Introduction
Osteosarcoma (OS) is a commonly and highly invasive
primary malignant mesenchymal tumors in bone, which usually
affecting the metaphysis of long bones of adolescents and young
adults (1,2). Over the past decades, although early
diagnosis and effective treatment have been achieved, neoadjuvant
and adjuvant chemotherapy coupled with limb-sparing surgery remains
the most effective regimens. The 5-year survival rate of patients
is still very poor due to recurrent or metastatic OS (3). Since the cause of OS is associated with
abnormal genetic and epigenetic changes, the identification of
potential effector molecules targeting key oncogenes are vital for
OS treatment (4,5). Several studies have investigated the
genes associated with proliferation and metastasis in OS. Among
them, CDH2, N-cadherin has become a new research hotspot in
gene therapy. Gain-of-function of N-cadherin was considered as a
universal hallmark of epithelial-mesenchymal transition (6,7).
N-cadherin was reported to promote tumorigenesis during different
cancer progression such as esophageal cancer (8), breast cancer (9), germ cell tumours (10), thyroid cancer (11). Although it is well known that
interfering of N-cadherin function may prove beneficial in multiple
cancers, the exact regulation about N-cadherin in OS remains
largely unknown.
SIRT6 is a member of the class III histone
deacetylase family (12,13). During idiopathic pulmonary fibrosis,
SIRT6 could inhibit epithelial to mesenchymal transition (14). SIRT6 was reported to function as a
tumor suppressor or oncogene depend on the types of cancer, which
indicated SIRT6 was at the crossroads of multiple pathways
(15–17). Until now, there is no report about the
function of SIRT6 in OS. In the present study, we revealed the
exact role of SIRT6 in the regulation of OS cell proliferation and
invasion in vitro. We also investigated the associated
underlying mechanisms, focusing on the repression of N-cadherin in
OS cells.
Materials and methods
Cell culture and reagents
The human OS cell lines, SAOS-2, MG-63, U2OS and one
osteoblastic cell line hFOB were purchased from the American Type
Culture Collection (Manassas, VA, USA) and incubated in RPMI-1640
medium (GE Healthcare, Chicago, IL, USA) supplement with 10% fetal
bovine serum (FBS; Thermo Fisher Scientific, Inc., Waltham, MA,
USA). 100 U/ml penicillin and 100 µg/ml streptomycin was added to
the medium. All cells were incubated in a humidified atmosphere at
37°C with 5% CO2. Lipofectamine 2000 (Thermo Fisher
Scientific, Inc.) was used for transfection, according to the
manufacturer's instructions.
Ethics statement and tissues
samples
The Medical Ethics Committee of Nanjing University
of Traditional Chinese Medicine had approved this research, and
written informed consent was obtained from all patients. Primary OS
tissues samples were collected from 112 patients who were diagnosed
with OS based on histopathological evaluation and underwent
surgical treatment in our hospital from 2008 to 2013 (Table I). The clinical stage was classified
according to the sixth edition of the TNM classification of the
International Union Against Cancer (UICC). Prior anticancer
treatment (radiotherapy or chemotherapy) were excluded, the
biopsies were frozen in liquid nitrogen immediately and stored at
−80°C until use. All OS patients were studied in a follow-up for
overall survival.
 | Table I.Clinicopathologic variables in 112
osteosarcoma cancer patients. |
Table I.
Clinicopathologic variables in 112
osteosarcoma cancer patients.
|
|
| SIRT6 expression |
|
|---|
|
|
|
|
|
|---|
| Variables | No. (n=112) | Low (n=72) | High (n=40) | P-value |
|---|
| Age (years) |
|
|
| 0.352 |
|
<40 | 55 | 33 | 22 |
|
| ≥40 | 57 | 39 | 18 |
|
| Sex |
|
|
| 0.284 |
| Male | 54 | 32 | 22 |
|
|
Female | 58 | 40 | 18 |
|
| Histological
grade |
|
|
| 0.002 |
| I–II | 48 | 23 | 25 |
|
| III | 64 | 49 | 15 |
|
| Enneking staging |
|
|
| 0.032 |
| I–II | 52 | 28 | 24 |
|
| III | 60 | 44 | 16 |
|
| Metastasis |
|
|
| <0.001 |
| Yes | 57 | 46 | 11 |
|
| No | 55 | 26 | 29 |
|
Lentiviral transduction
The relative lentivirus was purchased from Shanghai
Genepharma, and transduced into the target cells with 8 µg/ml
polybrene (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). 48 h
after incubation, the cells were selected using puromycin (2.0
µg/ml; Sigma-Aldrich; Merck KGaA). Following puromycin selection
for 48 h, the cells were harvest and used for RT-qPCR or western
blot analysis.
RNA extraction and RT-qPCR
Total RNA was extracted from the relative cells or
tissues using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions. 2
µg RNA was transcribed into cDNA using iScript cDNA Synthesis kit
(Bio-Rad Laboratories Inc., Hercules, CA, USA). RT-qPCR was
performed on the Applied Biosystems 7500 Real-Time PCR System
(Thermo Fisher Scientific, Inc.). The expression of GAPDH was used
as a control. The primers for N-cadherin were forward,
5′-GTCAGCAGAAGTTGAAGAAATAGTG-3′ and reverse,
5′-GCAAGTTGATTGGAGGGATG-3′. The primers for SIRT6 were forward,
5′-CTTTGTTACTTGTTTCTGTCCC-3′ and reverse,
5′-GACAACACAGCAAGTCAGAG-3′. The primers for GAPDH were forward,
5′-TGGGTGTGAACCATGAGAAGT-3′ and reverse,
5′-TGAGTCCTTCCACGATACCAA-3′. Relative gene expression was
calculated according to the 2−∆∆Cq method (18).
Western blott analysis
Proteins were extracted from cells using the lysis
buffer (Beyotime Institute of Biotechnology, Nanjing, China) in the
presence of a proteinase inhibitor cocktail (Complete Mini; Roche
Diagnostics, Basel, Switzerland). After centrifugation at 15,000 g
for 15 min at 4°C, the supernatants were quantified using Bradford
assay reagent (Thermo Fisher Scientific International Inc.) 30 µg
proteins in each group were separated on 10% SDS-PAGE and then
blotted onto a polyvinylidene difluoride (PVDF) membrane (GE
Healthcare). After blocked with 5% non-fat dried milk in TBST
(Sigma-Aldrich; Merck KGaA) overnight at 4°C. The membrane was
incubated with specific primary antibodies at room temperature for
3 h, respectively.
Anti-β-actin (cat no. 612656; 1:2,000; BD
Biosciences, Franklin Lakes, NJ, USA). Anti-SIRT6 (ab 62739;
1:1,000; polyclonal rabbit); Anti-N-cadherin (ab18203; 1:1,000;
polyclonal rabbit; both Abcam, Cambridge, MA, USA). HRP-linked goat
anti-rabbit (cat no. 7074) or goat anti-mouse secondary antibodies
(cat no. 7076), both from Cell Signaling Technology, Inc. were used
for incubation for 1 h at 37°C. The chemiluminescent horseradish
peroxidase substrate (Merck KGaA, Darmstadt, Germany) was used to
detect the signals according to the manufacturer's
instructions.
MTT assay
Cell proliferation was performed using MTT assay
(Dojindo, Kumamoto, Japan), according to the manufacturer's
instructions. Briefly, the cells were plated in 96-well plates.
5×103 cells each well were harvested at 24, 48, 72 and
96 h. 10 µl CCK-8 solution was added to each well with 100 µl
RPMI-1640. The plates were further incubated at 37°C for 2 h and
the absorbance at 490 nm was measured using a microplate reader
(Infinite Pro 2000; Tecan GmbH, Grödig, Austria). All experiments
were performed at least three times.
Cell invasion assay
To assess the role of SIRT6 in cell invasion,
Transwell assay was performed. Transwell chambers were purchased
from Merck KGaA (24-well; 8 µm). The filter of the upper chamber
was coated with 50 µl of diluted Matrigel (BD Biosciences),
according to the manufacturer's instructions. The lower chambers
were filled with 600 µl of RPMI-1640 with 10% FBS. A total of
1×105 cells were suspended in 100 µl serum free
RPMI-1640 medium and added into each top chamber. After the cells
were incubated for 10 h at 37°C in a 5% CO2 chamber, the
non-invaded cells were removed with a cotton swab.
The invasive cells on the lower surface of the
membrane were were fixed with 4% paraformaldehyde for 25 min and
then stained with 0.1% crystal violet for 10 min. The invasive
cells were counted under a microscope (Leica DM 5000 B; Leica
Microsystems, Wetzlar, Germany) in five random selected fields. The
experiments were repeated at least three times and results were
expressed as the relative fold change over vector.
qChIP assay
qChIP was performed using kit from Upstate
Biotechnology according to manufacturer's instructions, Sybr-Green
(Molecular Probes, Eugene, OR, USA) was used as a marker for DNA
amplification on the ABI 7500 system.
Luciferase reporter assay
Luciferase reporter assay was used to clarify
whether N-cadherin was a direct target of SIRT6. The promoter of
CDH2 was cloned into the pRL-TK report vector (Thermo Fisher
Scientific, Inc.). The relative cells were transfected with the
wild-type or mutant type promoter of CDH2 and SIRT6 or Si-SIRT6,
using Lipofectamine 2000, respectively. 48 h later, luciferase
activity was determined using a dual luciferase reporter assay
(Promega Corp., Madison, WI, USA) and LD400 luminometer (Beckman
Coulter, Inc., Brea, CA, USA). Data was presented as the ratio of
Renilla luciferase to that of the firefly luciferase. All
experiments were repeated at least three times.
Statistical analysis
SPSS 18.0 software (SPSS, Inc., Chicago, IL, USA)
was used to perform statistical analysis. Data was expressed as
mean ± standard deviation for at least three separate experiments.
P<0.05 was considered to indicate a statistically significant
difference. Student's t test and one-way analysis of variance
(ANOVA) were used to analyze the difference. Cox regression
(proportional hazards model) was used to perform multivariate
analysis of prognostic factors. The log-rank test was used to
determine the patient survival and the differences.
Results
SIRT6 was downregulated in the OS
tissues and cell lines
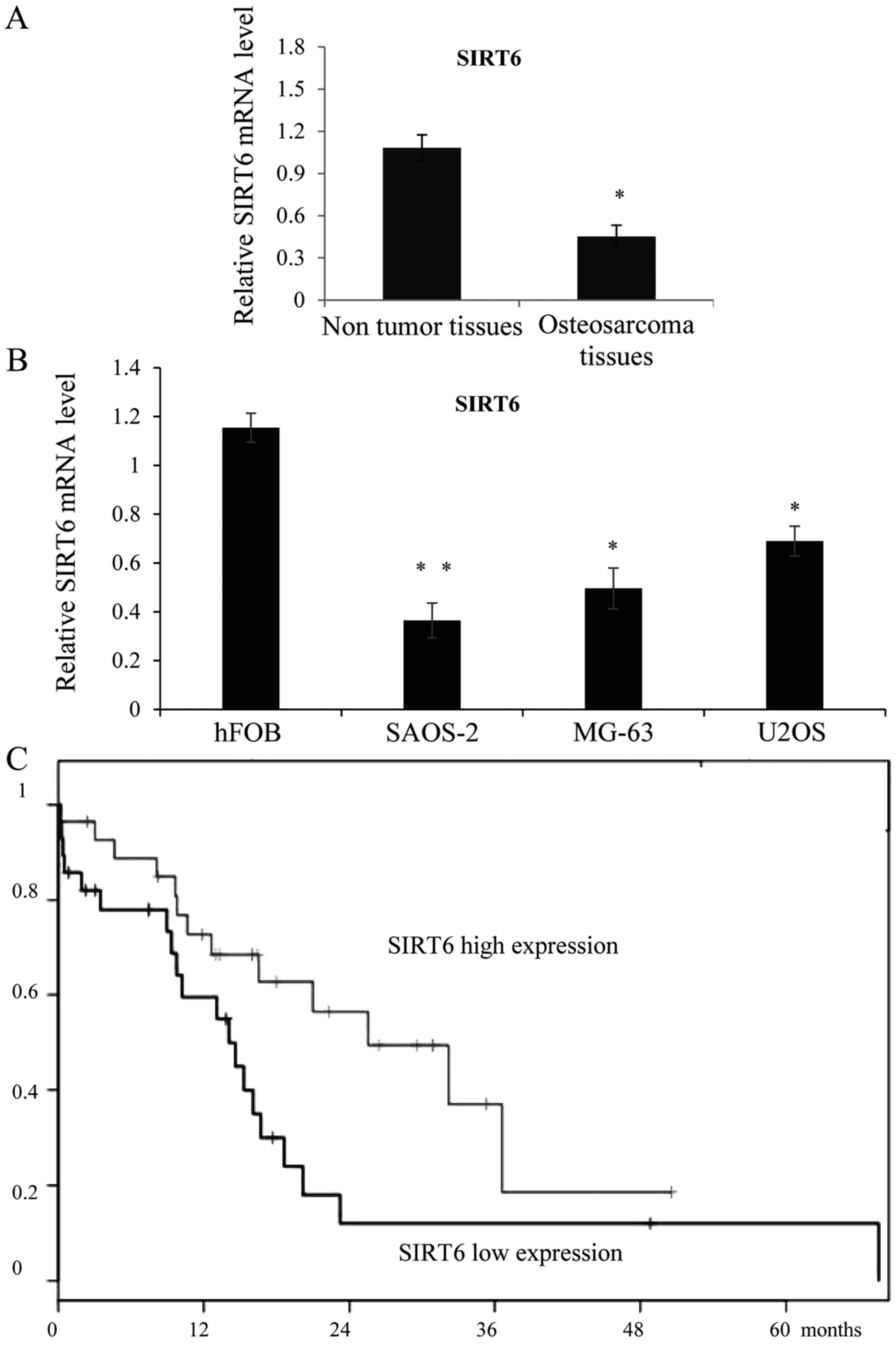
In order to study the role of SIRT6 in OS, we first
examined the expression of SIRT6 in OS tissues, as shown in
Fig. 1A, SIRT6 was downregulated in
the OS tissues than in the adjacent non-tumor soft tissues.
Meanwhile, the expression of SIRT6 was also lower in the OS cell
lines SAOS-2, MG-63 and U2OS compared with the osteoblastic cell
line hFOB. We then chose SAOS-2 and MG-63 to perform functional
analysis of SIRT6. As collected, the follow-up information was
available for all patients, Kaplan-Meier curve was analyzed to show
that the higher expression of SIRT6 was statistically correlated
with favorable overall survival rates in OS (Fig. 1C), (P=0,0381). With confidence
intervals (1.04~4.36) and Hazard Ratio=2.13.
SIRT6 inhibited the proliferation of
human OS cells
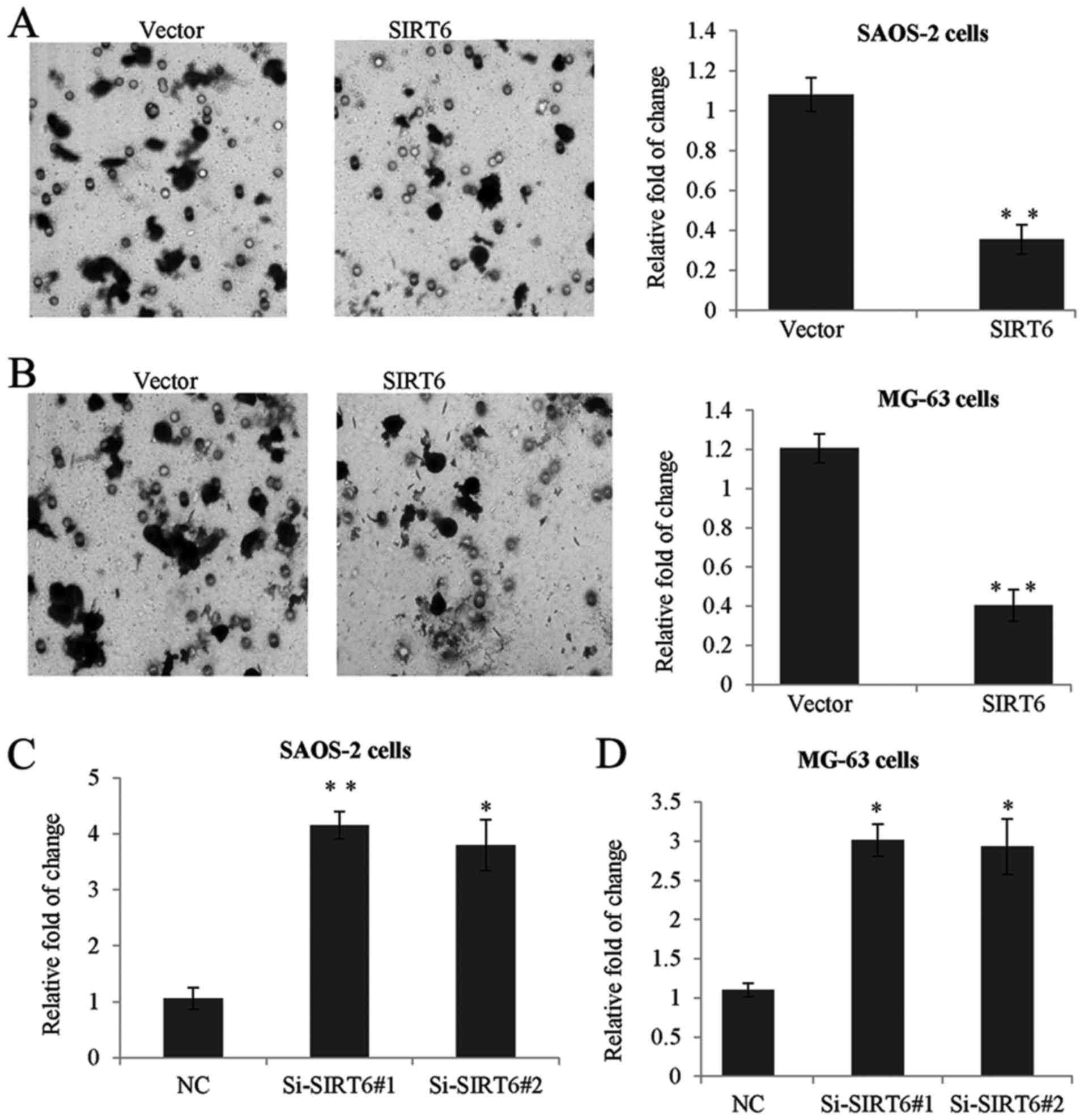
To confirm whether SIRT6 has any effect on the
proliferation of OS cells. We established SAOS-2 and MG-63
overexpression SIRT6 cell lines using lentivirus carrying SIRT6
construct. As shown in Fig. 2A, the
expression of SIRT6 was significantly increased after transfection
with SIRT6 construct compared to the control group (Fig. 2A). Furthermore, we used two specific
siRNA targeting SIRT6 to knockdown the expression of SIRT6. RT-qPCR
assay was performed, as expected, Si-SIRT6 could remarkably reduce
the level of SIRT6 (Fig. 2B).
Overexpression of SIRT6 decreased the proliferation rate of SAOS-2
and MG-63 cells, on the other hand, knockdown of SIRT6 dramatically
enhanced cell proliferation (Fig.
2C).
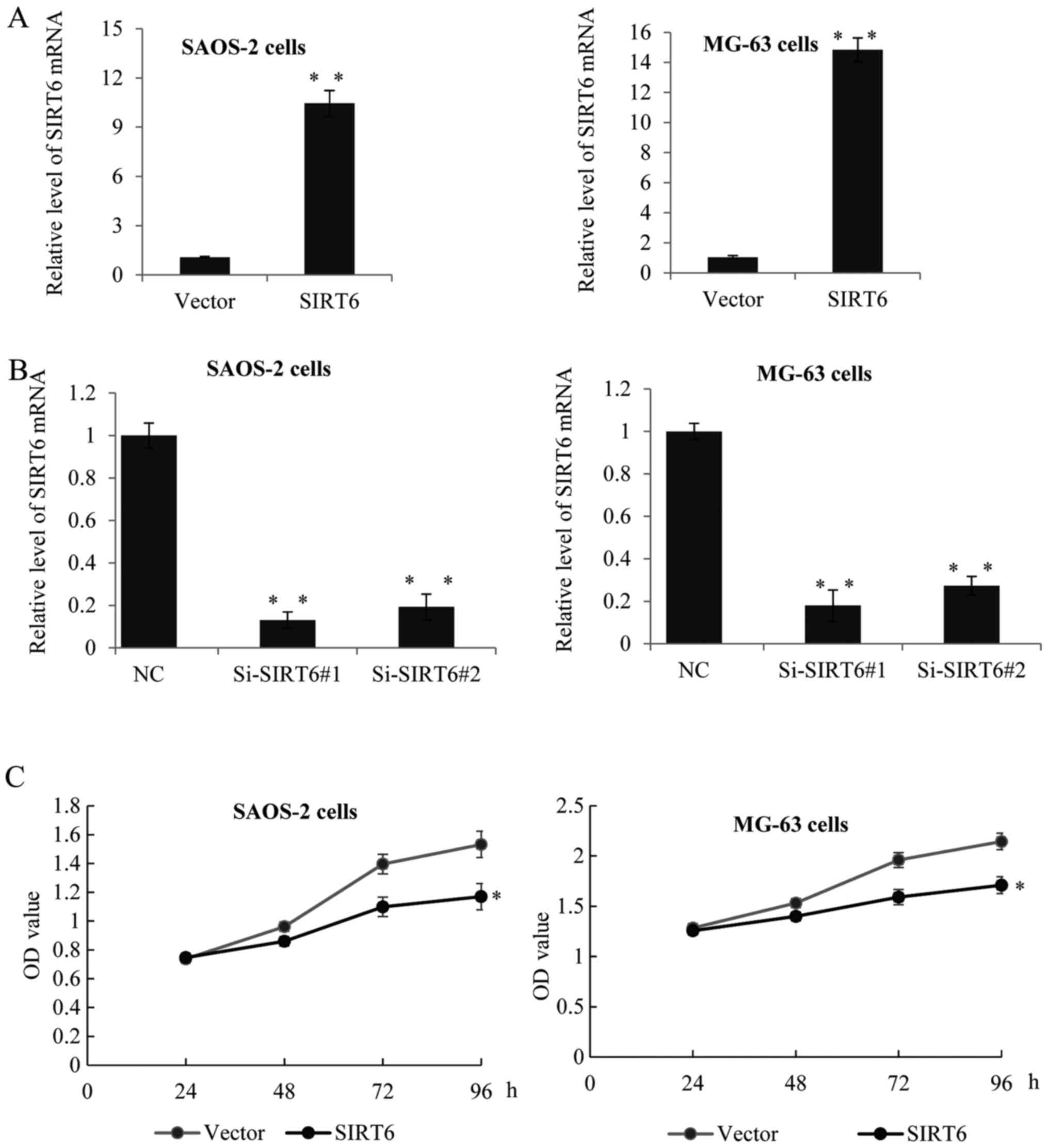 | Figure 2.SIRT6 inhibited the proliferation of
human osteosarcoma cells. (A) RT-qPCR was used to analyze SIRT6
expression in SAOS-2 and MG-63 cells transfected with SIRT6
construct or vector, or transfected with control siRNA or (B)
Si-SIRT6#1, or Si-SIRT6#2. GAPDH was used as an internal control.
Data was presented as means ± SD, of three independent experiments,
**P<0.01 vs. vector or NC. (C) MTT assay was performed in SAOS-2
cells or MG-63 cells transfected with control or SIRT6 construct,
or transfected with control siRNA or Si-SIRT6#1, or Si-SIRT6#2. The
absorbance at 490 nm was measured at 0, 24, 48, 72 and 96 h, of
three independent experiments, *P<0.05 vs. vector or NC. SIRT6,
sirtuin-6; RT-qPCR, reverse transcription-quantitative polymerase
chain reaction. |
SIRT6 inhibited the invasion of human
OS cells
In order to investigate the function of SIRT6 on the
invasion of OS cells, Transwell invasion assay was performed in the
above two human OS cells. As shown in Fig. 3A, the invasion capacity of SAOS-2
transfected with SIRT6 was significantly reduced, as compared with
the control groups. Moreover, SIRT6 overexpression also suppressed
the invasive capacity of MG-63 cells, when compared to the control
groups (Fig. 3B). By contrast, the
knockdown of SIRT6 had significantly promotion effect on the
invasion of the SAOS-2 cells and MG-63 cells (Fig. 3C and D).
Identification of N-cadherin as a
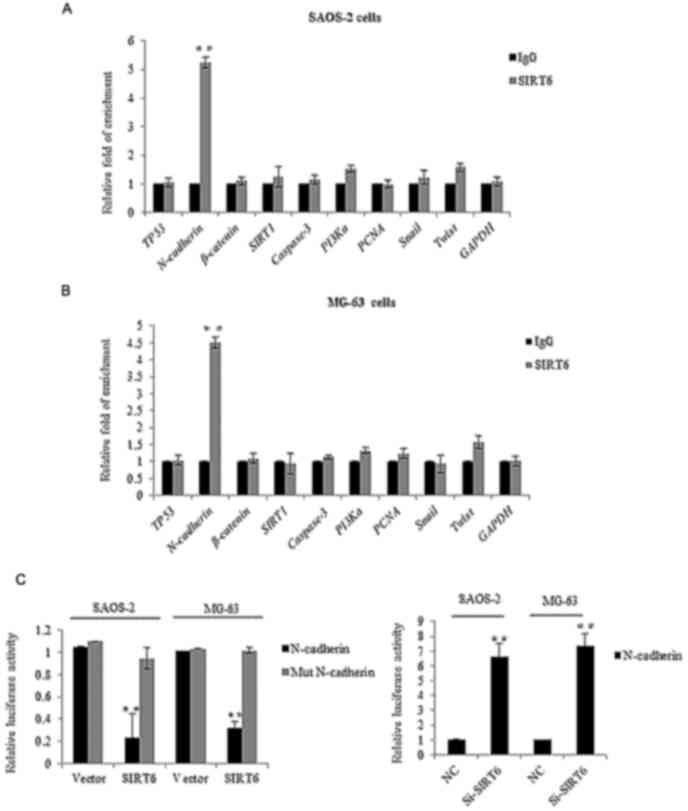
direct target of SIRT6
We next investigated the molecular mechanism of
SIRT6 mediated biological functions. We examined its potential
targets by searching quantitative ChIP assays in SAOS-2 cells and
MG-63 cells. Several key genes in different pathways were chosen,
as shown in Fig. 4A and B, on the
promoter of CDH2 (N-cadherin), there was obviously binding
enrichment of SIRT6. To further support the argument, luciferase
reporter activity assay was carried out, the SAOS-2 cells and MG-63
cells transfected with SIRT6 significantly repressed the luciferase
activity of the wild-type CDH2 promoter, whereas the mutation of
the promoter abolished this repression, oppositely, the knockdown
of SIRT6 resulted in the enhanced CDH2 reporter activity (Fig. 4C). Consistently, we detected the mRNA
and protein level of N-cadherin in the SAOS-2 cells and MG-63 cells
transfected with the SIRT6 overexpression plasmid. As shown in
Fig. 4D and E, overexpression of
SIRT6 inhibited the level of N-cadherin in the OS cells as compared
to the vector groups. Together, the above data demonstrated that
N-cadherin was a direct target of SIRT6, and SIRT6 reduced
N-cadherin expression.
N-cadherin was involved in SIRT6
mediated proliferation and invasion of OS cells
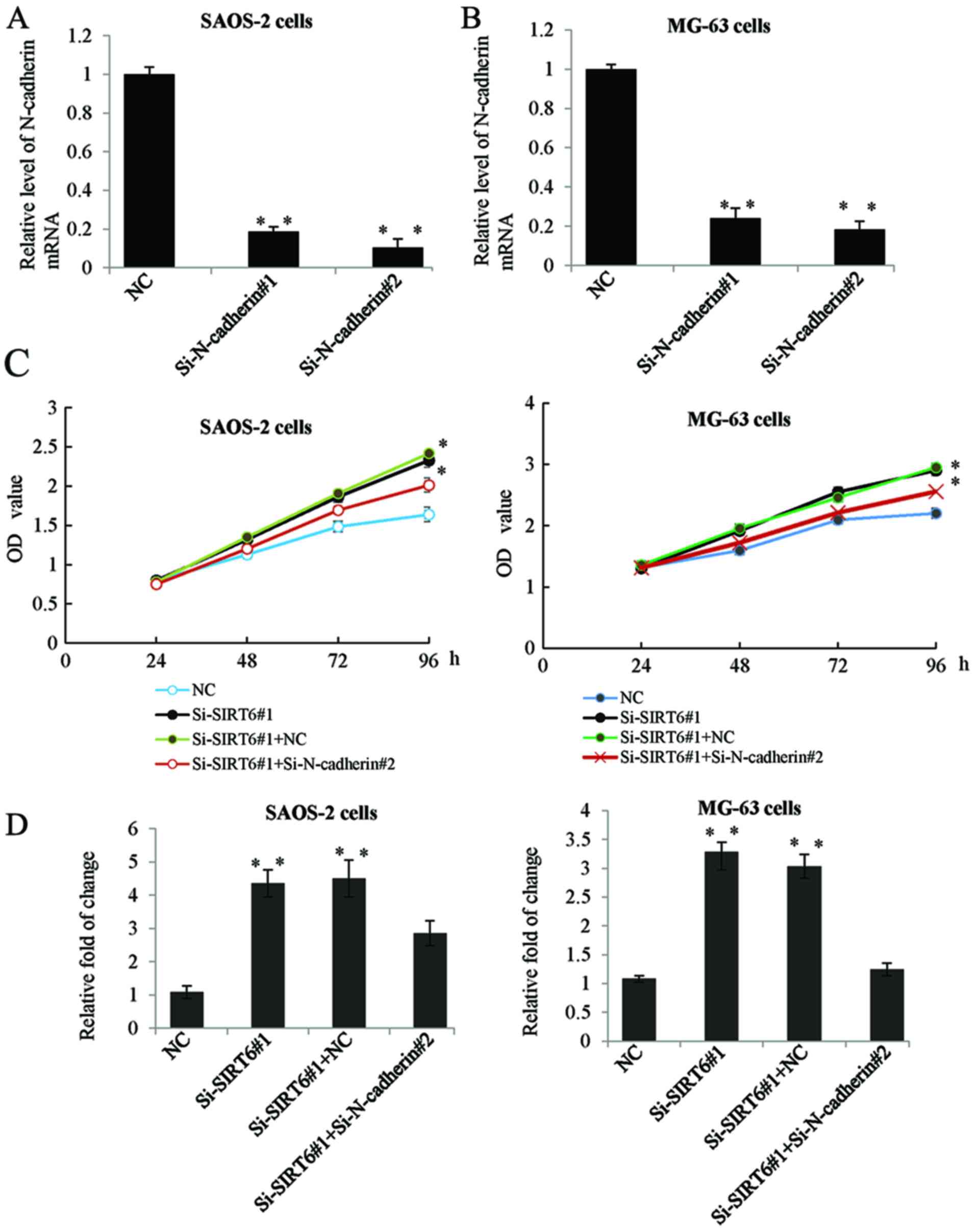
To further clarify whether the inhibition of
N-cadherin was as a downstream effector in SIRT6 mediated
proliferation and invasion of OS cells. Firstly, we determined the
mRNA expression of N-cadherin in the SAOS-2 and MG-63 cells
transfected with siRNA targeting N-cadherin. As shown in Fig. 5A and B, transfection with
Si-N-cadherin significantly inhibited the expression level of
N-cadherin. Furthermore, we transfected SAOS-2 and MG-63 cells with
SIRT6 siRNA or co-transfected with N-cadherin siRNA. MTT assay and
Transwell assay was performed respectively. While transfection with
Si-N-cadherin reduced the promotion effect of Si-SIRT6 on OS cells
proliferation and invasion (Fig. 5C and
D).
Discussion
SIRT6 was one of the NAD+-dependent deacetylase
sirtuin family. Sirtuins had seven members in mammals and were
widely expressed in different tissues. Among them, SIRT6 was a
broadly expressed, predominantly nuclear protein (19). As reported, the major function of
SIRT6 is to maintenance genomic integrity (20). SIRT6 was identified as a tumor
suppressor and regulated aerobic glycolysis in cancer cells
(13), furthermore, SIRT6 was
reported to cooperate with tumor suppressor p53 to regulate
gluconeogenesis (21). Marquardt
et al (22) showed SIRT6 was
significantly down-regulated in hepatocellular carcinoma and the
dysregulated genes by loss of SIRT6 displayed oncogenic effects in
HCC. Feng et al (23) reported
in glioma, the expression of SIRT6 was significantly lower in
glioblastoma multiform tissues and SIRT6 suppressed glioma cell
growth via induction of apoptosis by inhibition of JAK2/STAT3
activation. However, the correlation between SIRT6 and OS has not
been investigated.
Significant difference of SIRT6 expression was found
in OS tissues and adjacent non-cancerous tissues, as shown in
Table I. A higher expression of SIRT6
was associated with a higher histological grade and higher enneking
staging. Higher expression of SIRT6 indicative of better overall
survival times, at 24 months, the difference was major, however, at
36 months, since most patients died, the difference has vanished,
which indicated SIRT6 might be involved in the development of OS.
As known, during tumor progression, there were two essential steps
as uncontrolled cell proliferation and aggressive cell metastasis,
so in our study, we focus on whether SIRT6 has a function in the OS
tumor growth and metastasis. We also performed apoptosis analysis
in the preliminary experiments, but there is no obvious difference
between the SIRT6 group and the vector group, data was not shown.
Although Kok et al (24)
reported SIRT6 modulated hypoxia-induced apoptosis in osteoblasts,
but in OS, there is no exact evidence to confirm that. Furthermore,
as Sugatani et al (25)
reported, SIRT6 deficiency culminated in low-turnover osteopenia.
As Zhang et al (26) reported,
SIRT6 deficiency could cause senile osteoporosis. The above
observation together with us revealed multiple function of SIRT6
and OS, osteoporosis and osteopenia.
The results of the present study demonstrated that
SIRT6 inhibited the proliferation and invasion of the SAOS-2 and
MG-63 OS cell lines, further investigations such as qChIP and
luciferase reporter assay demonstrated that N-cadherin was a direct
target inhibited by SIRT6. The expression of N-cadherin was
opposite with the SIRT6 status from mRNA and protein level.
Therefore we made the hypothesis that N-cadherin might be involved
in the regulation by SIRT6 in the progression of OS cells. The
evidences were as follows: By reduced the expression of N-cadherin
under SIRT6 knockdown, the promotion phenotypes of Si-SIRT6 could
be almostly reduced. Although further investigation was required
for characterization of other targets involved in SIRT6 inhibiting
OS. The molecular mechanisms underlying SIRT6 as a tumor suppressor
gene in OS may prove to be a promising gene therapeutic agent.
Competing interests
The authors declare that they have no conflicts of
interest about this article.
References
|
1
|
Mirabello L, Troisi RJ and Savage SA:
Osteosarcoma incidence and survival rates from 1973 to 2004: Data
from the surveillance, epidemiology and end results program.
Cancer. 115:1531–1543. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bielack S, Carrle D and Casali PG: ESMO
Guidelines Working Group: Osteosarcoma: ESMO clinical
recommendations for diagnosis, treatment and follow-up. Ann Oncol.
20 Suppl 4:S137–S139. 2009. View Article : Google Scholar
|
|
3
|
Bielack SS, Kempf-Bielack B, Delling G,
Exner GU, Flege S, Helmke K, Kotz R, Salzer-Kuntschik M, Werner M,
Winkelmann W, et al: Prognostic factors in high-grade osteosarcoma
of the extremities or trunk: An analysis of 1,702 patients treated
on neoadjuvant cooperative osteosarcoma study group protocols. J
Clin Oncol. 20:776–790. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Marina N, Gebhardt M, Teot L and Gorlick
R: Biology and therapeutic advances for pediatric osteosarcoma.
Oncologist. 9:422–441. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kansara M, Teng MW, Smyth MJ and Thomas
DM: Translational biology of osteosarcoma. Nat Rev Cancer.
14:722–735. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nieman MT, Prudoff RS, Johnson KR and
Wheelock MJ: N-cadherin promotes motility in human breast cancer
cells regardless of their E-cadherin expression. J Cell Biol.
147:631–644. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Islam S, Carey TE, Wolf GT, Wheelock MJ
and Johnson KR: Expression of N-cadherin by human squamous
carcinoma cells induces a scattered fibroblastic phenotype with
disrupted cell-cell adhesion. J Cell Biol. 135:1643–1654. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nishitani S, Noma K, Ohara T, Tomono Y,
Watanabe S, Tazawa H, Shirakawa Y and Fujiwara T: Iron
depletion-induced downregulation of N-cadherin expression inhibits
invasive malignant phenotypes in human esophageal cancer. Int J
Oncol. 49:1351–1359. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Park KS, Dubon MJ and Gumbiner BM:
N-cadherin mediates the migration of MCF-10A cells undergoing bone
morphogenetic protein 4-mediated epithelial mesenchymal transition.
Tumour Biol. 36:3549–3556. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bremmer F, Schallenberg S, Jarry H, Küffer
S, Kaulfuss S, Burfeind P, Strauß A, Thelen P, Radzun HJ, Ströbel
P, et al: Role of N-cadherin in proliferation, migration, and
invasion of germ cell tumours. Oncotarget. 6:33426–33437. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Da C, Wu K, Yue C, Bai P, Wang R, Wang G,
Zhao M, Lv Y and Hou P: N-cadherin promotes thyroid tumorigenesis
through modulating major signaling pathways. Oncotarget.
8:8131–8142. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Michishita E, McCord RA, Berber E, Kioi M,
Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL,
Barrett JC, et al: SIRT6 is a histone H3 lysine 9 deacetylase that
modulates telomeric chromatin. Nature. 452:492–496. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sebastián C, Zwaans BM, Silberman DM,
Gymrek M, Goren A, Zhong L, Ram O, Truelove J, Guimaraes AR, Toiber
D, et al: The histone deacetylase SIRT6 is a tumor suppressor that
controls cancer metabolism. Cell. 151:1185–1199. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tian K, Chen P, Liu Z, Si S, Zhang Q, Mou
Y, Han L, Wang Q and Zhou X: Sirtuin 6 inhibits epithelial to
mesenchymal transition during idiopathic pulmonary fibrosis via
inactivating TGF-β1/Smad3 signaling. Oncotarget. 8:61011–61024.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bai L, Lin G, Sun L, Liu Y, Huang X, Cao
C, Guo Y and Xie C: Upregulation of SIRT6 predicts poor prognosis
and promotes metastasis of non-small cell lung cancer via the
ERK1/2/MMP9 pathway. Oncotarget. 7:40377–40386. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kugel S, Sebastián C, Fitamant J, Ross KN,
Saha SK, Jain E, Gladden A, Arora KS, Kato Y, Rivera MN, et al:
SIRT6 suppresses pancreatic cancer through control of Lin28b. Cell.
165:1401–1415. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Elhanati S, Ben-Hamo R, Kanfi Y, Varvak A,
Glazz R, Lerrer B, Efroni S and Cohen HY: Reciprocal regulation
between SIRT6 and miR-122 controls liver metabolism and predicts
hepatocarcinoma prognosis. Cell Rep. 14:234–242. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liszt G, Ford E, Kurtev M and Guarente L:
Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J
Biol Chem. 280:21313–21320. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mostoslavsky R, Chua KF, Lombard DB, Pang
WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy
MM, et al: Genomic instability and aging-like phenotype in the
absence of mammalian SIRT6. Cell. 124:315–329. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang P, Tu B, Wang H, Cao Z, Tang M,
Zhang C, Gu B, Li Z, Wang L, Yang Y, et al: Tumor suppressor p53
cooperates with SIRT6 to regulate gluconeogenesis by promoting
FoxO1 nuclear exclusion. Proc Natl Acad Sci USA. 111:10684–10689.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Marquardt JU, Fischer K, Baus K, Kashyap
A, Ma S, Krupp M, Linke M, Teufel A, Zechner U, Strand D, et al:
Sirtuin-6-dependent genetic and epigenetic alterations are
associated with poor clinical outcome in hepatocellular carcinoma
patients. Hepatology. 58:1054–1064. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Feng J, Yan PF, Zhao HY, Zhang FC, Zhao WH
and Feng M: SIRT6 suppresses glioma cell growth via induction of
apoptosis, inhibition of oxidative stress and suppression of
JAK2/STAT3 signaling pathway activation. Oncol Rep. 35:1395–1402.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kok SH, Hou KL, Hong CY, Chao LH,
Hsiang-Hua Lai E, Wang HW, Yang H, Shun CT, Wang JS and Lin SK:
Sirtuin 6 modulates hypoxia-induced apoptosis in osteoblasts via
inhibition of glycolysis: Implication for pathogenesis of
periapical lesions. J Endod. 41:1631–1637. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sugatani T, Agapova O, Malluche HH and
Hruska KA: SIRT6 deficiency culminates in low-turnover osteopenia.
Bone. 81:168–177. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang DM, Cui DX, Xu RS, Zhou YC, Zheng
LW, Liu P and Zhou XD: Phenotypic research on senile osteoporosis
caused by SIRT6 deficiency. Int J Oral Sci. 8:84–92. 2016.
View Article : Google Scholar : PubMed/NCBI
|