Introduction
Lung cancer accounts for 23% of all cancer-related
deaths in the world in 2012 in spite of decades-long advancement in
early detection, prevention, and drug development (1,2).
Molecularly, lung cancer development is caused by either loss of
tumor suppressor genes and/or activation of oncogenes (1,3). During
lung cancer progression, tumor cells gain the ability to migrate
and invade into the surrounding tissues and distant organs, like
most of other human cancers (4). To
date, most of lung cancer are diagnosed at advanced stages, which
limits the surgical option and most of such patients are also
resistant to routine chemoradiotherapy (4). Thus, identification of novel molecular
targets and/or biomarkers for early detection or to predict
prognosis of lung cancer could reduce disease burdens in
patients.
RNA-binding protein 10 (RBM10) contains an RNA
recognition motif and has been identified as a component of
spliceosome complex and functions to regulate pre-mRNA splicing in
the alternatively splicing pathway (5) and mRNA stabilization (6). RBM10 alteration is more frequently
associated with alternative exon skipping (7), while expression of more than 90% of
human genes, including tumorigenesis-related genes, is regulated by
the alternative gene splicing (8).
Moreover, alteration of mRNA alternative splicing, especially exon
skipping (7) or exon-inclusion of the
affected proteins (7,9), results in abnormal protein expression
and functions in cells. RBM10 knockdown provoked alterations in
10–20% of the pre-mRNA splicing events (10), while other studies showed that RBM10
is a tumor suppressor gene (11,12) Thus,
RBM10 may play an important role in suppression of human
tumorigenesis. Previous studies showed that RBM10 mutations
did occur in lung and pancreatic cancers (13,14) and
the Cancer Genome Atlas (TCGA) data showed that RBM10
mutations occurred in 8% of lung adenocarcinoma and is more common
in male patients. RBM10 mutations were also reported to be
more prevalent (76%) in the transversion-high group (C>T) of
lung adenocarcinoma in a male cohort (14) and in 12 out of 183 of lung
adenocarcinoma cases (15). Lung
adenocarcinoma makes up to approximately 40% of all lung cancer
cases (1). Thus, further
investigation of RBM10 and its functions in lung adenocarcinoma
could provide a better understanding of the role of RBM10
mutation in lung adenocarcinoma cell growth and invasion.
In the present study, we first assessed the clinical
significance of RBM10 exon 10 mutations in lung
adenocarcinoma for association with clinicopathological features,
such as tobacco smoking, age, gender, tumor histological grade,
AJCC stage, lymph node metastasis, and survival. We then
investigated the effect of RBM10 exon 10 mutations on
regulating lung adenocarcinoma cell proliferation, invasion, and
apoptosis as well as the underlying molecular events. Our study
expected to provide novel insightful information for RBM10 as a
novel biomarker or therapeutic target for lung adenocarcinoma.
Materials and methods
Patients and follow-up
In the present study, we retrospectively collected
paraffin-embedded tissue samples from 50 lung adenocarcinoma
patients who received surgical tumor resection from the Fourth
Hospital of Jinan (Shandong, China) and Shanxian Central Hospital
(Shandong, China) between April 2011 and January 2015. All patients
were histologically diagnosed with lung adenocarcinoma and the AJCC
stage was determined according to the Seventh Edition of the AJCC
Staging System for Lung Cancer (16).
Patients were followed up every three months during the first year
after surgery and then six months thereafter and the most recent
follow-up was October 30, 2017. The median follow-up period was
44.5 months (ranged between 18.0 and 60.0 months). A laboratory
protocol of this study was approved by the Fourth Hospital of Jinan
and Shanxian Central Hospital Review Board according to the
Declaration of Helsinki. Written informed consent was obtained from
all patients to allow utilizing their tissue specimens in the
present study.
Genomic DNA extraction
Paraffin-embedded tissue blocks from 50 lung
adenocarcinoma and paired adjacent non-cancerous lung tissues were
retrieved from the Department of Pathology of both hospitals and
sectioned into 4 µm-thick tissue sections for H&E staining and
confirmation of the diagnosis. After that, 10 µm tissue sections
were prepared and subjected to DNA extraction using the TIANquick
FFPE DNA Kit and TIANamp Genomic DNA kit (Tiangen Biotech Co.,
Ltd., Beijing, China) according to the manufacturer's protocol.
Analysis of RBM10 mutations
The RBM10 exon 10 was amplified using
polymerase chain reaction (PCR) with Hot Start Taq MasterMix
(Tiangen Biotech Co., Ltd.) and the specific primers
(5′-GGGGTGTCCTCTAACATTGG-3′ and 5′-ATGGTCTTGCCGTCGATAGT-3′). The
size of the expected amplicon is 486 bp. In brief, genomic DNA of
50–100 ng was amplified in a 50 µl reaction mixture containing 25
µl of Hot Start Taq MasterMix and 5 µl of 10 µM primer mix. The PCR
conditions were set to 95°C for 2 min and followed by 40 cycles of
94°C for 20 sec, 54°C for 20 sec, and 72°C for 20 sec with a final
extension at 72°C for 3 min. PCR products of 6 µl of each were
analyzed in 1.5% agarose gel with 50 bp ladder DNA markers. The
resulting PCR products were purified and sequenced in both
directions using Sanger sequencing (Sangon Biotech Co., Ltd.,
Shanghai, China).
Cell lines and culture
Human lung adenocarcinoma A549 and H1299 cell lines
were obtained from Chinese Academy of Sciences (Shanghai, China)
and cultured in RPMI 1640 Medium (GE Healthcare, Chicago, IL, USA)
supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), 1% penicillin-streptomycin
(Gibco; Thermo Fisher Scientific, Inc.) in a humidified atmosphere
with 5% CO2 at 37°C.
Plasmid construction and cell
transfection
The wild-type full-length RBM10 cDNA was
amplified using PCR according to the coding sequence of human RBM10
from GenBank, whereas RBM10 cDNA carrying exon 10 mutation
at c.763 C>T was amplified using PCR with specific primer sets.
These cDNA fragments were then cloned into pcDNA3.1 plasmid
(Invitrogen; Thermo Fisher Scientific, Inc.). After amplification
and DNA sequencing confirmation, these plasmids were named
pcDNA3.1-RBM10w and pcDNA3.1-RBM10m, respectively and used for cell
transfection.
For cell transfection, A549 and H1299 cells were
seeded into 6-well plates at a density of 1×106
cells/well and cultured in RPMI1640 with 10% FBS overnight to reach
approximately 70% confluency and then transfected with
Lipofectamine2000 (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. We prepared three groups
of transfections, i.e., the negative control (transfected with
pcDNA3.1 only), wild type (transfected with pcDNA3.1-RBM10w) and
the mutation group (transfected with pcDNA3.1-RBM10m). After that,
cell growth medium was refreshed with complete growth medium 6 h
after transfection and the cells were further incubated for 48
h.
Quantitative reverse
transcriptase-polymerase chain reaction
TRIzol (Thermo Fisher Scientific, Inc.) was used to
isolate RNA following the manufacturer's protocol. For reverse
transcription, the components in a total volume of 10 µl were used
as follows: 3 µg total RNA; 10 mM deoxyribonucleotide triphosphate;
0.5 µg oligo deoxythymine; 20 U RNasin®; 200 U Maloney
murine leukemia virus reverse transcriptase (Thermo Fisher
Scientific, Inc.). The primer sequences were as follows: RBM10
sense, 5′-GCACGACTATAGGCATGACAT-3′; antisense,
5′-AGTCAAACTTGTCTGCTCCA-3′; GAPDH sense, 5′-GAAGGTGAAGGTCGGAGTC-3′;
antisense, 5′-GAAGATGGTGATGGGATTTC-3′. PCR was performed with 25–30
cycles as follows: 95°C for 30 sec; 55°C for 30 sec; 72°C for 1
min. Densitometry analysis was performed with ImageMaster VDS-CL
Image Master 1.0.3.7 software (GE Healthcare, Chicago, IL,
USA).
Cell viability CCK-8 assay
Cell viability was assessed using the Cell Counting
Kit-8 (CCK-8; Beijing Solarbio Science & Technology Co., Ltd.,
Beijing, China). In brief, gene transfected A549 and H1299 cells
were detached from cell culture dishes using trypsin and re-seeded
into 96-well plates at a density of 1×104 per well and
cultured for 24 h, 48 h, and 72 h, respectively. At the end of each
experiment, 10 µl of CCK-8 reagent was added into each well and
cells were further incubated for 90 min. The absorbance values were
then measured at 450 nm using a plate reader (Eppendorf, Hamburg,
Germany) and the inhibitory rate was then calculated.
Flow cytometric apoptosis assay
The flow cytometer assay was performed to assess the
changed cell apoptosis after gene transfection. Specifically, A549
and H1299 growing in the exponential phase were transfected with
pcDNA3.1 plasmids carrying wild type RBM10 cDNA or RBM10 exon 10
mutation at c.763 C>T, or vector-only for 24 h. The cells were
then detached using trypsin and re-seeded into 6-well plates at a
density of 1×105 per well and cultured in a serum-free
medium for 24 h. After that, cells were harvested in ice-cold
phosphate buffered saline (PBS) and apoptotic cells were detected
using the FITC Annexin-FITC/PI Apoptosis Detection kit (cat. no:
4A; Biotech Co., Ltd., Beijing, China) according to the
manufacturer's protocol. The stained cells were then measured with
a flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA). The
experiments were in triplicate and repeated at least three
times.
Transwell tumor cell invasion
assay
Tumor cell invasion ability was assessed by using
the Transwell chamber assay (EMD Millipore, Billerica, MA, USA).
Briefly, A549 and H1299 cells were transfected with pcDNA3.1-RBM10
wild type or mutated cDNA or vector-only. After 24 h of incubation,
the cells were suspended in serum-free RPMI 1640 and plated on the
upper chamber of the polycarbonate Transwell filters that were
pre-coated with Matrigel (BD Biosciences), while the lower chamber
was filled with RPMI 1640 supplemented with 10% FBS. After 24 h
incubation, the non-invaded cells in the upper chamber were removed
using cotton swabs, whereas the cells invaded into the low surface
of the filters were fixed with 4% paraformaldehyde and stained with
0.1% crystal violet. Cells were photographed in five random fields
under a Nikon TS100-F inverted microscope (Nikon, Tokyo, Japan) and
then counted.
Western blot analysis
A549 and H1299 cells were transfected with
pcDNA3.1-RBM10 wild type or mutated cDNA or vector-only for 48 h.
Total cellular protein was then extracted using the
radioimmunoprecipitation assay lysis buffer and protein
concentrations were measured using bicinchoninic acid (BCA) kit
(CWBiotech, Beijing, China). Equal amounts of protein samples (20
µg each) were heated at 100°C for 10 min and then separated in 10%
sodium dodecyl sulfate polyacrylamide gel electrophoresis
(SDS-PAGE) gel and transferred onto a polyvinylidenedifluoride
(PVDF) membrane (EMD Millipore). The membranes were incubated in 5%
bovine serum albumin (BSA) for 2 h at the room temperature and then
incubated with primary antibodies at a dilution of 1:1,000 against
Numb (1:1,000; cat. no: 18701-1-AP), Notch-1 (1:1,000; cat. no:
10062-2-AP), Fas (1:1,000; cat. no: 13098-1-AP), E-cadherin
(1:1,000; cat. no: 20874-1-AP), and CyclinD1 (1:1,000; cat. no:
60186-1-Ig; all from ProteinTech Group, Inc., Chicago, IL, USA) at
4°C overnight. On the next day, the membranes were washed with
Tris-based saline-Tween 20 solution (TBST) three times and then
incubated with a secondary antibody at a dilution of 1:1,000
(1:1,000; cat. no: AP124P; EMD Millipore) for 1 h at the room
temperature. Protein bands were visualized using enhanced
chemiluminescence (ECL)-Plus western blotting detection reagents
(EMD Millipore).
Statistical analysis
Association of RBM10 mutations with
clinicopathological characteristics was assessed using the
Chi-square test. Association of RBM10 mutations with
five-year survival rate was assessed using Kaplan-Meier method and
the log-rank test and the multivariate analysis was performed using
Cox regression test. Moreover, all in vitro experiments were
in triplicates and repeated at least three times and the data were
expressed as a mean ± standard deviation. Student's t-test was used
to analyze the mean difference between two groups, while multi
group comparisons of the means were analyzed by using one-way
analysis of variance (ANOVA) test followed by the post-hoc
Student-Newman-Keuls test. A P<0.05 was considered to indicate a
statistically significant difference. All statistical analyses were
performed using the Statistical Package of Social Sciences (v19.0;
SPSS, Inc., Chicago, IL, USA).
Results
Patients' characteristics
In this study, we analyzed 50 lung adenocarcinoma
patients, including 30 males and 20 females with a median age of
59.0 years old (ranging from 42 to 77 years at the time of
diagnosis). Among these patients, there were 20 smokers and 30
non-smokers. Tumor stage T1, T2 and T3 were observed in 23 (46%),
21 (42%), and 6 (12%) patients, respectively, while 30, 8, and 12
patients had N0, N1 and N2 stage tumors, respectively. The AJCC
staging showed that there were 23 patients at stage I, 11 at stage
II, and 16 at stage III. Tumor differentiation was 12 well, 25
moderate, and 13 poor-differentiated cases (Table I). The last follow-up was conducted on
October 30, 2017 and these patients had a 5-year survival rate of
41.5%.
 | Table I.Association of RBM10 mutations with
clinicopathological characteristics of lung adenocarcinoma
patients. |
Table I.
Association of RBM10 mutations with
clinicopathological characteristics of lung adenocarcinoma
patients.
|
|
| RBM10 exon10
status |
|
|---|
|
|
|
|
|
|---|
| Variables | No. | Mutant | Wild-type | P-value |
|---|
| Age (years) |
|
|
| 0.728 |
|
≤60 | 27 | 5 | 22 |
|
|
>60 | 23 | 5 | 18 |
|
| Gender |
|
|
| 0.033 |
|
Female | 20 | 1 | 19 |
|
|
Male | 30 | 10 | 20 |
|
| Smoking status |
|
|
| 0.780 |
|
No-smoking | 30 | 7 | 23 |
|
|
Smoking | 20 | 4 | 16 |
|
|
Differentiation |
|
|
|
|
|
Poor-differentiated | 13 | 5 | 8 | 0.195 |
|
Moderate | 25 | 5 | 20 |
|
|
Well-differentiated | 12 | 1 | 11 |
|
| Tumor size |
|
|
| 0.793 |
| T1 | 23 | 5 | 18 |
|
| T2 | 21 | 4 | 17 |
|
| T3 | 6 | 2 | 4 |
|
| Lymphatic
metastasis |
|
|
| 0.012 |
| N0 | 30 | 3 | 27 |
|
|
Yes | 20 | 8 | 12 |
|
| AJCC stage |
|
|
| 0.005 |
| I | 23 | 2 | 21 |
|
| II | 11 | 1 | 10 |
|
|
III | 16 | 8 | 8 |
|
Association of RBM10 exon 10 (R241C)
mutation with clinicopathological characteristics of lung
adenocarcinoma patients
RBM10 exon 10 mutations were detected in 11
(22%) of 50 patients, whereas there was no mutation found in all
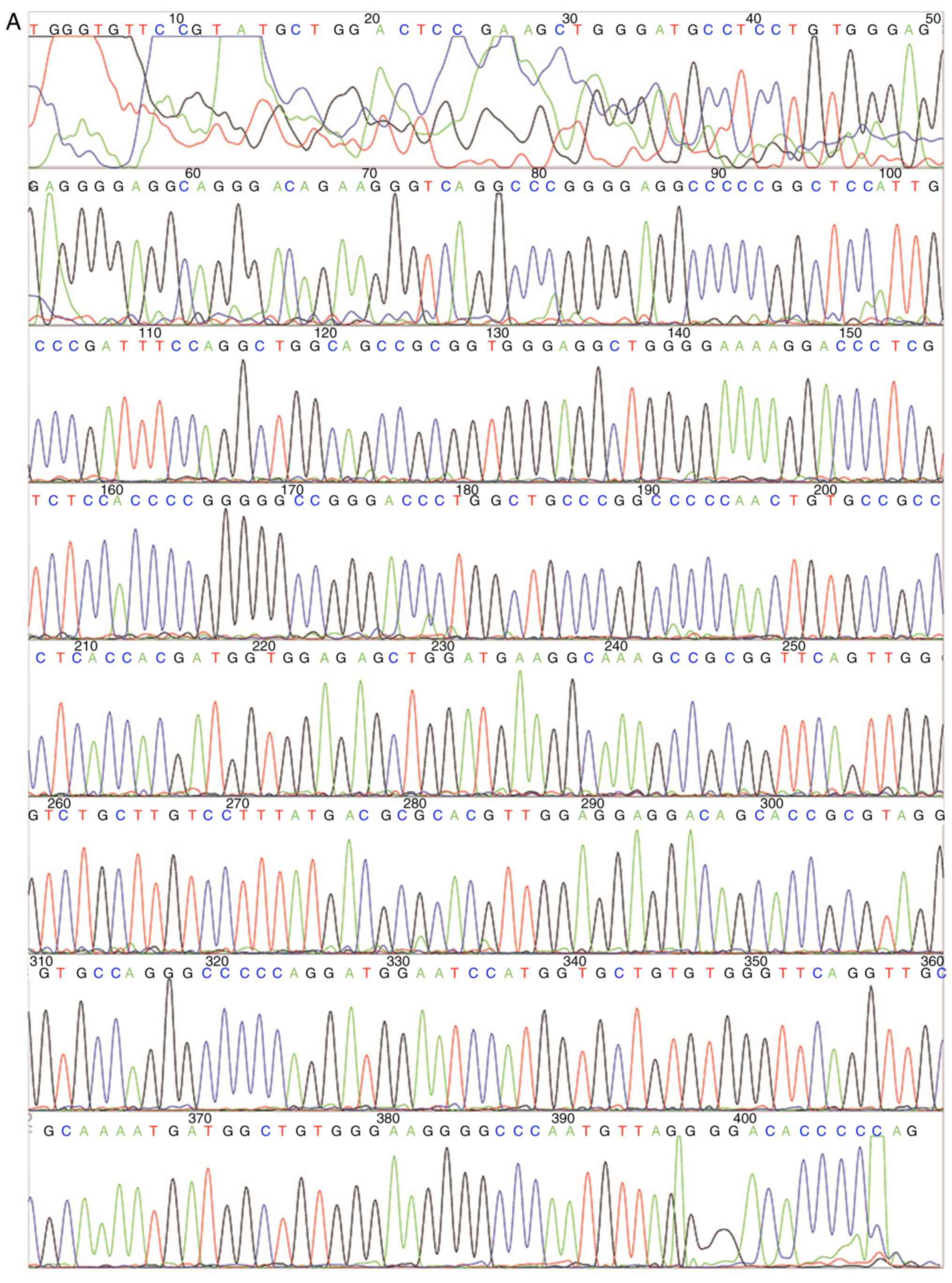
normal tissues (Fig. 1A and B). All
11 patients had RBM10 exon 10 mutation c.763 C>T, which
causes a replacement of arginine with cysteine at codon 241 (R241C)
(Fig. 1B).
We then associated the RBM10 (R241C) mutation with
the clinicopathological characteristics of lung adenocarcinoma
patients and found that RBM10 mutation was significantly
associated with the AJCC stage (χ2=10.751, P=0.005) and
lymph node metastasis (χ2=6.294, P=0.012), and male
patients (χ2=5.614, P=0.033). However, RBM10
(R241C) mutation was not associated with tobacco smoking, age of
patients and tumor size and differentiation (Tables I and II).
 | Table II.Univariate analysis of NSCLC patient
prognosis (n=50). |
Table II.
Univariate analysis of NSCLC patient
prognosis (n=50).
| Variables | No. | 5-year survival
rate (%) | P-value |
|---|
| Age (years) |
|
| 0.786 |
|
≤60 | 31 | 48.0 |
|
|
>60 | 29 | 42.7 |
|
| Gender |
|
| 0.875 |
|
Female | 20 | 48.2 |
|
|
Male | 30 | 32.7 |
|
| Smoking status |
|
| 0.780 |
|
Smoking | 20 | 36 |
|
|
No-smoking | 30 | 49.3 |
|
|
Differentiation |
|
| <0.001 |
|
Low | 13 | 13.5 |
|
|
Moderate | 25 | 48.9 |
|
|
High | 12 | 69.8 |
|
| Tumor size |
|
| 0.144 |
| T1 | 23 | 57.7 |
|
| T2 | 21 | 46.2 |
|
| T3 | 6 | 22.2 |
|
| Lymphatic
metastasis |
|
| <0.001 |
| N0 | 30 | 26.9 |
|
| N1 | 8 | 67.6 |
|
| N2 | 12 |
|
|
| AJCC stage |
|
| <0.001 |
| I | 23 | 72.4 |
|
| II | 11 | 49.9 |
|
|
III | 16 | 20.8 |
|
| RBM10 exon10 gene
status |
|
| 0.019 |
|
Mutation | 11 | 36.4 |
|
| Wild
type | 39 | 46.5 |
|
Association of RBM10 mutation (R241C)
with poor prognosis of patients
We then assessed the association of RBM10
exon 10 mutation (R241C) with 5-year survival rate of these
patients, As shown in Fig. 1C,
patients carrying the RBM10 (R241C) mutation had much
shorter 5-year survival rate (36.4% vs. 46.5 of RBM 10 wild type;
χ2=5.466, P=0.019; Fig. 1C
and Table II). Our multivariate
analysis revealed that RBM10 exon 10 mutation and tumor
differentiation and lymphatic metastasis were all independent
prognostic factors (Table III).
 | Table III.Multivariate analysis of NSCLC
patients' prognosis. |
Table III.
Multivariate analysis of NSCLC
patients' prognosis.
|
|
|
| 95% confidence
interval |
|---|
|
|
|
|
|
|---|
| Variables | P-value | HR | Lower | Upper |
|---|
| RBM10 mutation | 0.033 | 3.787 | 1.112 | 12.895 |
|
Differentiation | <0.001 | 0.03 | 0.153 | 0.909 |
| Lymphatic
metastasis | 0.029 | 4.306 | 1.165 | 15.91 |
RBM10 (R241C) mutation induction of
tumor cell proliferation and modulation of Numb and Notch
expression in vitro
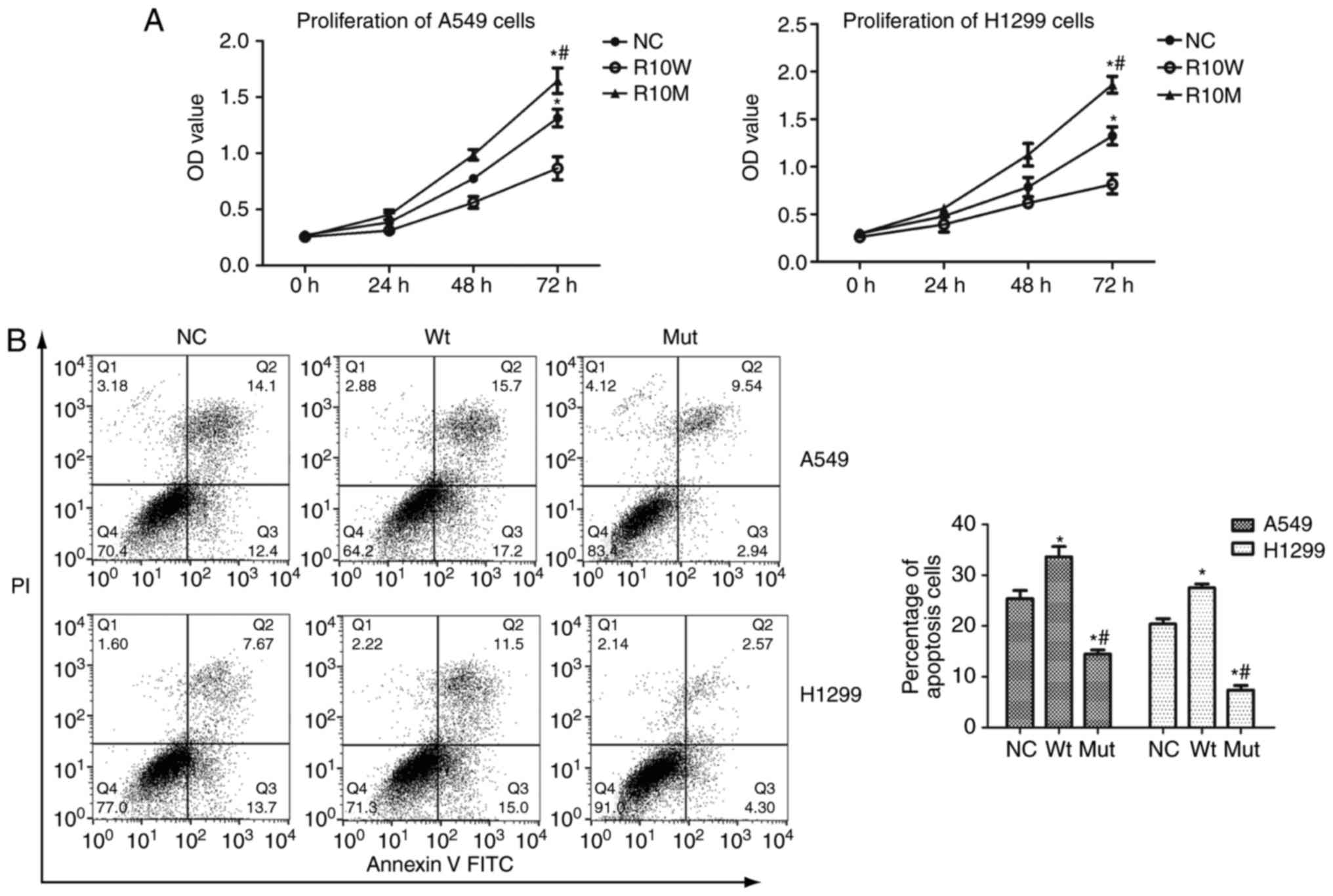
After that, we further assessed the effect of
RBM10 (R241C) mutation on regulation of lung cancer cell
proliferation and found that viability of A549 and H1299 cells
after transfection with wild type RBM10 was slightly inhibited
compared with that of blank plasmid (P<0.01; Fig. 2), whereas RBM10 (R241C)
mutation significantly induced A549 and H1299 cell viability
(Fig. 2). Furthermore, RBM10
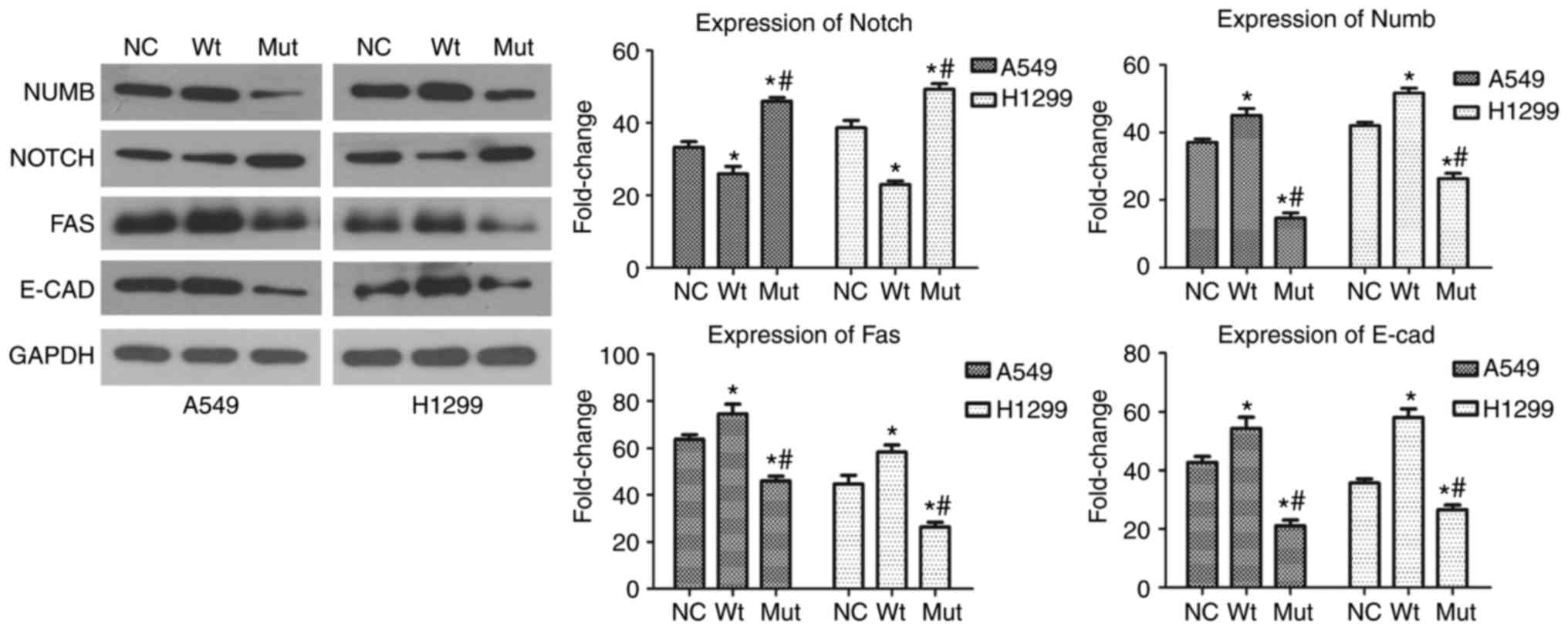
(R241C) mutation also modulated expression of Numb and Notch
proteins, which are the key regulatory proteins of NSCLC cell
proliferation (17). As shown in
Fig. 3, transfection with plasmid
carrying RBM10 (R241C) mutated cDNA was able to inhibit Numb
protein expression, which negatively regulates NSCLC cell
proliferation (17) (P<0.01;
Fig. 3). In contrast, Notch
expression in A549 and H1299 cells was significantly increased
after transfection with the plasmid carrying RBM10 (R241C)
mutated cDNA (P<0.01; Fig. 3).
RBM10 (R241C) mutation suppression of
A549 and H1299 cell apoptosis through Fas down-regulation
We performed FITC Annexin-FITC/PI Apoptosis assay to
assess the effect of RBM10 (R241C) mutation on A549 and H1299 cell
apoptosis. Our data showed that tumor cell apoptosis rate was
reduced after transfection with RBM10 (R241C) mutation
compared with wild type RBM10 (P<0.01; Fig. 2B). Moreover, expression of Fas protein
was decreased in tumor cells after transfection with RBM10 mutated
cDNA compared with that of wild type RBM10 (P<0.01;
Fig. 3).
RBM10 (R241C) mutation promotion of
A549 and H1299 cell invasion through downregulation of E-cadherin
protein
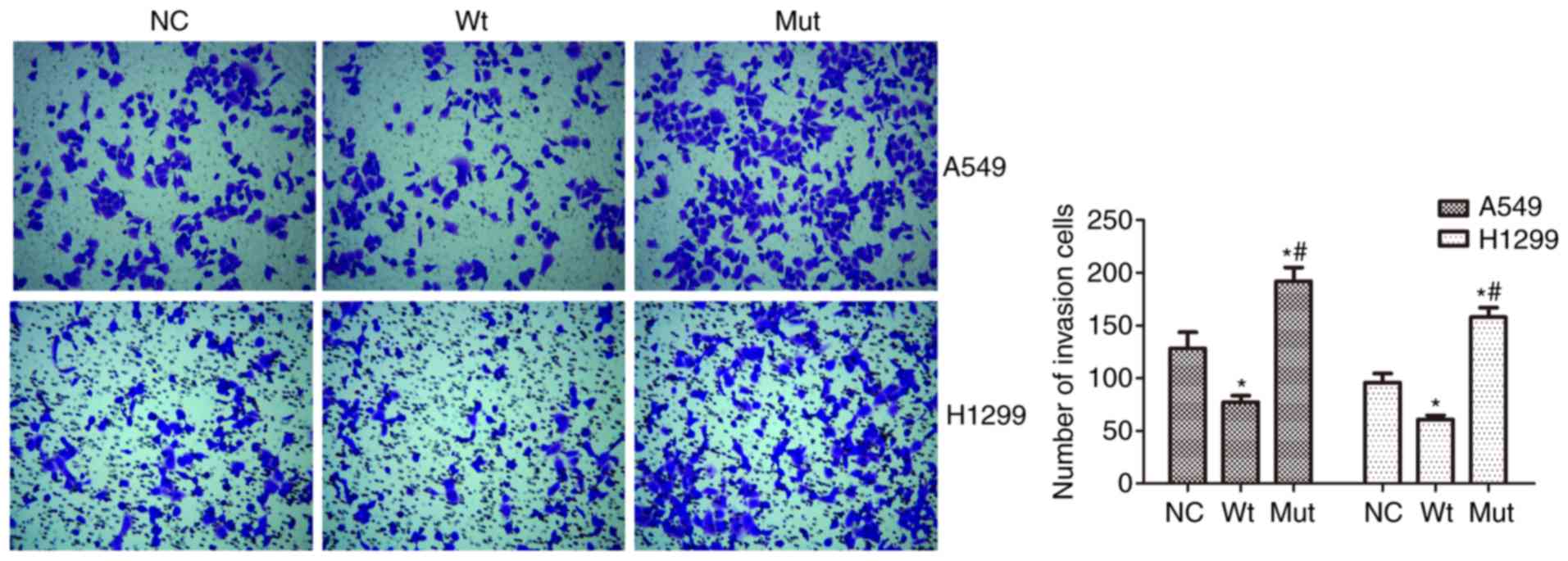
We also assessed the effect of RBM10 (R241C)
mutation on NSCLC cell invasion capacity. Our data showed that
RBM10 (R241C) mutation promoted A549 cell invasion capacity,
whereas RBM10 wild type inhibited tumor cell invasion
(P<0.01; Fig. 4). Moreover,
E-cadherin expression was downregulated in RBM10 (R241C)
mutation group compared with RBM10 wild type and blank
plasmid group (P<0.01; Fig.
3).
Discussion
RBM10 is localized at chromosome
Xp11.23-q13.3 (18), coding a protein
that contains two RNA recognition motifs, two Zinc fingers,
bipartite nuclear localization signals, a glycine (G)-patch, one
arginine/serine-rich domain, and an OCtamerREpeat (OCRE) domain
(19). RBM10 is able to regulate mRNA
alternative splicing and affect protein expression (7,9). Aberrant
RBM10 expression could alter exon-skipping or exon-inclusion of the
affected proteins (7,9). Previous studied showed disruption of
alternative splicing of mRNA in promotion of cancer progression due
to gene transcripts involving in cell proliferation, apoptosis,
DNA-damage response, angiogenesis, and metastasis (9,20). In our
current study, we found that RBM10 exon 10 mutations
occurred in 11 of 50 lung adenocarcinomas and was associated with
tumor stages, lymph node metastasis, and poor survival. Our data
also showed that RBM10 exon 10 mutation at c.763 C>T
significantly promoted lung adenocarcinoma cell proliferation and
invasion capacity in vitro. Future studies are warranted to
investigate RBM 10 protein as a predicative biomarker for
lung adenocarcinoma progression and prognosis.
Indeed, recent studies have shown that RBM10
depletion or mutation was associated with human cancer development,
including lung and pancreatic cancers (13,15).
RBM10 missense somatic and frame shift mutations were
identified in lung adenocarcinoma (15) with a frequency of 8% (14). An in vitro study demonstrated
that RBM10 exon 10 mutation led to enhancement of lung
adenocarcinoma cell proliferation (19). Our current study further confirmed
these findings; however, our data showed a higher frequency of
RBM10 exon 10 mutation (R241C) in lung adenocarcinoma
patients (22%) and such a mutation was associated with advanced
tumor stage, lymph node metastasis, and poor survival of patients.
Furthermore, the TCGA data showed that RBM10 mutations were
enriched in male lung adenocarcinoma (14). In the present study, we found that 10
out of these 11 patients carrying this mutation were male,
confirming the previous data (14).
RBM10 mutations contributed to lung cancer
development (19), but the mechanisms
by which RBM10 mutation promotes lung adenocarcinoma development
remain to be elucidated. To explore the effect of mutant RBM10 on
regulating lung adenocarcinoma cell growth and invasion capacity,
we constructed plasmids carrying mutant RBM10 (R241C) or wild type
RBM10 cDNA and transfected them into lung adenocarcinoma A549 and
H1299 cells. Our data showed that RBM10 exon 10 (R241C) mutation
significantly promoted tumor cell growth and invasion capacity
compared with controls. Lung cancer A549 cells showed activation of
the Notch pathway, which was responsible for increased tumor cell
malignant behaviors (9). However,
loss of Numb resulted in progression of NSCLC through the
activation of Notch protein (17).
We, therefore, investigated the effect of RBM10 mutation on
expression of Notch and Numb proteins and found that Notch
expression in A549 and H1299 cells after transfected with RBM10
mutated cDNA was upregulated compared with that of RBM10 wild type
and vector-only. In contrast, Numb expression was downregulated in
cells transfected with RBM10 mutation as compared with that of with
wild type and blank plasmid. Based on these results, we concluded
that RBM10 mutation promoted NSCLC cell proliferation through
modulation of Numb and Notch expression.
Furthermore, the irrational increase in cell number
is considered one of the hallmarks of carcinogenesis, which occurs
due to either the increase in cell proliferation or defect in cell
death (21). A previous study
reported that p53, Fas, tumor necrosis factor (TNF)-α, and death
receptor (DR)-5 were able to activate an alternative apoptotic
pathway distinct from the well-established apoptotic pathway that
is mediated by cytochrome C (22).
Our current results indicated that the apoptotic rates of RBM10
(R241C) transfected A549 and H1299 cells were significantly
decreased as compared to that of RBM10 wild type and vector-only.
Downregulation of Fas protein expression in tumor cell surface
resulted in evasion of apoptosis (22). For example, in HeLa cells, a cervical
cancer cell line, RBM10 deletion resulted in apoptosis inhibition
and contributed to carcinogenesis and tumor progression by
alternative splicing of Fas (23). In
our current study, we found that expression of Fas protein was
lower in A549 and H1299 cells after transfected with RBM10 (R241C)
compared to that of wild type and vector-only. Although our current
study is just a proof-of-principle, Fas downregulation may be
related to the RBM10 exon 10 mutation and the inhibition of lung
adenocarcinoma cells apoptosis.
In addition, tumor cell migration and invasion
capacity is one of the characteristics during tumor progression and
metastasis (9). During tumorigenesis,
cells lose normal homeostasis control and transform into
premalignant or malignant phenotypes. These alterations are gained
through mutations or loss of cell growth-critical genes or through
epigenetic changes in genomic DNA that could lead to the silence of
tumor suppressor genes or the activation of certain oncogenes
(9). In our current study, we
assessed the significance of RBM10 (R241C) in promoting lung
adenocarcinoma invasion. We confirmed that RBM10 (R241C)
significantly increased lung adenocarcinoma invasion compared to
those transfected with RBM10 wild type and vector-only. At the gene
level, our data showed that E-cadherin expression was reduced after
RBM10 (R241C) transfection in lung adenocarcinoma cells, suggesting
that RBM10 regulated E-cadherin, further confirmed that decrease in
E-cadherin expression contributed to NSCLC cell migration (24).
However, there are several limitations in the
current study; for example, we didn't assess protein levels after
transfection of the wt and mutant RBM10; we didn't investigate the
effect of the RBM10 mutant in alternative splicing although we
detected expression of related proteins.
Our current study confirmed that RBM10 exon 10
(R241C) mutation at p.763 C>T frequently occurred in lung
adenocarcinoma, especially in male patients. RBM10 (R241C) mutation
was associated with AJCC stage, lymph node metastasis, and shorter
5-year survival rate of lung adenocarcinoma patients. Our in
vitro data demonstrated that RBM10 (R241C) mutation induced
lung adenocarcinoma cell proliferation, blocked tumor cell
apoptosis, and promoted tumor cell invasion. However, it remains
unknown why RBM10 mutation frequently occurred in male lung
adenocarcinoma patients. It may be related to the chromosomal
localization of RBM10 gene at Xp11.23-q13.3 (10), since man only carries one X chromosome
and any alteration could show a dominant phenotype.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
XWW conceived and designed the experiments. LLY and
XWW prepared the manuscript. LLY, XMW and ML performed the
experiments. YMX and XFZ interpreted the results. LLY and JL
analyzed the data.
Ethics approval and consent to
participate
The study protocol was approved by the Fourth
Hospital of Jinan and Shanxian Central Hospital Review Board
according to the Declaration of Helsinki. Written informed consent
was obtained from all patients.
Patient consent for publication
Written informed consent was obtained from all
patients for the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Herbst RS, Heymach JV and Lippman SM: Lung
cancer. N Engl J Med. 359:1367–1380. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cooper WA, Lam DC, O'Toole SA and Minna
JD: Molecular biology of lung cancer. J Thorac Dis. 5 Suppl
5:S479–S490. 2013.PubMed/NCBI
|
|
4
|
Hong WK, Bast RC, Hait WN, Kufe DW,
Pollock RE, Weichselbaum RR, Holland JE and Frei E: Chapter 78:
Cancer of the Lung. People's Medical Publishing House; 2010
|
|
5
|
Glisovic T, Bachorik JL, Yong J and
Dreyfuss G: RNA-binding proteins and post-transcriptional gene
regulation. FEBS Lett. 582:1977–1986. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mueller CF, Berger A, Zimmer S, Tiyerili V
and Nickenig G: The heterogenous nuclear riboprotein S1-1 regulates
AT1 receptor gene expression via transcriptional and
posttranscriptional mechanisms. Arch Biochem Biophys. 488:76–82.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang Y, Gogol-Döring A, Hu H, Fröhler S,
Ma Y, Jens M, Maaskola J, Murakawa Y, Quedenau C, Landthaler M, et
al: Integrative analysis revealed the molecular mechanism
underlying RBM10-mediated splicing regulation. EMBO Mol Med.
5:1431–1442. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pan Q, Shai O, Lee LJ, Frey BJ and
Blencowe BJ: Deep surveying of alternative splicing complexity in
the human transcriptome by high-throughput sequencing. Nat Genet.
40:1413–1415. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
David CJ and Manley JL: Alternative
pre-mRNA splicing regulation in cancer: Pathways and programs
unhinged. Genes Dev. 24:2343–2364. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sutherland LC, Thibault P, Durand M,
Lapointe E, Knee JM, Beauvais A, Kalatskaya I, Hunt SC, Loiselle
JJ, Roy JG, et al: Splicing arrays reveal novel RBM10 targets,
including SMN2 pre-mRNA. BMC Mol Biol. 18:192017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ji Y, Xie S, Jiang L, Liu L, Li L, Luo L,
Chen Y, Zhang J, Yu L, Zhang Y, et al: Increased cell apoptosis in
human lung adenocarcinoma and in vivo tumor growth inhibition by
RBM10, a tumor suppressor gene. Oncol Lett. 14:4663–4669. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hernández J, Bechara E, Schlesinger D,
Delgado J, Serrano L and Valcárcel J: Tumor suppressor properties
of the splicing regulatory factor RBM10. RNA Biol. 13:466–472.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Witkiewicz AK, McMillan EA, Balaji U, Baek
G, Lin WC, Mansour J, Mollaee M, Wagner KU, Koduru P, Yopp A, et
al: Whole-exome sequencing of pancreatic cancer defines genetic
diversity and therapeutic targets. Nat Commun. 6:67442015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cancer Genome Atlas Research Network:
Comprehensive molecular profiling of lung adenocarcinoma. Nature.
511:543–550. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Imielinski M, Berger AH, Hammerman PS,
Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M,
Sivachenko A, et al: Mapping the hallmarks of lung adenocarcinoma
with massively parallel sequencing. Cell. 150:1107–1120. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ganti AK, Huang CH, Klein MA, Keefe S and
Kelley MJ: Lung cancer management in 2010. Oncology (Williston
Park). 25:64–73. 2011.PubMed/NCBI
|
|
17
|
Westhoff B, Colaluca IN, D'Ario G,
Donzelli M, Tosoni D, Volorio S, Pelosi G, Spaggiari L, Mazzarol G,
Viale G, et al: Alterations of the Notch pathway in lung cancer.
Proc Natl Acad Sci USA. 106:22293–22298. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Johnston JJ, Teer JK, Cherukuri PF, Hansen
NF, Loftus SK; NIH Intramural Sequencing Center (NISC), ; Chong K,
Mullikin JC and Biesecker LG: Massively parallel sequencing of
exons on the X chromosome identifies RBM10 as the gene that causes
a syndromic form of cleft palate. Am J Hum Genet. 86:743–748. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhao J, Sun Y, Huang Y, Song F, Huang Z,
Bao Y, Zuo J, Saffen D, Shao Z, Liu W and Wang Y: Functional
analysis reveals that RBM10 mutations contribute to lung
adenocarcinoma pathogenesis by deregulating splicing. Sci Rep.
7:404882017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kaida D, Schneider-Poetsch T and Yoshida
M: Splicing in oncogenesis and tumor suppression. Cancer Sci.
103:1611–1616. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Israels LG and Israels ED: Apoptosis.
Oncologist. 4:332–339. 1999.PubMed/NCBI
|
|
23
|
Inoue A, Yamamoto N, Kimura M, Nishio K,
Yamane H and Nakajima K: RBM10 regulates alternative splicing. FEBS
Lett. 588:942–947. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ma K, Fan Y, Dong X, Dong D, Guo Y, Wei X,
Ning J, Geng Q, Wang C, Hu Y, et al: MTA1 promotes epithelial to
mesenchymal transition and metastasis in non-small-cell lung
cancer. Oncotarget. 8:38825–38840. 2017.PubMed/NCBI
|