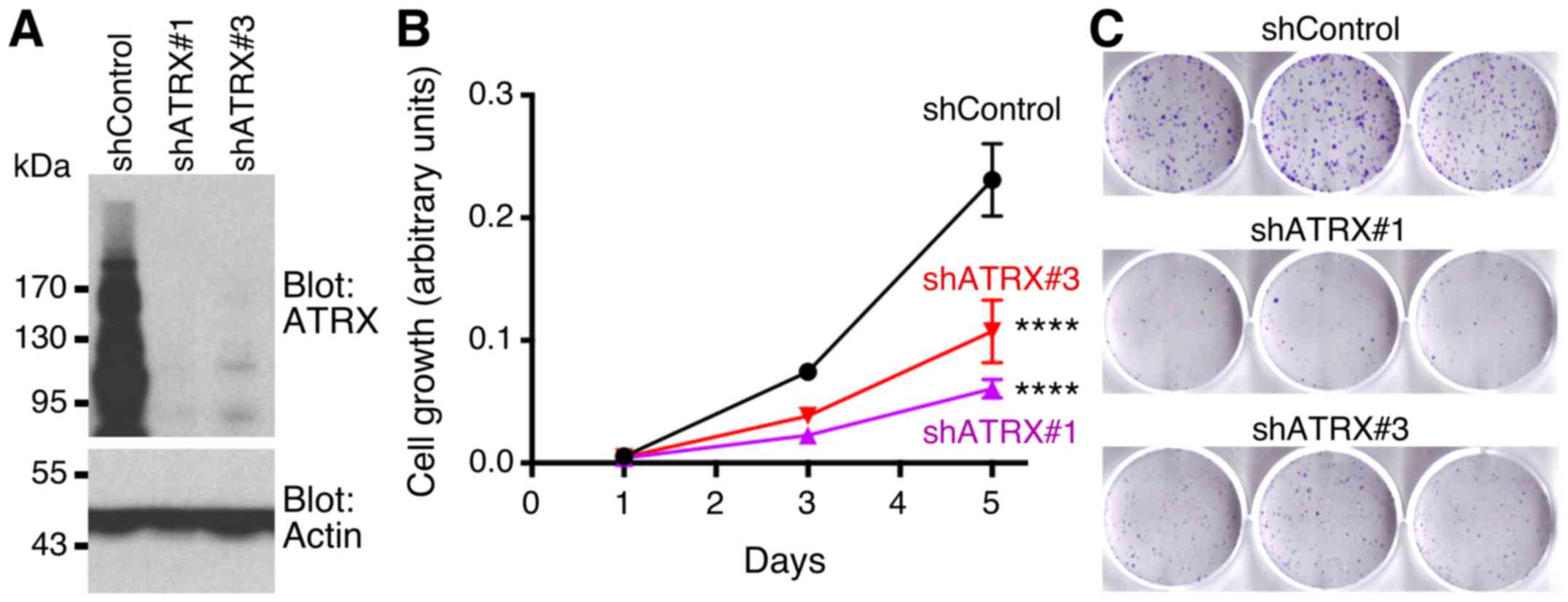
We then downregulated ATRX with two different shRNAs
in HCT116 cells (Fig. 8A). This led
to a significant reduction of HCT116 cell growth (Fig. 8B). In addition, both ATRX shRNAs
caused a reduction in clonogenic activity of HCT116 cells (Fig. 8C). These data are further supporting
the notion that ATRX is a promoter of colorectal cancer.
Discussion
In the current study, we provided evidence that
JMJD1A is overexpressed at the mRNA and protein level in colorectal
tumors and is associated with worse clinical outcomes, the latter
being consistent with a previous report (18). Further, we demonstrated that JMJD1A
has no or a minimal, effect on the in vitro growth of HCT116
cells. This result is consistent with data published by Krieg et
al (17), but in contrast to
Uemura et al (18) who
reported that JMJD1A downregulation led to basically complete loss
of in vitro HCT116 proliferation. However, it has to be
noted that only one JMJD1A siRNA was utilized in the latter study,
whose potential off-target effects might have caused the dramatic
phenotype observed. Yet, both of our JMJD1A shRNAs caused a robust
reduction in HCT116 clonogenic activity, which is the ability of
single cells to form colonies that can be associated with the
seeding of tumors and which is reliant on cancer stem cell
properties. Hence, JMJD1A may be more important for promoting
cancer stem cells than for stimulating the growth rate of tumors.
Consistent with this concept, JMJD1A has been reported to foster
cancer stemness in a variety of different tumors (15,16,19,56).
JMJD1A gene transcription is upregulated upon oxygen
depletion by the transcription factor HIF-1, the master regulator
of hypoxia (57–59). Given that most tumors are in a hypoxic
environment, JMJD1A is therefore destined to become overexpressed
in cancer cells. Further, JMJD1A can form complexes with the HIF-1
protein, which may particularly be important for the regulation of
glycolytic enzymes and adaptation of cancer cells to a hypoxic
environment (60,61). In how far JMJD1A's role in colorectal
cancer is related to hypoxia and whether hypoxia is the driving
force behind its overexpression remains to be studied.
Previous studies have found a predominantly nuclear
localization of JMJD1A (1,3,62). In
contrast, our cell fractionation experiments with two different
colorectal cancer cell lines indicated that JMJD1A protein levels
are comparable in the cytoplasm and cell nucleus. This result
suggests that JMJD1A may perform non-nuclear functions that are
independent of its histone demethylase activity. Interestingly,
hypoxia was reported to reduce cytoplasmic residence of JMJD1A
(62), implicating that the
intracellular localization of JMJD1A and hence its epigenetic
nuclear function are likely regulated through environmental
cues.
Bioinformatic analyses revealed that JMJD1A
downregulation affects multiple pathways in HCT116 colorectal
cancer cells, suggesting that JMJD1A may perform pleiotropic
functions. Notably, JMJD1A downregulation led to stimulation of
TP53- and TGF-β1-regulated and inhibition of MYC- and MYCN-driven
pathways (Fig. 4D). Both TP53 and
TGF-β1 are tumor suppressors and mutations in respective pathways
are commonly observed upon the progression of colorectal adenomas
to early carcinomas (63). On the
other hand, MYC and MYCN are prominent oncoproteins and
transcription factors, whose overexpression is an underlying cause
of cancer development in many different tissues (64,65).
Further, the peroxisome proliferator-activated receptor α (PPARA)
upstream regulator pathway was the most significantly stimulated
pathway upon JMJD1A shRNA expression (Fig. 4D). While PPARA is known to
tissue-specifically act as a tumor promoter or suppressor, current
evidence points to PPARA as an inhibitor of colon cancer
development, possibly by curtailing inflammation (66). Lastly, the estrogen biosynthesis
pathway was enhanced upon JMJD1A downregulation (Fig. 4C). Estrogen in colonic tissue is
thought to activate estrogen receptor-β, which seems to prevent
colorectal cancer formation and may account for the fact that women
have a lower risk for colon cancer than men (67). All of the above described
transcriptional changes upon JMJD1A downregulation likely reduce
the oncogenic potential of HCT116 cells. This conversely outlines
potential mechanisms by which JMJD1A overexpression promotes
tumorigenesis.
In addition, we discovered that ATRX levels were
positively regulated by JMJD1A, which likely entailed activation of
the ATRX gene promoter with concurrent removal of dimethylation on
histone H3 lysine 9. ATRX has been shown to be a chromatin
remodeler, which includes its role in the deposition of histone
variant H3.3 at repetitive regions such as telomeres and
pericentric heterochromatin, binding to and likely resolving
G-quadruplexes, potentially evicting histone variant macroH2A1 and
also promoting homologous recombination after DNA double-strand
breaks (54,68). Interestingly, while ATRX was mostly
localized to intergenic regions in mouse embryonic stem cells, the
majority of ATRX was bound to promoters and gene bodies in
neuroepithelial progenitors. Accordingly, knockout of ATRX could
result in pleiotropic changes of the gene expression program
through altering chromatin accessibility, implying that ATRX can
epigenetically impact on many genes (69). In vivo, ATRX is essential for
development, as respective knockout mice died midway through
embryogenesis (70). Also, mutations
in ATRX can cause α-thalassemia and mental retardation, a syndrome
that manifests predominantly in males due to the fact that the ATRX
gene is encoded on the X chromosome (71). Mutations in the ATRX gene thought to
inactivate its function have also been found in various cancers,
especially in pancreatic neuroendocrine tumors, pediatric
glioblastoma multiforme and adult low-grade gliomas (72). This implicated that ATRX is a tumor
suppressor. Consistently, ablation of ATRX accelerated tumor growth
in a glioblastoma model (73).
In contrast, our data are indicative of a
tumor-promoting role of ATRX. We demonstrate that ATRX is
overexpressed in colorectal cancer, and high ATRX mRNA levels are
positively correlated with a worse outcome of this disease. In line
with the idea of ATRX stimulating tumorigenesis, ATRX
downregulation led to significantly reduced HCT116 cell growth and
clonogenic activity. Similar to our results with HCT116 cancer
cells, knockout of ATRX in mouse embryonic stem cells disadvantaged
their growth (70). Please note that
JMJD1A knockdown had only a negligible effect on HCT116 cell growth
(Fig. 3B) despite the fact that this
concurrently led to a reduction in ATRX expression (Fig. 5A and B). However, the degree of ATRX
downregulation was much lower upon JMJD1A knockdown compared to
when we utilized ATRX shRNA (compare Figs. 5B to 8A), and this relatively weaker degree of
ATRX downregulation may have been insufficient to robustly impair
cell growth. Another possibility is that JMJD1A knockdown
compensated by an unknown mechanism for the reduction of ATRX
levels. Finally, it remains to be studied if ATRX has the
capability to stimulate cancer cells also in tissues other than the
colon and rectum.
Altogether, our data suggest that JMJD1A and ATRX
may act in common to promote colon cancer. In addition, to our
knowledge, our data for the first time provide considerable
evidence that ATRX is a tumor promoter, which challenges the dogma
of ATRX solely being a tumor suppressor.
Acknowledgements
Not applicable.
Funding
The present study was in part funded by a Team
Science Seed grant from the Stephenson Cancer Center (received by
WMF and RJ), a grant from the National Institutes of Health (grant
no. P30 AG050911; received by WMF), and also financially supported
by the Graduate School of Jilin University and China-Japan Union
Hospital of Jilin University (received by XL).
Availability of data and materials
RNA sequencing data have been deposited in the NCBI
BioProject database under accession no. PRJNA453378 and can be
freely downloaded from the NCBI Sequence Read Archive (accession
nos. SRX3992514, SRX3992513 and SRX3992472).
Authors' contributions
XL, SO, HS and RJ designed and performed
experiments. XL, SO, HS, SS, BZ, WMF and RJ analyzed and
interpreted data. RJ supervised the whole study and wrote the
manuscript with input from all other authors. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yamane K, Toumazou C, Tsukada Y,
Erdjument-Bromage H, Tempst P, Wong J and Zhang Y: JHDM2A, a
JmjC-containing H3K9 demethylase, facilitates transcription
activation by androgen receptor. Cell. 125:483–495. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kooistra SM and Helin K: Molecular
mechanisms and potential functions of histone demethylases. Nat Rev
Mol Cell Biol. 13:297–311. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Knebel J, De Haro L and Janknecht R:
Repression of transcription by TSGA/Jmjd1a, a novel interaction
partner of the ETS protein ER71. J Cell Biochem. 99:319–329. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lockman K, Taylor JM and Mack CP: The
histone demethylase, Jmjd1a, interacts with the myocardin factors
to regulate SMC differentiation marker gene expression. Circ Res.
101:e115–e123. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fan L, Peng G, Sahgal N, Fazli L, Gleave
M, Zhang Y, Hussain A and Qi J: Regulation of c-Myc expression by
the histone demethylase JMJD1A is essential for prostate cancer
cell growth and survival. Oncogene. 35:2441–2452. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tee AE, Ling D, Nelson C, Atmadibrata B,
Dinger ME, Xu N, Mizukami T, Liu PY, Liu B, Cheung B, et al: The
histone demethylase JMJD1A induces cell migration and invasion by
up-regulating the expression of the long noncoding RNA MALAT1.
Oncotarget. 5:1793–1804. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sechler M, Parrish JK, Birks DK and
Jedlicka P: The histone demethylase KDM3A, and its downstream
target MCAM, promote Ewing sarcoma cell migration and metastasis.
Oncogene. 36:4150–4160. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Abe Y, Rozqie R, Matsumura Y, Kawamura T,
Nakaki R, Tsurutani Y, Tanimura-Inagaki K, Shiono A, Magoori K,
Nakamura K, et al: JMJD1A is a signal-sensing scaffold that
regulates acute chromatin dynamics via SWI/SNF association for
thermogenesis. Nat Commun. 6:70522015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kuroki S, Matoba S, Akiyoshi M, Matsumura
Y, Miyachi H, Mise N, Abe K, Ogura A, Wilhelm D, Koopman P, et al:
Epigenetic regulation of mouse sex determination by the histone
demethylase Jmjd1a. Science. 341:1106–1109. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Okada Y, Scott G, Ray MK, Mishina Y and
Zhang Y: Histone demethylase JHDM2A is critical for Tnp1 and Prm1
transcription and spermatogenesis. Nature. 450:119–123. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tateishi K, Okada Y, Kallin EM and Zhang
Y: Role of Jhdm2a in regulating metabolic gene expression and
obesity resistance. Nature. 458:757–761. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Inagaki T, Tachibana M, Magoori K, Kudo H,
Tanaka T, Okamura M, Naito M, Kodama T, Shinkai Y and Sakai J:
Obesity and metabolic syndrome in histone demethylase
JHDM2a-deficient mice. Genes Cells. 14:991–1001. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Abe Y, Fujiwara Y, Takahashi H, Matsumura
Y, Sawada T, Jiang S, Nakaki R, Uchida A, Nagao N, Naito M, et al:
Histone demethylase JMJD1A coordinates acute and chronic adaptation
to cold stress via thermogenic phospho-switch. Nat Commun.
9:15662018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Loh YH, Zhang W, Chen X, George J and Ng
HH: Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate
self-renewal in embryonic stem cells. Genes Dev. 21:2545–2557.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ramadoss S, Guo G and Wang CY: Lysine
demethylase KDM3A regulates breast cancer cell invasion and
apoptosis by targeting histone and the non-histone protein p53.
Oncogene. 36:47–59. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ramadoss S, Sen S, Ramachandran I, Roy S,
Chaudhuri G and Farias-Eisner R: Lysine-specific demethylase KDM3A
regulates ovarian cancer stemness and chemoresistance. Oncogene.
36:1537–1545. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Krieg AJ, Rankin EB, Chan D, Razorenova O,
Fernandez S and Giaccia AJ: Regulation of the histone demethylase
JMJD1A by hypoxia-inducible factor 1 alpha enhances hypoxic gene
expression and tumor growth. Mol Cell Biol. 30:344–353. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Uemura M, Yamamoto H, Takemasa I, Mimori
K, Hemmi H, Mizushima T, Ikeda M, Sekimoto M, Matsuura N, Doki Y
and Mori M: Jumonji domain containing 1A is a novel prognostic
marker for colorectal cancer: In vivo identification from hypoxic
tumor cells. Clin Cancer Res. 16:4636–4646. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li J, Yu B, Deng P, Cheng Y, Yu Y, Kevork
K, Ramadoss S, Ding X, Li X and Wang CY: KDM3 epigenetically
controls tumorigenic potentials of human colorectal cancer stem
cells through Wnt/β-catenin signalling. Nat Commun. 8:151462017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rhodes DR, Kalyana-Sundaram S, Mahavisno
V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ,
Kincead-Beal C, Kulkarni P, et al: Oncomine 3.0: Genes, pathways,
and networks in a collection of 18,000 cancer gene expression
profiles. Neoplasia. 9:166–180. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Uhlén M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C,
Sjöstedt E, Asplund A, et al: Proteomics. Tissue-based map of the
human proteome. Science. 347:12604192015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim TD, Oh S, Lightfoot SA, Shin S, Wren
JD and Janknecht R: Upregulation of PSMD10 caused by the JMJD2A
histone demethylase. Int J Clin Exp Med. 9:10123–10134.
2016.PubMed/NCBI
|
|
24
|
Kim TD, Oh S, Shin S and Janknecht R:
Regulation of tumor suppressor p53 and HCT116 cell physiology by
histone demethylase JMJD2D/KDM4D. PLoS One. 7:e346182012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mooney SM, Grande JP, Salisbury JL and
Janknecht R: Sumoylation of p68 and p72 RNA helicases affects
protein stability and transactivation potential. Biochemistry.
49:1–10. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dowdy SC, Mariani A and Janknecht R:
HER2/Neu- and TAK1-mediated up-regulation of the transforming
growth factor beta inhibitor Smad7 via the ETS protein ER81. J Biol
Chem. 278:44377–44384. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bosc DG, Goueli BS and Janknecht R:
HER2/Neu-mediated activation of the ETS transcription factor ER81
and its target gene MMP-1. Oncogene. 20:6215–6224. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shin S, Kim TD, Jin F, van Deursen JM,
Dehm SM, Tindall DJ, Grande JP, Munz JM, Vasmatzis G and Janknecht
R: Induction of prostatic intraepithelial neoplasia and modulation
of androgen receptor by ETS variant 1/ETS-related protein 81.
Cancer Res. 69:8102–8110. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Berry WL, Shin S, Lightfoot SA and
Janknecht R: Oncogenic features of the JMJD2A histone demethylase
in breast cancer. Int J Oncol. 41:1701–1706. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Berry WL, Kim TD and Janknecht R:
Stimulation of β-catenin and colon cancer cell growth by the KDM4B
histone demethylase. Int J Oncol. 44:1341–1348. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shin S, Oh S, An S and Janknecht R: ETS
variant 1 regulates matrix metalloproteinase-7 transcription in
LNCaP prostate cancer cells. Oncol Rep. 29:306–314. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kim TD, Shin S and Janknecht R: ETS
transcription factor ERG cooperates with histone demethylase KDM4A.
Oncol Rep. 35:3679–3688. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kim TD, Shin S, Berry WL, Oh S and
Janknecht R: The JMJD2A demethylase regulates apoptosis and
proliferation in colon cancer cells. J Cell Biochem. 113:1368–1376.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Oh S, Shin S, Lightfoot SA and Janknecht
R: 14-3-3 proteins modulate the ETS transcription factor ETV1 in
prostate cancer. Cancer Res. 73:5110–5119. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shin S, Bosc DG, Ingle JN, Spelsberg TC
and Janknecht R: Rcl is a novel ETV1/ER81 target gene upregulated
in breast tumors. J Cell Biochem. 105:866–874. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kim TD, Fuchs JR, Schwartz E, Abdelhamid
D, Etter J, Berry WL, Li C, Ihnat MA, Li PK and Janknecht R:
Pro-growth role of the JMJD2C histone demethylase in HCT-116 colon
cancer cells and identification of curcuminoids as JMJD2
inhibitors. Am J Transl Res. 6:236–247. 2014.PubMed/NCBI
|
|
37
|
Goel A and Janknecht R: Concerted
activation of ETS protein ER81 by p160 coactivators, the
acetyltransferase p300 and the receptor tyrosine kinase HER2/Neu. J
Biol Chem. 279:14909–14916. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Papoutsopoulou S and Janknecht R:
Phosphorylation of ETS transcription factor ER81 in a complex with
its coactivators CREB-binding protein and p300. Mol Cell Biol.
20:7300–7310. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mooney SM, Goel A, D'Assoro AB, Salisbury
JL and Janknecht R: Pleiotropic effects of p300-mediated
acetylation on p68 and p72 RNA helicase. J Biol Chem.
285:30443–30452. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wu J and Janknecht R: Regulation of the
ETS transcription factor ER81 by the 90-kDa ribosomal S6 kinase 1
and protein kinase A. J Biol Chem. 277:42669–42679. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li X, Moon G, Shin S, Zhang B and
Janknecht R: Cooperation between ETS variant 2 and Jumonji
domain-containing 2 histone demethylases. Mol Med Rep.
17:5518–5527. 2018.PubMed/NCBI
|
|
42
|
Janknecht R: Regulation of the ER81
transcription factor and its coactivators by mitogen- and
stress-activated protein kinase 1 (MSK1). Oncogene. 22:746–755.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kim TD, Jin F, Shin S, Oh S, Lightfoot SA,
Grande JP, Johnson AJ, van Deursen JM, Wren JD and Janknecht R:
Histone demethylase JMJD2A drives prostate tumorigenesis through
transcription factor ETV1. J Clin Invest. 126:706–720. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Oh S and Janknecht R: Histone demethylase
JMJD5 is essential for embryonic development. Biochem Biophys Res
Commun. 420:61–65. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Goel A and Janknecht R:
Acetylation-mediated transcriptional activation of the ETS protein
ER81 by p300, P/CAF, and HER2/Neu. Mol Cell Biol. 23:6243–6254.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kim TD, Shin S and Janknecht R: Repression
of Smad3 activity by histone demethylase SMCX/JARID1C. Biochem
Biophys Res Commun. 366:563–567. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Janknecht R and Hunter T: Activation of
the Sap-1a transcription factor by the c-Jun N-terminal kinase
(JNK) mitogen-activated protein kinase. J Biol Chem. 272:4219–4224.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Goueli BS and Janknecht R: Regulation of
telomerase reverse transcriptase gene activity by upstream
stimulatory factor. Oncogene. 22:8042–8047. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Goueli BS and Janknecht R: Upregulation of
the catalytic telomerase subunit by the transcription factor ER81
and oncogenic HER2/Neu, Ras, or Raf. Mol Cell Biol. 24:25–35. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kim J, Shin S, Subramaniam M, Bruinsma E,
Kim TD, Hawse JR, Spelsberg TC and Janknecht R: Histone demethylase
JARID1B/KDM5B is a corepressor of TIEG1/KLF10. Biochem Biophys Res
Commun. 401:412–416. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Cancer Genome Atlas Natwork: Comprehensive
molecular characterization of human colon and rectal cancer.
Nature. 487:330–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Smith JJ, Deane NG, Wu F, Merchant NB,
Zhang B, Jiang A, Lu P, Johnson JC, Schmidt C, Bailey CE, et al:
Experimentally derived metastasis gene expression profile predicts
recurrence and death in patients with colon cancer.
Gastroenterology. 138:958–968. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Tsuji S, Midorikawa Y, Takahashi T, Yagi
K, Takayama T, Yoshida K, Sugiyama Y and Aburatani H: Potential
responders to FOLFOX therapy for colorectal cancer by Random
Forests analysis. Br J Cancer. 106:126–132. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ratnakumar K and Bernstein E: ATRX: The
case of a peculiar chromatin remodeler. Epigenetics. 8:3–9. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Lin YH, Friederichs J, Black MA, Mages J,
Rosenberg R, Guilford PJ, Phillips V, Thompson-Fawcett M, Kasabov
N, Toro T, et al: Multiple gene expression classifiers from
different array platforms predict poor prognosis of colorectal
cancer. Clin Cancer Res. 13:498–507. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Nakatsuka T, Tateishi K, Kudo Y, Yamamoto
K, Nakagawa H, Fujiwara H, Takahashi R, Miyabayashi K, Asaoka Y,
Tanaka Y, et al: Impact of histone demethylase KDM3A-dependent AP-1
transactivity on hepatotumorigenesis induced by PI3K activation.
Oncogene. 36:6262–6271. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wellmann S, Bettkober M, Zelmer A, Seeger
K, Faigle M, Eltzschig HK and Bührer C: Hypoxia upregulates the
histone demethylase JMJD1A via HIF-1. Biochem Biophys Res Commun.
372:892–897. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Pollard PJ, Loenarz C, Mole DR, McDonough
MA, Gleadle JM, Schofield CJ and Ratcliffe PJ: Regulation of
Jumonji-domain-containing histone demethylases by hypoxia-inducible
factor (HIF)-1alpha. Biochem J. 416:387–394. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Beyer S, Kristensen MM, Jensen KS,
Johansen JV and Staller P: The histone demethylases JMJD1A and
JMJD2B are transcriptional targets of hypoxia-inducible factor HIF.
J Biol Chem. 283:36542–36552. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Mimura I, Nangaku M, Kanki Y, Tsutsumi S,
Inoue T, Kohro T, Yamamoto S, Fujita T, Shimamura T, Suehiro J, et
al: Dynamic change of chromatin conformation in response to hypoxia
enhances the expression of GLUT3 (SLC2A3) by cooperative
interaction of hypoxia-inducible factor 1 and KDM3A. Mol Cell Biol.
32:3018–3032. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wan W, Peng K, Li M, Qin L, Tong Z, Yan J,
Shen B and Yu C: Histone demethylase JMJD1A promotes urinary
bladder cancer progression by enhancing glycolysis through
coactivation of hypoxia inducible factor 1α. Oncogene.
36:3868–3877. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Sar A, Ponjevic D, Nguyen M, Box AH and
Demetrick DJ: Identification and characterization of demethylase
JMJD1A as a gene upregulated in the human cellular response to
hypoxia. Cell Tissue Res. 337:223–234. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Jones S, Chen WD, Parmigiani G, Diehl F,
Beerenwinkel N, Antal T, Traulsen A, Nowak MA, Siegel C, Velculescu
VE, et al: Comparative lesion sequencing provides insights into
tumor evolution. Proc Natl Acad Sci USA. 105:4283–4288. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Dang CV: MYC on the path to cancer. Cell.
149:22–35. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Rickman DS, Schulte JH and Eilers M: The
expanding world of N-MYC-driven tumors. Cancer Discov. 8:150–163.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Gao J, Yuan S, Jin J, Shi J and Hou Y:
PPARα regulates tumor progression, foe or friend? Eur J Pharmacol.
765:560–564. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Williams C, DiLeo A, Niv Y and Gustafsson
JÅ: Estrogen receptor beta as target for colorectal cancer
prevention. Cancer Lett. 372:48–56. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Juhász S, Elbakry A, Mathes A and Löbrich
M: ATRX promotes DNA repair synthesis and sister chromatid exchange
during homologous recombination. Mol Cell. 71:11–24.e7. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Danussi C, Bose P, Parthasarathy PT,
Silberman PC, Van Arnam JS, Vitucci M, Tang OY, Heguy A, Wang Y,
Chan TA, et al: Atrx inactivation drives disease-defining
phenotypes in glioma cells of origin through global epigenomic
remodeling. Nat Commun. 9:10572018. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Garrick D, Sharpe JA, Arkell R, Dobbie L,
Smith AJ, Wood WG, Higgs DR and Gibbons RJ: Loss of Atrx affects
trophoblast development and the pattern of X-inactivation in
extraembryonic tissues. PLoS Genet. 2:e582006. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Gibbons RJ, Picketts DJ, Villard L and
Higgs DR: Mutations in a putative global transcriptional regulator
cause X-linked mental retardation with alpha-thalassemia (ATR-X
syndrome). Cell. 80:837–845. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Dyer MA, Qadeer ZA, Valle-Garcia D and
Bernstein E: ATRX and DAXX: Mechanisms and mutations. Cold Spring
Harb Perspect Med. 7:pii: a0265672017. View Article : Google Scholar
|
|
73
|
Koschmann C, Calinescu AA, Nunez FJ,
Mackay A, Fazal-Salom J, Thomas D, Mendez F, Kamran N, Dzaman M,
Mulpuri L, et al: ATRX loss promotes tumor growth and impairs
nonhomologous end joining DNA repair in glioma. Sci Transl Med.
8:328ra282016. View Article : Google Scholar : PubMed/NCBI
|