Introduction
Breast cancer (BC) is the most frequently diagnosed
cancer and the major cause of cancer-related deaths in women
(1). The establishment of adjuvant
therapies including several drugs and radiation has improved the
prognosis of patients with BC. Adjuvant drug therapies are selected
according to the immunohistochemical detection of relevant target
molecules such as estrogen receptor (ER), progesterone receptor
(PgR), and anti-human epidermal growth factor 2 (HER2) in
surgically resected specimens. Although several multigene
expression assays are available to predict patient prognosis and to
evaluate the necessity of adjuvant chemotherapy (2), identifying new molecules related to
conventional biomarkers could contribute to the selection of more
precise treatment strategies.
RAS and EF-hand domain-containing (RASEF) is
a member of the Rab family of GTPases (3). The regulatory mechanism of RASEF
and its role in malignancies have been reported for melanoma
(4,5),
lung cancer (6), esophageal cancer
(7), and myeloid leukemia (8,9). These
studies demonstrated that RASEF has inconsistent roles
depending on tumor type: It can function as an oncogene (6,7) or as a
tumor-suppressor gene (5,8,9). Rab
protein members govern the transportation of substances between
cellular compartments to influence various cell functions (10). In malignant tumors, several Rab
proteins have been reported as important factors in cancer
development and progression (11). In
BC, for example, increased RAB25 was associated with lymphatic
metastasis and poor prognosis (12,13), and
RAB31 is elevated in BC to promote its progression (11). Although various Rab proteins have been
studied in BC, there are no reports that describe the roles of
RASEF.
In the present study, we aimed to investigate the
importance of RASEF expression in BC by evaluating
RASEF mRNA expression in BC cell lines and patient
specimens.
Materials and methods
Sample collection
Thirteen BC cell lines (BT-20, BT-474, BT-549,
HCC1419, HCC1954, Hs578T, MCF7, MDA-MB-231, MDA-MB-361, MDA-MB-415,
MDA-MB-468, SK-BR-3, and ZR-75-1) and two non-cancerous breast
epithelial cell lines (MCF-10A, and MCF-12A) were used in this
study. We purchased BT-549, HCC1419, HCC1954, and Hs578T cell lines
from the Japanese Collection of Research Bioresources Cell Bank
(Osaka, Japan), and BT-474, MCF-7 and MCF-12A were kindly gifted
from Prof. David Sidransky of Johns Hopkins University (Baltimore,
MD, USA). All other cell lines were purchased from the American
Type Culture Collection (Manassas, VA, USA). All cell lines were
cultured in RPMI 1640 (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) supplemented with 10% fetal bovine serum and incubated in
an atmosphere with 5% CO2 at 37°C (14,15).
BC patients who underwent surgery at Nagoya
University Hospital from March 2002 to November 2009 and whose
surveillance data for more than five years after surgery were
available were selected for this study. We collected primary BC
specimens and clinical data from 167 patients in total. The
specimens were resected approximately to 1.5 mm in diameter, and
frozen immediately at −80°C. We resected non-cancerous specimens at
>3 cm away from the edge of the tumor. The resected BC specimens
were diagnosed histologically as BC and classified using the Union
for International Cancer Control (UICC) staging system for BC (7th
edition). Administration of adjuvant medication therapy was
determined by physician discretion considering each patient's
general condition, pathological feature and subtype (16,17).
The present study complies with the Declaration of
Helsinki and was approved by our institutional review board
(approval no: 2016-0224). Participants granted written informed
consent for use of clinical samples and data.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
We evaluated RASEF mRNA expression levels by
RT-qPCR. RNA was extracted from cell lines (8.0×106
cells per cell line), and BC and non-cancerous specimens from 167
patients. cDNA was synthesized as previously described (16,17).
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA levels
were evaluated for normalizing RASEF mRNA expression levels.
RASEF-specific primers were: Forward
5′-ATCAGACTTCAAAGCACAGAAATGG-3′ and reverse
5′-TTCCTCTTCCAACTCACTCAACTG-3′, which generated a 96-bp product.
GAPDH-specific primers were: Forward
5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse 5′-GAAGATGGTGATGGGATTTC-3′,
which generated a 226-bp product. We used a SYBR Green PCR core
reagents kit (Applied Biosystems; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) for RT-qPCR with these cycling conditions: One
cycle at 95°C for 10 min, followed by 40 cycles at 95°C for 5 sec,
and 60°C for 60 sec, using an ABI StepOnePlus real-time PCR System
(Applied Biosystems; Thermo Fisher Scientific, Inc.). All samples
were assayed in triplicate. The mRNA expression level of
RASEF in each sample was obtained from the RASEF
value divided by GAPDH value (18,19).
Statistical analysis
Differences in the levels of RASEF mRNA
between two groups were evaluated with the Mann-Whitney test. When
they were compared between multiple groups, ANOVA with Tukey's
post-hoc test was performed. We analyzed the association between
RASEF mRNA expression levels and patient clinicopathological
factors using the χ2 test. We utilized the Kaplan-Meier
method for evaluating disease-free survival (DFS) and overall
survival (OS) rates; the survival curves were compared using the
log-rank test. Patients' RASEF expression levels were
divided into quartiles with low RASEF levels being taken as
the lowest quartile. JMP 12 (SAS Institute, Inc., Cary, NC, USA)
was exploited for the statistical analysis, and P<0.05 was
considered to indicate a statistically significant difference.
Results
RASEF mRNA expression levels in BC
cell lines
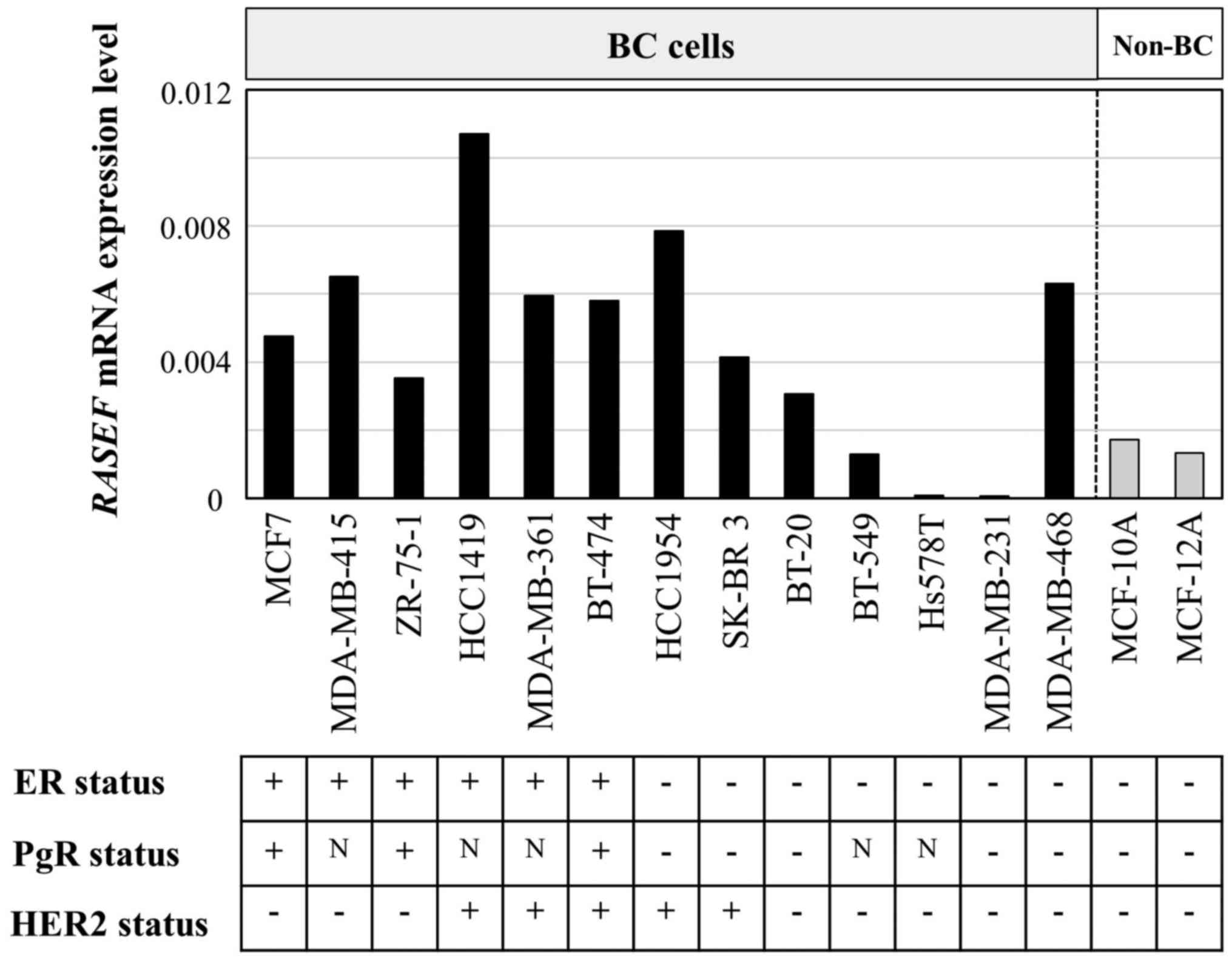
We evaluated the levels of RASEF mRNA
expression in 13 BC cell lines and two non-cancerous cell lines of
mammary gland (Fig. 1). The ER, PgR,
and HER2 statuses of the cell lines have been evaluated in previous
studies (20,21). All ER-positive BC cell lines expressed
higher levels of RASEF mRNA than non-BC cell lines. Although
RASEF mRNA expression levels did not significantly differ
among ER-positive and -negative (P=0.083), PgR-positive and
-negative (P=0.833), or HER2-positive and -negative (P=0.053)
cells, the expression levels in ER-negative/HER2-negative BC cell
lines were lower than in other cell lines (P=0.014).
Patient characteristics
A total of 167 BC patients were enrolled in the
present study and all were women. The mean age (± standard
deviation) was 54.4±11.6 years (range, 26–78 years). The UICC stage
distribution was as follows: Stage 0, seven patients; stage I, 47
patients; stage II, 78 patients; stage III, 34 patients; and stage
IV, one patient. The median follow-up duration was 100.0 months
(range, 8–155 months) or until death. The conventional biomarkers
status determined from immunohistochemistry tests in primary tumors
was as follows: ER-positive, n=127; ER-negative, n=40;
PgR-positive, n=115; PgR-negative, n=52; HER2-positive, n=39;
HER2-negative, n=119 (HER2 data missing for nine patients);
triple-negative, n=18; and non-triple-negative, n=148 (data missing
for one patient). The patients who expressed at least one molecule
among ER, PgR, and HER2 were defined as ‘non-triple-negative’.
Because eight patients among nine whose HER2 statuses were unknown
showed ER-positivity, they were categorized as
non-triple-negative.
Association between RASEF mRNA
expression level and patient clinicopathological factors
In 78 (47%) of the 167 patients, BC specimens
expressed lower RASEF mRNA levels than non-cancerous
specimens. RASEF mRNA expression levels did not differ
between Tis (carcinoma in situ)/T1 (n=77) and T2/T3/T4
(n=90; P=0.337), lymph node metastasis-positive (n=82) and
-negative (n=85; P=0.326), or stage 0/I (n=54) and stage II/III/IV
(n=113; P=0.075) disease.
When we investigated conventional biomarkers, we
found that ER-negative specimens (n=40) exhibited significantly
lower RASEF mRNA expression levels than ER-positive
specimens (n=127; P<0.001; Fig.
2A). PgR-negative specimens (n=52) also exhibited lower
RASEF mRNA expression levels than PgR-positive specimens
(n=115; P<0.001). Additionally, RASEF mRNA expression
levels were significantly lower in triple-negative specimens (n=18)
than in non-triple-negative specimens (n=148; P<0.001; data
missing for one patient). The expression levels between
HER2-positive (n=39) and -negative specimens (n=119) did not differ
significantly (P=0.180; data missing for nine patients). When we
focused on RASEF mRNA levels among ER-positive/PgR-positive
(n=115), ER-positive/PgR-negative (n=12), and
ER-negative/PgR-negative specimens (n=40), we found that
ER-negative/PgR-negative specimens exhibited significantly lower
RASEF mRNA expression levels than ER-positive/PgR-positive
specimens (P<0.001; Fig. 2B).
ER-positive/PgR-negative specimens tended to have lower
RASEF mRNA expression levels than ER-positive/PgR-positive
specimens, although there was no significant difference
(P=0.086).
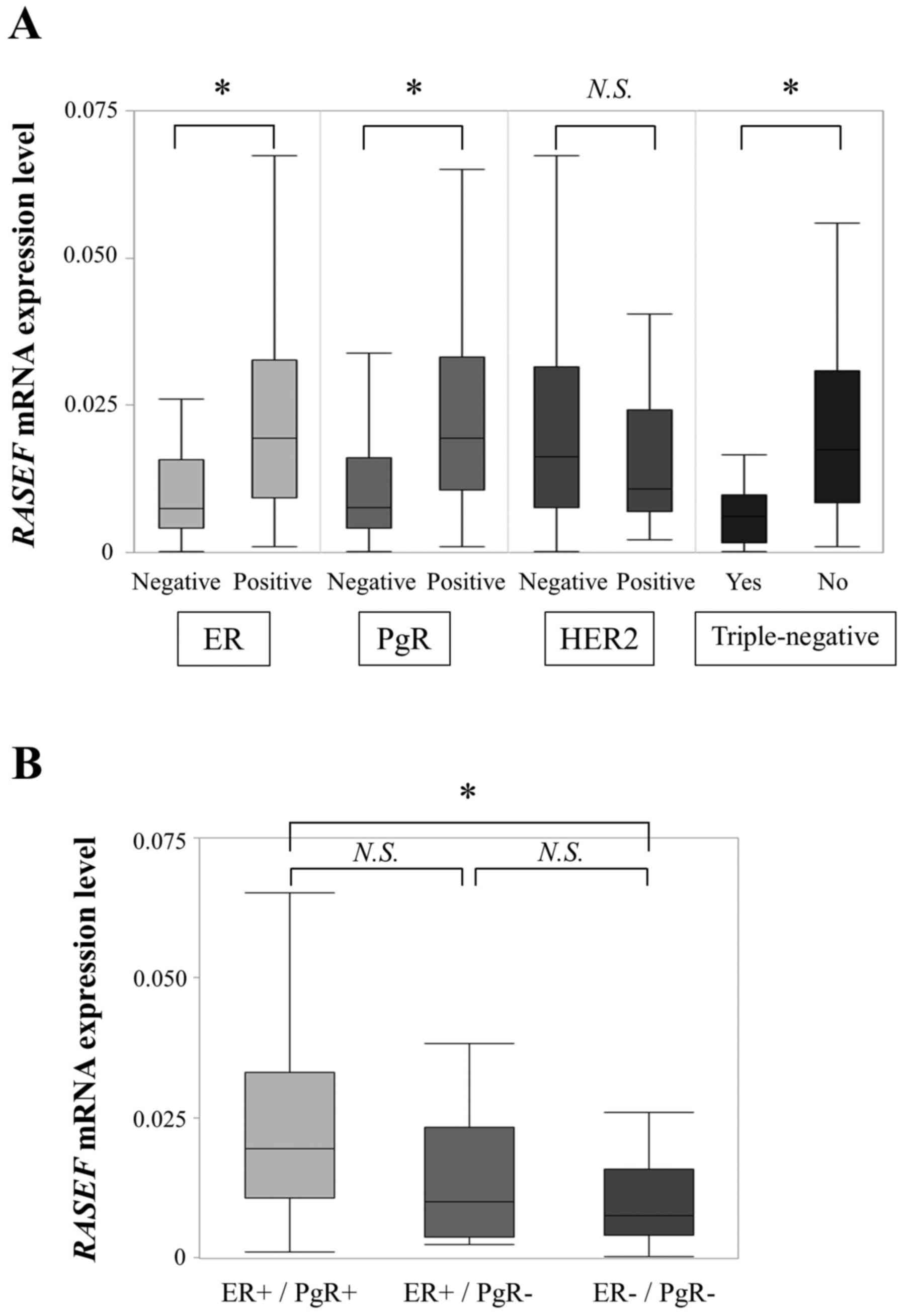 | Figure 2.(A) Correlation between expression of
RASEF mRNA and conventional biomarkers. RASEF mRNA
levels were significantly lower in ER-negative, PgR-negative, and
triple-negative specimens than in ER-positive, PgR-positive, and
non-triple-negative specimens. (B) Comparison of RASEF mRNA
levels among ER-positive/PgR-positive, ER-positive/PgR-negative,
and ER-negative/PgR-negative groups. ER-negative/PgR-negative
specimens exhibited significantly lower RASEF mRNA
expression than ER-positive/PgR-positive specimens. *P<0.001.
RASEF, RAS and EF-hand domain-containing; N.S, not
significant; ER, estrogen receptor; PgR, progesterone receptor;
HER2, human epidermal growth factor 2. |
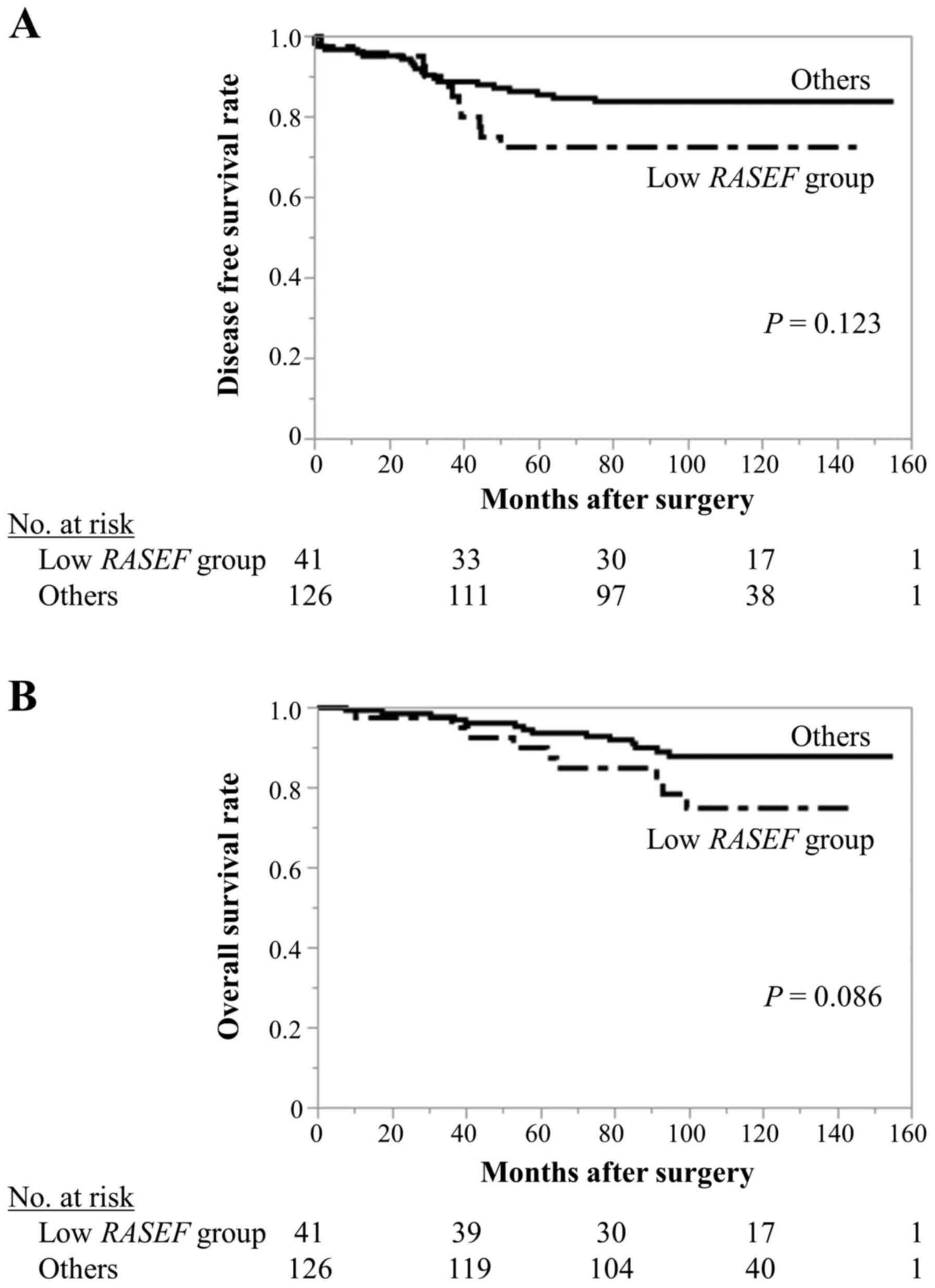
Patients with RASEF expression levels in the
lowest quartile were distributed into a ‘low RASEF group’
(n=41), and the remaining patients were designated as ‘others’
(n=126). The low RASEF group was associated with more
advanced UICC T factor (P=0.031; Table
I), and ER-negative (P<0.001), PgR-negative (P<0.001),
and triple-negative status (P<0.001). Although the differences
were not statistically significant, the low RASEF group
tended to have poorer DFS (5-year DFS rates, low RASEF
group: 72.6%; others: 85.6%; P=0.123; Fig. 3A) and OS (5-year OS rates, low
RASEF group: 90.1%; others: 93.5%; P=0.086; Fig. 3B).
 | Table I.Associations between RASEF
mRNA expression and clinicopathological characteristics of 167
patients with BC. |
Table I.
Associations between RASEF
mRNA expression and clinicopathological characteristics of 167
patients with BC.
| Clinicopathological
parameters | Low RASEF group
(n=41) | Others (n=126) | P-value |
|---|
| Age (years) |
|
| 0.847 |
|
≤60 | 26 | 82 |
|
|
>60 | 15 | 44 |
|
| Histology |
|
| 0.231 |
|
DCIS | 1 | 6 |
|
|
IDC | 39 | 109 |
|
|
ILC | 0 | 6 |
|
|
Others | 1 | 5 |
|
| UICC T factor |
|
| 0.031a |
|
Tis/T1 | 13 | 64 |
|
|
T2/T3/T4 | 28 | 62 |
|
| Node status |
|
| 0.164 |
|
Negative | 17 | 68 |
|
|
Positive | 24 | 58 |
|
| UICC pathological
stage |
|
| 0.413 |
|
0/I/II | 34 | 97 |
|
|
III/IV | 7 | 29 |
|
| ER status |
|
|
<0.001a |
|
Positive | 21 | 106 |
|
|
Negative | 20 | 20 |
|
| PgR status |
|
|
<0.001a |
|
Positive | 17 | 98 |
|
|
Negative | 24 | 28 |
|
| HER2 status |
|
| 0.873 |
|
Positive | 10 | 29 |
|
|
Negative | 29 | 90 |
|
|
Unknown | 2 | 7 |
|
|
Triple-negative |
|
|
<0.001a |
|
Yes | 13 | 5 |
|
| No | 27 | 121 |
|
|
Unknown | 1 | 0 |
|
| Adjuvant
therapy |
|
|
0.005a |
|
Endocrine therapy alone | 7 | 50 |
|
|
Chemotherapy alone | 14 | 16 |
|
|
Endocrine and
chemotherapy | 15 | 49 |
|
|
None | 5 | 11 |
|
Discussion
In this study, we demonstrated that low RASEF
mRNA expression levels were associated with negative hormone
receptor status.
RASEF is a member of the Rab GTPase protein family
and contains a Rab GTPase domain in its C-terminal region.
Uniquely, RASEF has 2 EF-hand domains in the N-terminal region,
which are important for binding to calcium ions, and an internal
coiled-coil motif (3). The Rab
protein family consists of 70 Rab proteins, and they govern the
transportation of various molecules among cellular compartments
(10). Recently, several Rab proteins
have been revealed to contribute to cancer development and
progression, and some have been focused on as novel therapeutic
targets (11). In BC, RAB25 was shown
to promote epithelial-mesenchymal transition (22), and its expression was associated with
more aggressive stage and poor prognosis (12,13). In
addition, FIP1C, an effector of RAB11, promoted lysosomal
degradation of HER2 to suppress tumor progression. Despite these
studies of Rab proteins, there are no reports that describe RASEF
in BC. Several previous reports have described the roles of
RASEF in malignant tumors. Interestingly, RASEF has
shown inconsistent behavior in different studies: It has been
reported to function as an oncogene and as a tumor suppressor gene.
Oshita et al (6), showed that
RASEF protein expression was positively associated with poor
prognosis in non-small cell lung cancer. They demonstrated that
RASEF interacted with extracellular signal-regulated kinase (ERK)
1/2 and enhanced ERK 1/2 signaling. Another study used cDNA
microarray to demonstrate that RASEF was overexpressed in
esophageal squamous carcinoma compared with non-cancerous tissues
(7). Conversely, Maat et al
(5) identified RASEF as a
tumor suppressor regulated by epigenetic mechanisms in uveal
melanoma. They revealed that missense mutation and methylation of
the RASEF gene is related to poor survival. In the present
study, we aimed to clarify the significance of RASEF
expression in BC patients.
Regarding RASEF mRNA expression levels in BC
and non-cancerous mammary cell lines, ER-positive BC cell lines
expressed higher RASEF mRNA levels than non-BC cell lines,
and ER-negative/HER2-negative BC cell lines expressed low
RASEF mRNA levels. Because of the small sample size, there
were no significant differences between ER-positive and ER-negative
BC cell lines or PgR-positive and PgR-negative BC cell lines.
When patient data was analyzed, although there were
no significant differences, patients with RASEF expression
levels in the lowest quartile (designated the ‘low RASEF
group’) tended to experience poorer DFS and OS. In this study,
adjuvant therapy was administered to most patients, which might
have abated the impact of RASEF mRNA expression. As a
possible explanation for poor prognosis, we found that low
RASEF expression correlated with ER-negative, PgR-negative
and triple-negative status. These cancers are known to be more
aggressive and to result in poorer survival than ER-positive,
PgR-positive, and non-triple-negative cancers (23–26).
Nakamura et al (8), suggested
that RASEF overexpression induced caspases-3 and −9, and increased
p38 phosphorylation levels, which induced apoptosis and inhibited
proliferation of chronic myeloid leukemia progenitor cells. Among
these molecules, caspase-9 is the apoptotic initiator protease of
the apoptotic pathway (27). p38, a
mitogen-activated protein kinase, is an important mediator of
signal transduction for cell survival and apoptosis (28). PgR-positive status in BC has been
reported to correlate with high phosphorylated p38 expression
(29). In our results, low
RASEF expression was associated with advanced T-stage.
RASEF may play tumor suppressive roles by suppressing the
proliferation and promoting the apoptosis of BC cells.
ER-positive/PgR-negative specimens tended to exhibit
lower RASEF mRNA levels than ER-positive/PgR-positive
specimens, although this difference was not significant.
ER-positive/PgR-positive BC is likely to belong to the ‘luminal
A-like’ subtype, and ER-positive/PgR-negative BC tends to be belong
to the ‘luminal B-like’ subtype (2).
Recently, Ki-67 has been widely used to distinguish these two
subtypes. However, the threshold for Ki-67 scoring remains
controversial. RASEF expression might help to discriminate
the ‘luminal A-like’ and ‘luminal B-like’ subtypes.
This is the first study to demonstrate an
association between RASEF mRNA expression and
clinicopathological characteristics in BC patients. These findings
may potentially be applied to clinical use in the future. For
example, RASEF levels in surgically resected samples might
aid evaluation of BC subtypes and facilitate selection of adjuvant
medication. However, this study has some limitations. Because the
functional role of RASEF in BC cells has not been
elucidated, further in vitro experiments are needed to
determine how RASEF interacts with hormone receptor status.
Additionally, this is a retrospective study. Evaluation of a large
number of patients or a prospective study is warranted to
investigate the potential clinical applications of our
findings.
To conclude, RASEF mRNA expression levels of
cell lines and the association between RASEF and BC patient
specimens were evaluated in this study. We demonstrated an
association between RASEF mRNA expression levels and hormone
receptor status in BC specimens. Low RASEF mRNA expression
is likely to reflect ER-negative and PgR-negative status.
Acknowledgements
The authors would like to thank Professor David
Sidransky, the director of the Otolaryngology Department (Head and
Neck Surgery of Johns Hopkins University School of Medicine,
Baltimore, MD, USA) for providing the BT-474, MCF-7, and MCF-12A
cell lines.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
MK conceived and designed this study. MS conducted
the experiments, analyzed the data and wrote the manuscript. MH
provided cell lines. TI, NM, YA, YT, KN, DT, SN and TK collected
the patients’ samples and acquired clinical data. DS, HT, SU, TM,
MH, YK and TK interpreted the experimental data and revised the
manuscript.
Ethics approval and consent to
participate
The present study was approved by the institutional
review board of Nagoya University Graduate School of Medicine
(reference number: 2016-0224). Written informed consent was
obtained from participants for the use of samples and data.
Patient consent for publication
Participants in this study granted written informed
consent for publication required by the institutional review
board.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
BC
|
breast cancer
|
|
DCIS
|
ductal carcinoma in situ
|
|
DFS
|
disease-free survival
|
|
ERK
|
extracellular signal-regulated
kinase
|
|
ER
|
estrogen receptor
|
|
GAPDH
|
glyceraldehyde-3-phosphate
dehydrogenase
|
|
HER2
|
human epidermal growth factor 2
|
|
IDC
|
invasive ductal carcinoma
|
|
ILC
|
invasive lobular carcinoma
|
|
OS
|
overall survival
|
|
PgR
|
progesterone receptor
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
RASEF
|
RAS and EF-hand domain-containing
|
|
Tis
|
carcinoma in situ
|
|
UICC
|
Union for International Cancer
Control
|
References
|
1
|
Jemal A, Center MM, DeSantis C and Ward
EM: Global patterns of cancer incidence and mortality rates and
trends. Cancer Epidemiol Biomarkers Prev. 19:1893–1907. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Coates AS, Winer EP, Goldhirsch A, Gelber
RD, Gnant M, Piccart-Gebhart M, Thürlimann B and Senn HJ; Panel
Members, . Tailoring therapies-improving the management of early
breast cancer: St Gallen International Expert Consensus on the
Primary Therapy of Early Breast Cancer 2015. Ann Oncol.
26:1533–1546. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shintani M, Tada M, Kobayashi T, Kajiho H,
Kontani K and Katada T: Characterization of Rab45/RASEF containing
EF-hand domain and a coiled-coil motif as a self-associating
GTPase. Biochem Biophys Res Commun. 357:661–667. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kaplon J, Hömig-Hölzel C, Gao LD, Meissl
K, Verdegaal EME, van der Burg SH, Doorn RV and Peeper DS:
Near-genomewide RNAi screening for regulators of
BRAFV600E-induced senescence identifies RASEF, a gene
epigenetically silenced in melanoma. Pigment Cell Melanoma Res.
27:2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Maat W, Beiboer SH, Jager MJ, Luyten GP,
Gruis NA and van der Velden PA: Epigenetic regulation identifies
RASEF as a tumor-suppressor gene in uveal melanoma. Invest
Ophthalmol Vis Sci. 49:1291–1298. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Oshita H, Nishino R, Takano A, Fujitomo T,
Aragaki M, Kato T, Akiyama H, Tsuchiya E, Kohno N, Nakamura Y and
Daigo Y: RASEF is a novel diagnostic biomarker and a therapeutic
target for lung cancer. Mol Cancer Res. 11:937–951. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang X, Lin P, Zhu ZH, Long H, Wen J,
Yang H, Zhang X, Wang DF, Fu JH, Fang Y and Rong TH: Expression
profiles of early esophageal squamous cell carcinoma by cDNA
microarray. Cancer Genet Cytogenet. 194:23–29. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nakamura S, Takemura T, Tan L, Nagata Y,
Yokota D, Hirano I, Shigeno K, Shibata K, Fujie M, Fujisawa S and
Ohnishi K: Small GTPase RAB45-mediated p38 activation in apoptosis
of chronic myeloid leukemia progenitor cells. Carcinogenesis.
32:1758–1772. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sweetser DA, Peniket AJ, Haaland C,
Blomberg AA, Zhang Y, Zaidi ST, Dayyani F, Zhao Z, Heerema NA,
Boultwood J, et al: Delineation of the minimal commonly deleted
segment and identification of candidate tumor-suppressor genes in
del(9q) acute myeloid leukemia. Genes Chromosomes Cancer.
44:279–291. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang S, Hu C, Wu F and He S: Rab25 GTPase:
Functional roles in cancer. Oncotarget. 8:64591–64599.
2017.PubMed/NCBI
|
|
11
|
Chua CE and Tang BL: The role of the small
GTPase Rab31 in cancer. J Cell Mol Med. 19:1–10. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cheng KW, Lahad JP, Kuo WL, Lapuk A,
Yamada K, Auersperg N, Liu J, Smith-McCune K, Lu KH, Fishman D, et
al: The RAB25 small GTPase determines aggressiveness of ovarian and
breast cancers. Nat Med. 10:1251–1256. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yin YX, Shen F, Pei H, Ding Y, Zhao H,
Zhao M and Chen Q: Increased expression of Rab25 in breast cancer
correlates with lymphatic metastasis. Tumour Biol. 33:1581–1587.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kanda M, Shimizu D, Fujii T, Tanaka H,
Shibata M, Iwata N, Hayashi M, Kobayashi D, Tanaka C, Yamada S, et
al: Protein arginine methyltransferase 5 is associated with
malignant phenotype and peritoneal metastasis in gastric cancer.
Int J Oncol. 49:1195–1202. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kanda M, Shimizu D, Nomoto S, Takami H,
Hibino S, Oya H, Hashimoto R, Suenaga M, Inokawa Y, Kobayashi D, et
al: Prognostic impact of expression and methylation status of
DENN/MADD domain-containing protein 2D in gastric cancer. Gastric
Cancer. 18:288–296. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shibata M, Kanda M, Shimizu D, Tanaka H,
Umeda S, Hayashi M, Inaishi T, Miyajima N, Adachi Y, Takano Y, et
al: Expression of regulatory factor X1 can predict the prognosis of
breast cancer. Oncol Lett. 13:4334–4340. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shibata M, Kanda M, Tanaka H, Umeda S,
Miwa T, Shimizu D, Hayashi M, Inaishi T, Miyajima N, Adachi Y, et
al: Overexpression of Derlin 3 is associated with malignant
phenotype of breast cancer cells. Oncol Rep. 38:1760–1766. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kanda M, Nomoto S, Oya H, Takami H,
Shimizu D, Hibino S, Hashimoto R, Kobayashi D, Tanaka C, Yamada S,
et al: The expression of melanoma-associated antigen D2 both in
surgically resected and serum samples serves as clinically relevant
biomarker of gastric cancer progression. Ann Surg Oncol. 23 Suppl
2:S214–S221. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kanda M, Shimizu D, Fujii T, Sueoka S,
Tanaka Y, Ezaka K, Takami H, Tanaka H, Hashimoto R, Iwata N, et al:
Function and diagnostic value of Anosmin-1 in gastric cancer
progression. Int J Cancer. 138:721–730. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Finn RS, Dering J, Conklin D, Kalous O,
Cohen DJ, Desai AJ, Ginther C, Atefi M, Chen I, Fowst C, et al: PD
0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially
inhibits proliferation of luminal estrogen receptor-positive human
breast cancer cell lines in vitro. Breast Cancer Res. 11:R772009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Subik K, Lee JF, Baxter L, Strzepek T,
Costello D, Crowley P, Xing L, Hung MC, Bonfiglio T, Hicks DG and
Tang P: The expression patterns of ER PR, HER2, CK5/6, EGFR, Ki-67
and AR by immunohistochemical analysis in breast cancer cell lines.
Breast Cancer (Auckl). 4:35–41. 2010.PubMed/NCBI
|
|
22
|
Mitra S, Federico L, Zhao W, Dennison J,
Sarkar TR, Zhang F, Takiar V, Cheng KW, Mani S, Lee JS and Mills
GB: Rab25 acts as an oncogene in luminal B breast cancer and is
causally associated with Snail driven EMT. Oncotarget.
7:40252–40265. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Agurs-Collins T, Dunn BK, Browne D,
Johnson KA and Lubet R: Epidemiology of health disparities in
relation to the biology of estrogen receptor-negative breast
cancer. Semin Oncol. 37:384–401. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cui XJ, Schiff R, Arpino G, Osborne CK and
Lee AV: Biology of progesterone receptor loss in breast cancer and
its implications for endocrine therapy. J Clin Oncol. 23:7721–7735.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Thakkar JP and Mehta DG: A review of an
unfavorable subset of breast cancer: Estrogen receptor positive
progesterone receptor negative. Oncologist. 16:276–285. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yao H, He G, Yan S, Chen C, Song L, Rosol
TJ and Deng X: Triple-negative breast cancer: Is there a treatment
on the horizon? Oncotarget. 8:1913–1924. 2017.PubMed/NCBI
|
|
27
|
Kim B, Srivastava SK and Kim SH: Caspase-9
as a therapeutic target for treating cancer. Expert Opin Ther
Targets. 19:113–127. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pearson G, Robinson F, Beers Gibson T, Xu
BE, Karandikar M, Berman K and Cobb MH: Mitogen-activated protein
(MAP) kinase pathways: Regulation and physiological functions.
Endocr Rev. 22:153–183. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang B, Jiang H, Ma N and Wang Y:
Phosphorylated-p38 mitogen-activated protein kinase expression is
associated with clinical factors in invasive breast cancer.
Springerplus. 5:9342016. View Article : Google Scholar : PubMed/NCBI
|