Introduction
Primary liver cancer is one of the most aggressive
types of neoplasm, characterized by high morbidity and mortality
rates (1–3). Although the prevalence of liver cancer
is highest in the Far East and Sub-Saharan Africa, its incidence
has increased in Western countries (4), and it is predicted that the incidence of
liver cancer in the United States will continue to increase in the
next two decades (5). At present,
surgical intervention (resection or transplant) is the first line
therapeutic method for cases of primary liver cancer at an early
stage. However, systemic metastasis and postsurgical recurrence
negatively affect the prognosis of patients with liver cancer
(2). Unfortunately, the majority of
liver cancer cases are diagnosed at at an advanced stage, by which
metastasis has occurred and no curative treatment is available
(6). Therefore, metastasis is the
leading factor that results in the high mortality rate and poor
5-year survival rate (<9%) in patients with liver cancer
(7).
Tumor metastasis is a highly integrated, multistep
process which is closely associated with a series of molecules,
including Rho GTPase activation proteins (Rho GAPases), matrix
metalloproteinases (MMPs), vascular endothelial growth factor
(VEGF), plasminogen activator (PA) etc (8–13). Rho
GTPases belong to the Ras super-family, including RhoA, RhoB and
RhoC (14). Previous studies have
demonstrated that Rho GTPases serve a vital role in tumor
progression and metastasis by regulating cell proliferation, the
actin cytoskeleton and cell adhesion (15). Furthermore, Rho GTPases often appear
to be elevated or to have their activity increased in several human
tumors by changing three major classes of regulators including Rho
GAPs, GEFs and GDIs (16). Notably,
RhoA was frequently found to be over-expressed in liver cancer and
which is associated with tumor metastasis, a lower survival rate
and a higher chance of tumor recurrence (17). As important negative regulators of Rho
signaling, numerous RhoGAPs have been demonstrated to be
downregulated in liver cancer, including p190RhoGAP, Deleted in
liver cancer (DLC) family and Rho GTPase activating protein 9
(ARHGAP) (18), which indicated the
association of RhoGAPs and liver cancer. RhoGAPs are emerging as a
novel cancer-related biomarker in recent years (19). ARHGAP9 is a member of the RhoGAP
family, and also named RhoGAP9 (20).
It has been demonstrated that ARHGAP9 inhibits Erk2 and p38α
activation in Swiss 3T3 fibroblasts by binding to mitogen-activated
protein kinases (MAPKs), which serve pivotal roles in cell
proliferation and metastasis (21).
ARHGAP9 reduced the adhesion of hematopoietic cells to the
extracellular matrix and the migration of vero cells (22). Studies using bioinformatics determined
that ARHGAP9 is a cancer-associated gene (23), however, no studies have reported an
association between ARHGAP9 and cancer.
Ginsenoside Rg3, one of the bioactive ginsenosides
extracted from Panax ginseng C.A. Meyer, displays
significant anti-metastasis effects in numerous types of cancer
models. For example, Rg3 was reported to effectively inhibit lung
cancer metastasis by suppressing epithelial-mesenchymal transition
(24,25). Rg3-induced downregulation of MMPs has
been demonstrated to be associated with the decreased invasive
capacity of ovarian cancer SKov3 cells and colon cancer SW480 cells
(26,27). Rg3 was also demonstrated to inhibit
the migration of Eca-109 and 786-0 cells through suppressing VEGF
expression (28). The aim of the
present study was to investigate the potential functions and
molecular mechanisms of Rg3 against liver cancer cell
metastasis.
Materials and methods
Reagents
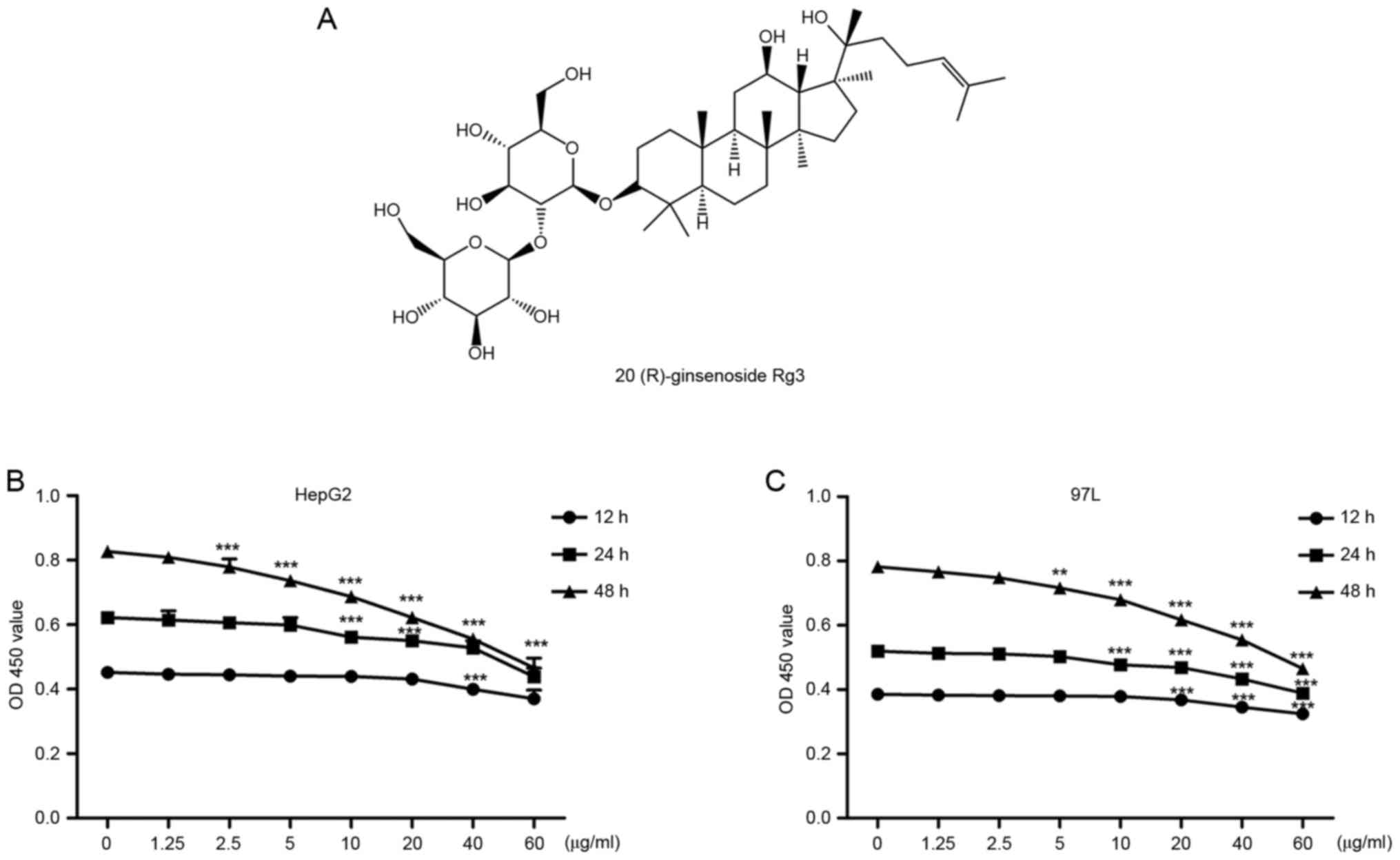
20(R)-ginsenoside Rg3 (98% pure, Fig. 1A) was purchased from the Nanjing
Zelang Biological Technology Company (Jiangsu, China). It was
dissolved in dimethyl sulfoxide (DMSO) and stored at −20°C.
Anti-ARHGAP9 was purchased from Abcam (Cambridge, UK; cat. no.
Ab101361) and anti-GAPDH was purchased from Cell Signalling
Technology, Inc. (Danvers, MA, USA; cat. no. 5174).
Cell culture
The human liver cancer cell lines, HepG2, SK-Hep1,
MHCC-97L, MHCC-97H, SMMC-7721and BEL-7404, were provided by the
Shanghai Cell Bank, Chinese Academy of Sciences (Shanghai, China)
(http://www.cellbank.org.cn/index.asp). HepG2 is a
human hepatoblastoma cell line, commonly misidentified as
hepatocellular carcinoma (29).
SK-Hep1 and the remaining cell lines were cultured in RPMI-1640
medium and Dulbecco's modified Eagle's medium (DMEM), respectively,
which were supplemented with 10% fetal bovine serum (FBS),100
units/ml penicillin and 100 µg/ml streptomycin at 37°C in 5%
CO2.
Cell viability analysis
Cells were seeded in 96-well plates at a density of
2,000 cells per well, incubated overnight, and treated with
different concentrations of Rg3 (0, 1.25, 2.5, 5, 10, 20, 40 and 60
µg/ml) for 12, 24 and 48 h. The cell proliferation was quantitated
using Cell Counting kit-8 (CCK-8; Dojindo Laboratories, Japan),
according to the manufacturer's recommendations. Control cells were
treated with culture media containing 0.15% (v/v) DMSO.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from the cells with TRIzol
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and
reverse transcribed to cDNA with oligo(dT) primers (Fermentas; cat.
no. K1622; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. SYBR Green-based RT-qPCR (Maxima
SYBR-Green/ROX qPCR Master Mix 2X; cat. no. K0223; Thermo Fisher
Scientific, Inc.) was performed to examine the relative ARHGAP9
mRNA level, which was normalized to GAPDH. The primer sequences
were as follows: ARHGAP9 forward, 5′-GAAGAGACCGCCCTTACAAAGC-3′ and
reverse, 5′-GCTCACCCGATAAATGCCATCC-3′; GAPDH forward,
5′-CACCCACTCCTCCACCTTTG-3′ and reverse,
5′-CCACCACCCTGTTGCTGTAG-3′.
RNA interference
A total of 3 small interfering RNAs (siRNAs)
targeting ARHGAP9 mRNA were synthesized: siRNA1,
5′-GACGCUGCUUCUACAUAAA-3′; siRNA2, 5′-GUGGUGUUAACGGGUAACA-3′, and
siRNA3, 5′-GCGUGCGCAACAAACUAAA-3′, and a control siRNA (siNC). The
siRNAs were transiently transfected into MHCC-97L cells using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's instructions. The most effective
siRNAs were selected according to the results of RT-qPCR and
western blotting after 48 h. Then MHCC-97L cells were transfected
with siRNA3 and a 24-h Rg3 treatment followed after 24 h. The
effect of ARHGAP9 silencing on the anti-invasive properties of Rg3
was assessed by Transwell assay.
Western blot analysis
The protein concentration was analyzed by
Bicinchoninic Acid Protein Assay Reagent (Sangon Biotech Co.,
Shanghai, China), according to the manufacturer's protocols.
Soluble lysates containing 20 µg proteins per sample were separated
via SDS-PAGE on a 10% gel and subsequently transferred onto
polyvinylidene fluoride membranes. Subsequent to blocking with 5%
bovine serum albumin at 4°C for 1 h, membranes were incubated with
primary antibodies anti-ARHGAP9 (dilution, 1:1,000; cat. no.
Ab101361; Abcam) and anti-GAPDH (dilution, 1:1,500; cat. no. 5174;
Cell Signalling Technology) at 4°C overnight. Following washing
with TBS Tween-20, the membranes were incubated with secondary
antibody horse-radish peroxidase-labeled goat anti-rabbit IgG
(dilution, 1:1,000; cat. no. A0208; Beyotime Institute of
Biotechnology, Shanghai, China) at room temperature for 1 h. The
membrane signals were detected using an Enhanced Chemiluminescent
Western Blotting Detection System (Bio-Rad Laboratories, Hercules,
CA, USA), according to the manufacturer's protocols. Quantification
of protein expression was performed by Image J 1.6 software
(National Institutes of Health, Bethesda, MD, USA).
Cell migration and invasion
assays
Cell migration was examined using a Transwell
migration assay. Cells were suspended in serum-free DMEM medium and
5×105 cells/ml were seeded into the upper chamber, with 8-µm pores
(Corning Incorporated, Corning, NY, USA) while DMEM with 10% FBS
was added to the lower chamber. After a 24-h incubation,
non-migrated cells were removed with cotton swabs, and microscopic
images of the migrated cells were captured in 5 random random
fields (magnification, ×200). The migration capability of the cells
was measured by counting the number of cells present. The data are
expressed as the mean number of cells/field ± standard
deviation.
For the invasion assay, 1 mg/ml Matrigel (BD
Biosciences, Franklin Lakes, NJ, USA) was pre-coated onto the
Transwell membrane, and the remaining steps were performed as per
the migration assay.
In vivo tumorigenesis
Male 5-week old BALB/c nude mice were purchased from
SLAC Laboratory Animal Center (Shanghai, China), anesthetized and
5×106 HepG2 cells or 5×106 MHCC-97L cells in 200 µl PBS were
injected subcutaneously into the right forelimb. Rg3 was
administrated to the rats by oral gavage, and distilled water with
0.5% CMC-Na was used as a control. After 21 days the nude mice were
sacrificed by cervical dislocation and the tumor tissues were
excised and weighed. The experiments were approved by the
Experimental Animal Ethical Committee of Seventh People's Hospital
of Shanghai University of Traditional Chinese Medicine (Shanghai,
China).
Statistical analysis
In vivo data are presented as the mean ±
standard error of the mean and in vitro data are presented
as the mean ± standard deviation. Student's t-test was used to
compare the difference between the two groups. One-way analysis of
variance followed by Dunnet's test was performed for comparisons
between multiple groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
The anti-proliferative effects of
Rg3
The effects of Rg3 on liver cancer cell viability
were investigated by CCK-8 assay. As demonstrated by Fig. 1B and C, Rg3 exhibited a dose- and
time-dependent anti-proliferative effect on HepG2 and MHCC-97L
cells. Cell viability decreased with longer duration or/and
increased concentration of Rg3. There was no noticeable influence
on cell viability with Rg3 at low concentrations (1.25, 2.5 or 5
µg/ml) for 12 or 24 h.
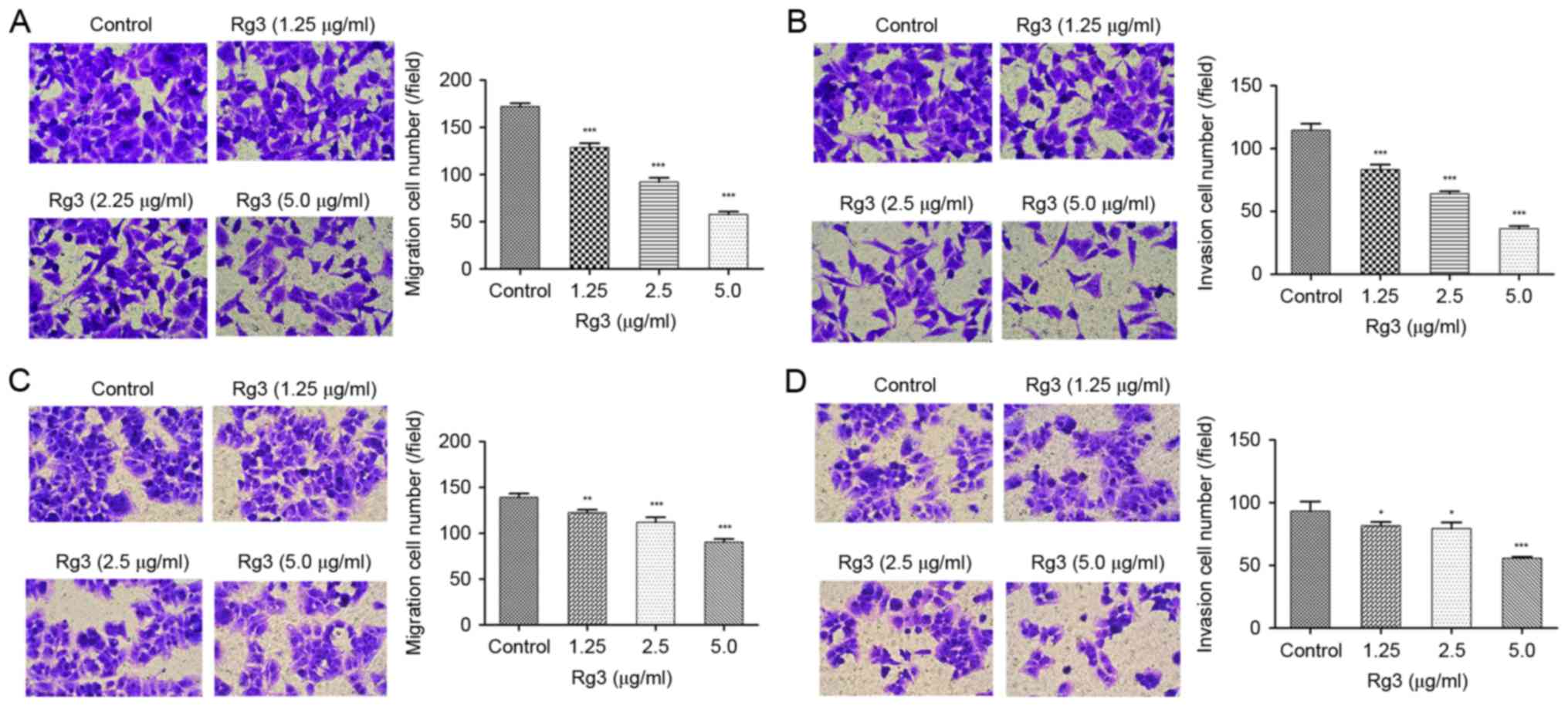
The anti-migration and anti-invasion
effects of Rg3
To evaluate the effect of Rg3 on the metastasis of
liver cancer cells, Transwell assays of HepG2 and MHCC-97L cells
were performed. After a 24-h incubation of Rg3 at concentrations of
1.25, 2.5 and 5 µg/ml, it was demonstrated that the number of
migratory and invasive HepG2 and 97L cells was significantly
reduced compared with those of the control group (Fig. 2).
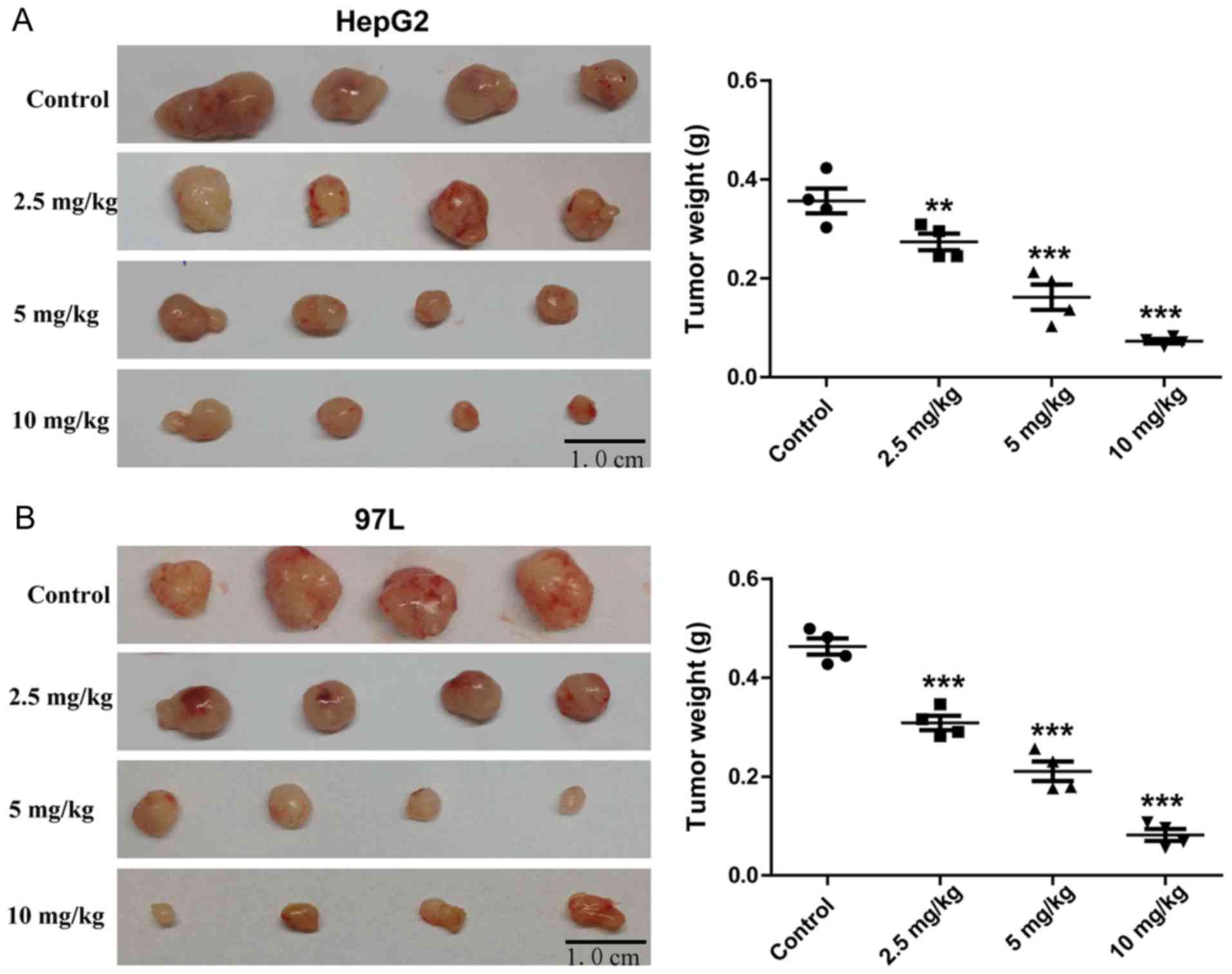
Rg3 suppresses tumor growth of liver
cancer cells in nude mice
BALB/c nude mice administered with Rg3 for 21 days
exhibited significant decrease in tumor size compared with the
control mice (Fig. 3A and B). The
weight of the tumors decreased with increasing Rg3 dose.
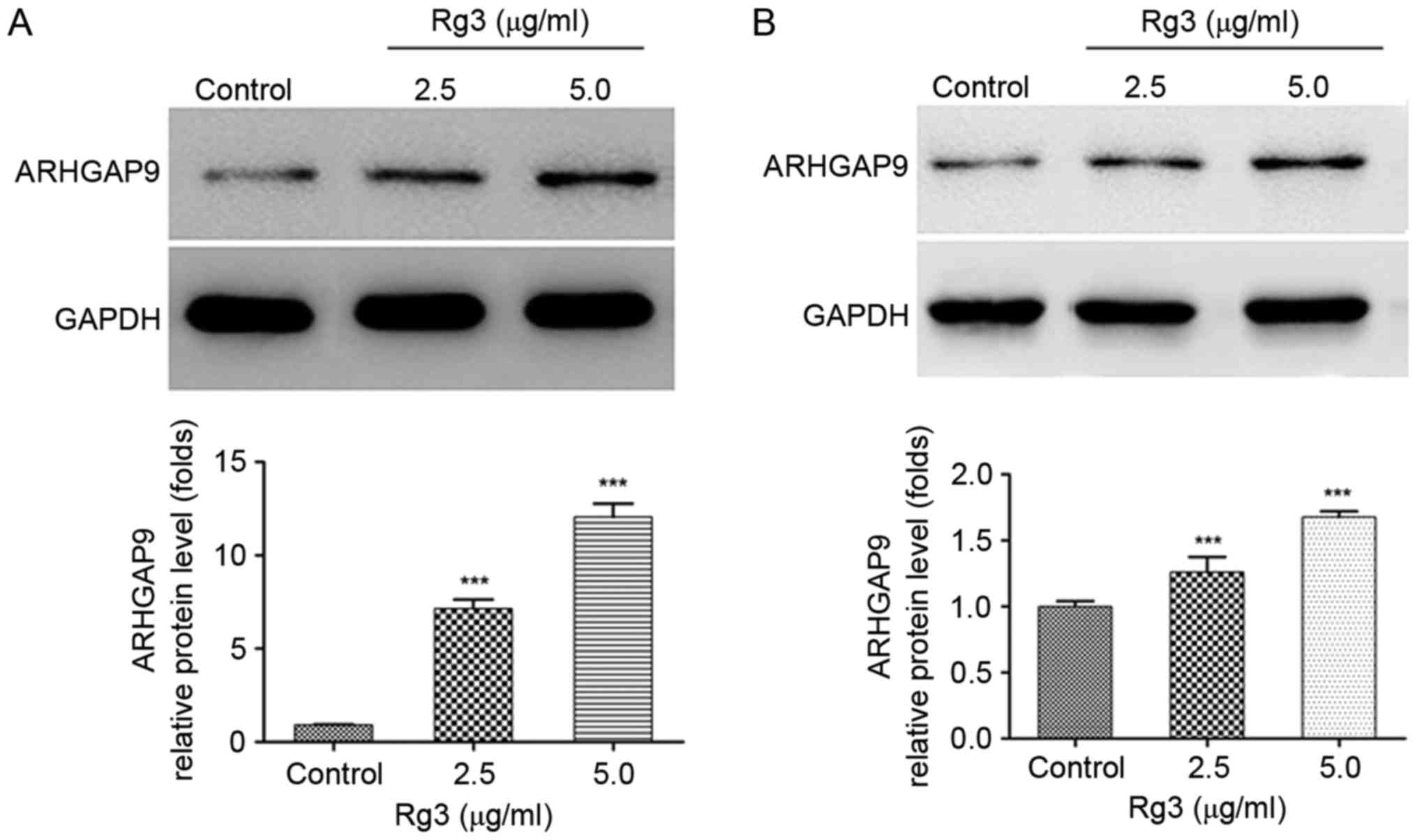
The increased expression of ARHGAP9
induced by Rg3
ARHGAP9, as a member of the RhoGAP family, has been
reported to be associated with cell migration in previous studies
(20–22). Bioinformatics investigation revealed
that ARHGAP9 is a cancer-associated gene (23). Whether the expression of ARHGAP9 was
regulated by Rg3 in liver cancer cells was investigated. Western
blot analysis demonstrated that the expression of ARHGAP9 protein
were significantly increased in a dose-dependent manner following
treatment of HepG2 and MHCC-97L cells with Rg3 (Fig. 4A and B).
Knockdown of ARHGAP9 attenuates the
anti-metastatic effect of Rg3
Rg3 inhibits the migration and invasion of liver
cancer cells, as well as increases the expression of ARHGAP9.
Whether the anti-migration and anti-invasion effects of Rg3 were
mediated by ARHGAP9 was explored.
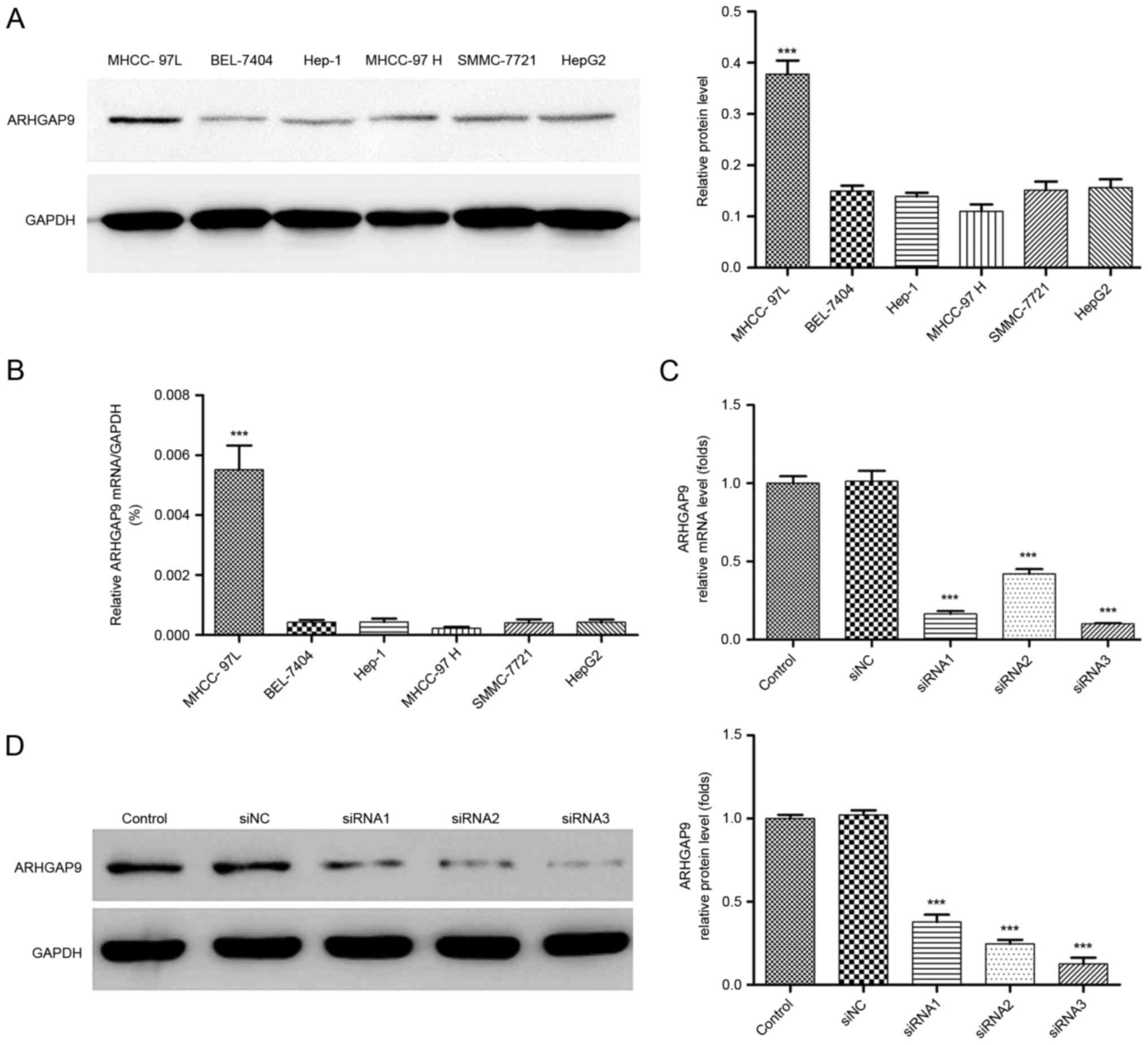
To better understand the role of ARHGAP9 in
anti-metastatic behavior of Rg3 in HCC cells, the expression levels
of ARHGAP9 were measured in six human liver cancer cell lines
(HepG2, SK-Hep1, MHCC-97L, MHCC-97H, SMMC-7721 and BEL-7404). The
expression of ARHGAP9 in the MHCC-97L cell line was higher than
that in the other cell lines (Fig. 5A and
B). siRNAs (siRNA1, siRNA2 and siRNA3) or control siRNA (siNC)
were transfected into MHCC-97L cells to knockdown ARHGAP9. As
demonstrated in Fig. 5C and D, all
three siRNAs efficiently reduced the expression of ARHGAP9, among
which siRNA3 was the most efficient, and selected for further
experiments.
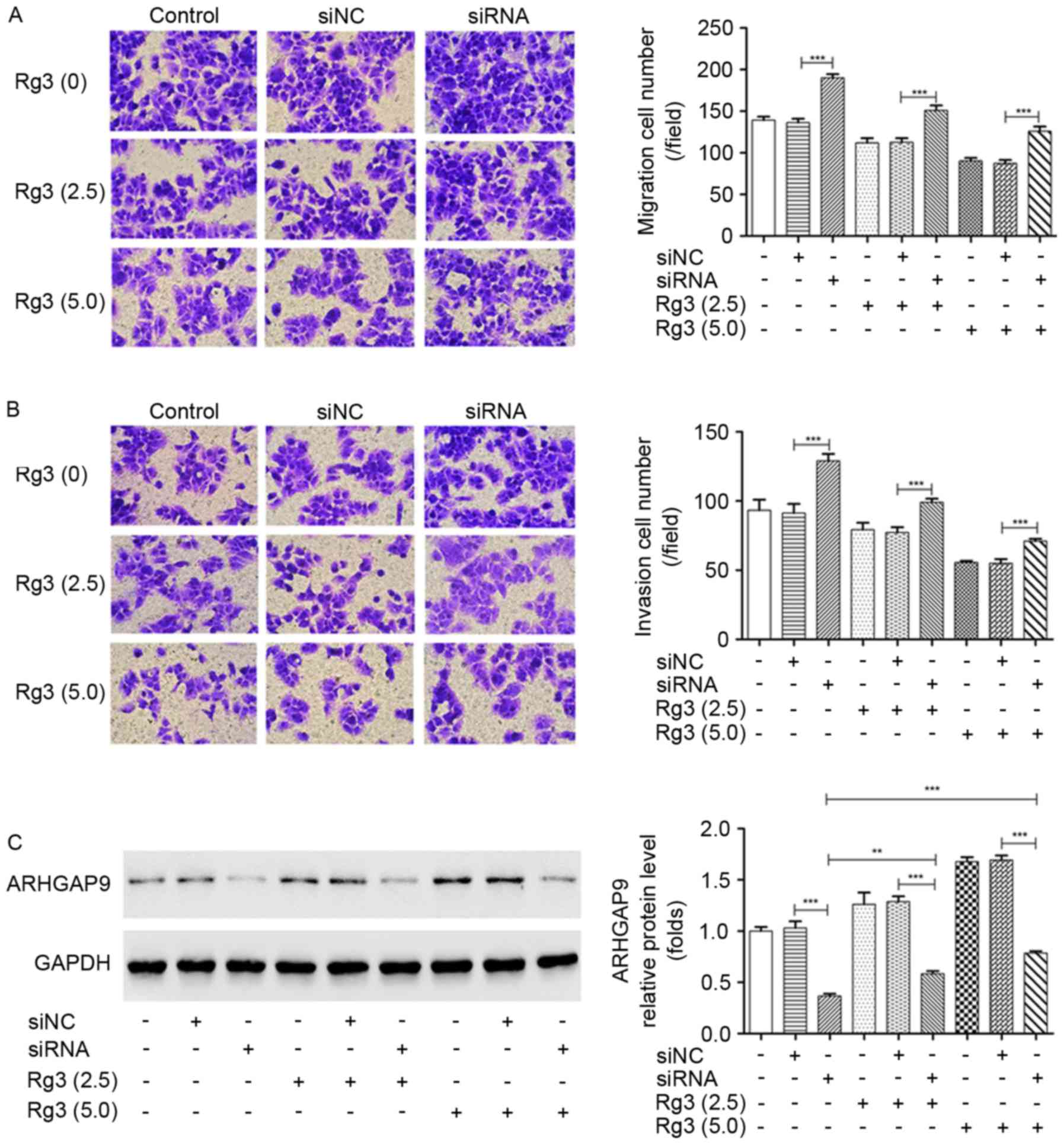
The essential aspects of ARHGAP9 function in cell
metastasis with or without Rg3 treatment were explored by Transwell
assays. It was demonstrated that the transfection of siRNA3
targeting ARHGAP9 significantly increased the migration and
invasion rates of MHCC-97L cells, and attenuated the inhibition of
cell migration and invasion induced by Rg3 (Fig. 6A and B). The increased expression of
ARHGAP9 induced by Rg3 was markedly suppressed by the transfection
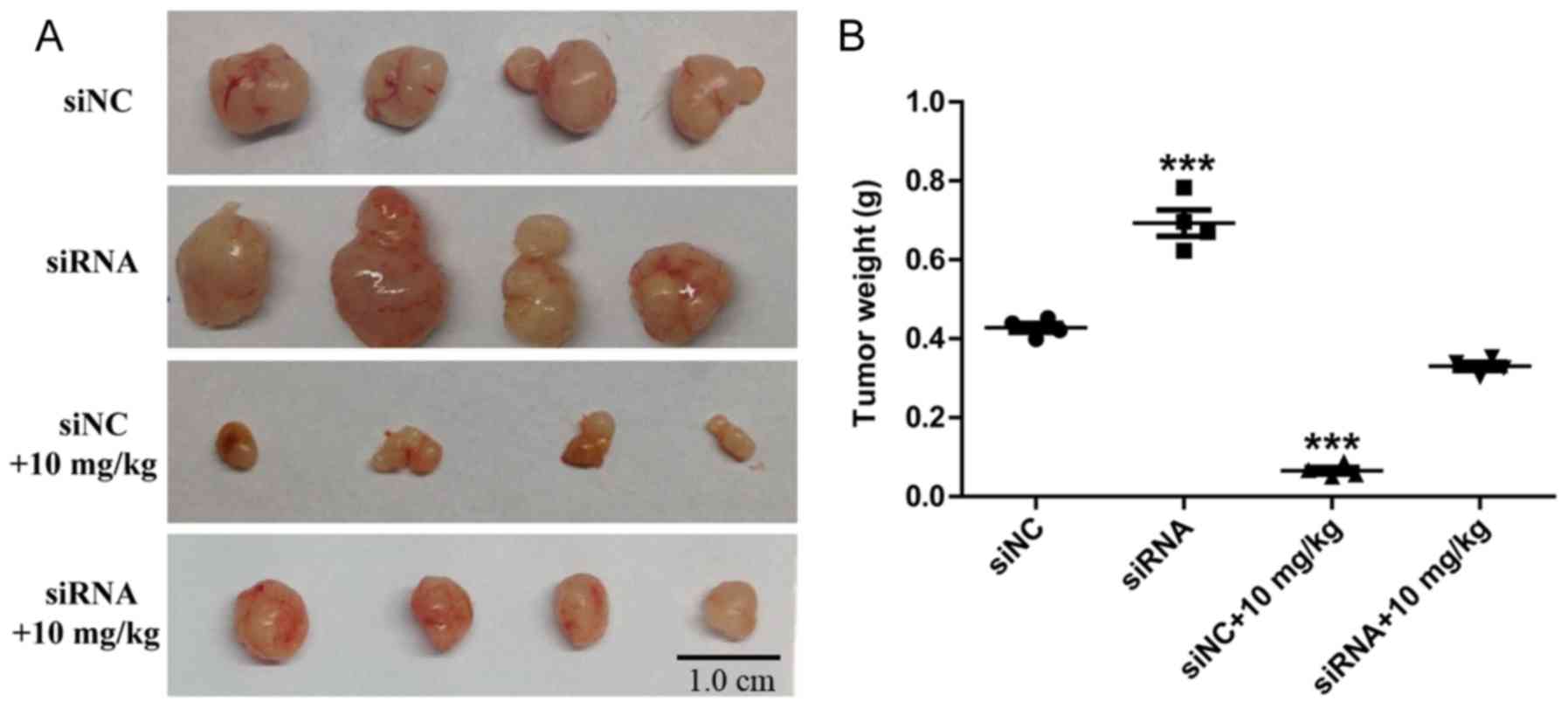
of siRNA3 targeting ARHGAP9 compared with the siNC (Fig. 6C). In vivo, following treatment
with Rg3 for 21 days, the siRNA3 targeting ARHGAP9 (siRNA) group
tumors were larger than those in the siNC group (Fig. 7A). Compared with siNC+10 mg/kg Rg3
group, a significant increase in tumor weight was observed in the
siRNA+10 mg/kg Rg3 group (Fig. 7B),
and the average tumor diameters are listed in Table I. No BALB/c nude mice presented
multiple tumors. The anti-metastasis experiments in vivo
will be the next stage of our work. These results suggest that
ARHGAP9 serves a key role in the metastasis-suppressive function of
Rg3 in liver cancer.
 | Table I.Tumor diameter measurements. |
Table I.
Tumor diameter measurements.
|
| Average diameter of
tumor (cm) |
|---|
|
|
|
|---|
| Time (day) | siNC | siRNA | siNC+10 mg/kg | siRNA+10 mg/kg |
|---|
| 9 | 0.34±0.06 | 0.42±0.06 |
0.08±0.01a,b |
0.22±0.04b |
| 12 | 0.57±0.05 | 0.73±0.08 |
0.12±0.02a,b |
0.34±0.05b |
| 15 | 0.77±0.07 |
0.96±0.09a |
0.21±0.04a,b |
0.47±0.07b |
| 18 | 0.90±0.08 |
1.29±0.10a |
0.30±0.05a,b |
0.53±0.09b |
| 21 | 1.02±0.11 |
1.65±0.14a |
0.39±0.05a |
0.62±0.12b |
Discussion
In the present study, it was demonstrated that Rg3
significantly reduces the migration and invasion capacity of liver
cancer cells. The increased ARHGAP9 expression induced by Rg3
contributes to the anti-migration and anti-invasion effects of Rg3.
The results revealed novel insight into the biological function of
ARHGAP9 and suggest clinical application of Rg3 in liver
cancer.
Ginseng, the root of Panax ginseng C.A.
Meyer, has been widely used in East Asian countries for
thousands years as a natural tonic (30). The triterpene glycosides known as
ginsenosides serve key roles in numerous medicinal effects of
ginseng (31). Over 100 ginsenosides
have been isolated from ginseng (32). Among these ginsenosides, ginsenoside
Rg3, rich in red ginseng, has been demonstrated to be relatively
safe medicinally, with significant anticancer activity in cancer
models both in vitro and in vivo (33).
Despite liver cancer therapy, metastasis remains a
clincial challenge (34). In recent
years, Shenyi capsule (ginsenoside Rg3 monomer preparation) was
approved by the China Food and Drug Administration for cancer
therapy (35). Aside from its
significant anti-cancer effects (36–38), Rg3
is a beneficial adjuvant to conventional cancer therapies due to
its capacity to improve chemosensitivity, reduce adverse effects
and enhance the immunity of tumor-bearing models (39–41). In
the present study, the impact of Rg3 on liver cancer cell survival,
migration and invasion, and growth of tumors in nude mice were
explored. The results of cell viability and Transwell assays
indicated that low concentrations (1.25, 2.5, 5 µg/ml) of Rg3 had
no effect on the viability of HepG2 or MHCC-97L cells, but
significantly inhibited the migration and invasion of tumor cells.
Thus, Rg3 is a potential anti-metastatic agent for liver cancer.
Combined administration of low-concentration Rg3 and conventional
cancer therapies may have efficacy due to synergistic effects.
To further investigate the mechanism of Rg3 in liver
cancer, the expression of ARHGAP9 protein was measured in
vitro. It was demonstrated that Rg3 significantly upregulated
ARHGAP9 expression in a dose-dependent manner. It was also
confirmed that siRNA targeting ARHGAP9 transfection markedly
reduced ARHGAP9 expression at the protein level, increased the
migration and invasion rates of MHCC-97L cells, and attenuated the
anti-migration and anti-invasive effects of Rg3. These findings
suggest that Rg3 suppressed the migration and invasion of liver
cancer cells by upregulating the expression of ARHGAP9.
ARHGAP9 belongs to the Rho GAPase family, which
regulate cell migration in cancer progression by promoting GTP
hydrolysis to inactivate small GTPases through various downstream
signaling pathways (42). Numerous
GAPs have implicated in various types of cancer, including the
tumor suppressor, DLC1, also known as ARHGAP7, which encodes
Rho-GAP deleted frequently as p53 in liver, breast and lung cancer
(43). Bioinformatics studies have
confirmed that ARHGAP family genes (including ARHGAP9) are
cancer-associated genes, as their genetic alterations are
associated with carcinogenesis (23,44).
RhoGAP6 isoform 1 variant was identified as a biomarker for the
development and progression of CRC (45). ARHGAP6 exerts a tumor-suppressive
function via binding to Rac3, and may serve as a potential
biomarker in cervical cancer (46).
ARHGAP10 expression was downregulated in ovarian tumors and
ARHGAP10 has been indicated to ameliorate ovarian cancer by
inhibiting Cdc42 activation (47).
Additionally, ARHGAP24 is implicated in breast cancer cell invasion
and migration, RCC development and malignant lymphoma prognosis
(48–50). Although ARHGAP9 has been associated
with cancer in bioinformatics studies, no experiments have
validated the association between ARHGAP9 and cancer. In the
present study, it was demonstrated that ARHGAP9 exerted suppressive
function in HCC migration and invasion, consistent with previous
studies suggesting that RhoGAPs are tumor suppressors (43,49).
However, the exact mechanism by which ARHGAP9 exerts its function
remains to be further investigated.
It is notable that, although GAPs have been
increasingly reported to be involved in major aspects of cancer
development, the mechanism of GAP regulation and alterations by
GAPs in different types of cancer remains unknown. It has been
suggested that the association of RhoGAPs with cancer could be
mediated by the Rho pathway and Rho-independent pathways. A number
of RhoGAPs have been reported to directly bind to p53 tumor
suppressor protein, and regulate the cell cycle and apoptosis
(51). RhoGAPs may serve as a key
crossroad between the cell-migration and proliferation pathways.
Thus, an improved understanding of the molecular mechanisms of
ARHGAP9 underlying the carcinogenesis of liver cancer is required,
which may allow the development of novel therapeutic
strategies.
In summary, the present study indicated that ARHGAP9
may serve an important role in the metastasis of liver cancer, and
that Rg3 inhibited the migration and invasion of liver cancer by
upregulating the expression of ARHGAP9. ARHGAP9 may provide a novel
strategy for treating liver cancer. The signaling pathway of
Rg3-induced ARHGAP9 and the association between ARHGAP9 expression
and clinicopathological features of patients with liver cancer will
be conducted to fully investigate these possibilities.
Acknowledgements
The authors would like to thank Dr Jianyong Zhu
(Central Laboratory, Seventh People's Hospital of Shanghai
University of Traditional Chinese Medicine, China) for providing
writing advice and improving the present manuscript.
Funding
The present study was supported by funds from the
National Natural Science Foundation of China (grant nos. 81502515,
81503332, 81703755 and 81773941), Outstanding Leaders Training
Program of Pudong Health Bureau of Shanghai (grant no.
PWR12015-05), Key Disciplines Group Construction Project of Pudong
Health Bureau of Shanghai (grant no. PWZxq2014-12 and PWZx2014-17)
and the Talents Training Program of Seventh People's Hospital of
Shanghai University of TCM (grant no. XX2017-01).
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HZ and LJZ conceived and designed the study. MYS
performed the experiments and collected data. YNS, MZ and CYZ
performed the data analysis. All authors discussed and interpreted
the data. MYS wrote the manuscript. All authors reviewed and edited
the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The use of animals in the present study was approved
by the Experimental Animal Ethical Committee of Seven People's
Hospital, Shanghai University of Traditional Chinese Medicine.
Patient consent for publication
Not applicable.
Competing interest
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CCK-8
|
Counting kit-8
|
|
DLC
|
Deleted in liver cancer
|
|
DMEM
|
Dulbecco's modified Eagle medium
|
|
DMSO
|
dimethyl sulfoxide
|
|
FBS
|
fetal bovine serum
|
|
HCC
|
hepatocellular carcinoma
|
|
MAPK
|
Mitogen-activated protein kinases
|
|
MMPs
|
Matrix metalloproteinases
|
|
PA
|
plasminogen activator
|
|
Rho GAPases
|
Rho GTPase activation proteins
|
|
VEGF
|
vascular endothelial growth
factor.
|
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Maluccio M and Covey A: Recent progress in
understanding, diagnosing, and treating hepatocellular carcinoma.
CA Cancer J Clin. 62:394–399. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Han W, Fu X, Xie J, Meng Z, Gu Y, Wang X,
Li L, Pan H and Huang W: MiR-26a enhances autophagy to protect
against ethanol-induced acute liver injury. J Mol Med.
93:1045–1055. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Palmer DH, Hussain SA, Smith AJ,
Hargreaves S, Ma YT, Hull D, Johnson PJ and Ross PJ: Sorafenib for
advanced hepatocellular carcinoma (HCC): Impact of rationing in the
United Kingdom. Br J Cancer. 109:888–890. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tanaka Y, Hanada K, Mizokami M, Yeo AE,
Shih JW, Gojobori T and Alter HJ: A comparison of the molecular
clock of hepatitis C virus in the United States and Japan predicts
that hepatocellular carcinoma incidence in the United States will
increase over the next two decades. Proc Natl Acad Sci USA.
99:15584–15589. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Forner A, Hessheimer AJ, Real Isabel M and
Bruix J: Treatment of hepatocellular carcinoma. Crit Rev Oncol
Hematol. 60:89–98. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sherman M: Hepatocellular carcinoma:
Epidemiology, risk factors, and screening, Seminars in liver
disease. Semin Liver Dis. 25:143–154. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Althoff TF and Offermanns S:
G-protein-mediated signaling in vascular smooth muscle
cells-implications for vascular disease. J Mol Med. 93:973–981.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huh YH, Oh S, Yeo YR, Chae IH, Kim SH, Lee
JS, Yun SJ, Choi KY, Ryu JH and Jun CD: Swiprosin-1 stimulates
cancer invasion and metastasis by increasing the Rho family of
GTPase signaling. Oncotarget. 6:13060–13071. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Deryugina EI and Quigley JP: Matrix
metalloproteinases and tumor metastasis. Cancer Metastasis Rev.
25:9–34. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen CK, Yu WH, Cheng TY, Chen MW, Su CY,
Yang YC, Kuo TC, Lin MT, Huang YC, Hsiao M, et al: Inhibition of
VEGF165/VEGFR2-dependent signaling by LECT2 suppresses
hepatocellular carcinoma angiogenesis. Sci Rep. 6:313982016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lampelj M, Arko D, Cas-Sikosek N, Kavalar
R, Ravnik M, Jezersek-Novakovic B, Dobnik S, Dovnik NF and Takac I:
Urokinase plasminogen activator (uPA) and plasminogen activator
inhibitor type-1 (PAI-1) in breast cancer - correlation with
traditional prognostic factors. Radiol Oncol. 49:357–364. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guo H, Xing Y, Mu A, Li X, Li T, Bian X,
Yang C, Zhang X, Liu Y and Wang X: Correlations between EGFR gene
polymorphisms and pleural metastasis of lung adenocarcinoma. Onco
Targets Ther. 9:5257–5270. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Etienne-Manneville S and Hall A: Rho
GTPases in cell biology. Nature. 420:629–635. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Vega FM and Ridley AJ: Rho GTPases in
cancer cell biology. FEBS Lett. 582:2093–2101. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Haga RB and Ridley AJ: Rho GTPases:
Regulation and roles in cancer cell biology. Small GTPases.
7:207–221. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wong CC, Wong CM, Au SL and Ng IO:
RhoGTPases and Rho-effectors in hepatocellular carcinoma
metastasis: ROCK N' Rho move it. Liver Int. 30:642–656. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Grise F, Bidaud A and Moreau V: Rho
GTPases in hepatocellular carcinoma. Biochim Biophys Acta.
1795:137–151. 2009.PubMed/NCBI
|
|
19
|
Kandpal RP: Rho GTPase activating proteins
in cancer phenotypes. Curr Protein Pept Sci. 7:355–365. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Furukawa Y, Kawasoe T, Daigo Y, Nishiwaki
T, Ishiguro H, Takahashi M, Kitayama J and Nakamura Y: Isolation of
a novel human gene, ARHGAP9, encoding a rho-GTPase activating
protein. Biochem Biophys Res Commun. 284:643–649. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ang BK, Lim CY, Koh SS, Sivakumar N, Taib
S, Lim KB, Ahmed S, Rajagopal G and Ong SH: ArhGAP9, a novel MAP
kinase docking protein, inhibits Erk and p38 activation through WW
domain binding. J Mol Signal. 2:12007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Takefuji M, Asano H, Mori K, Amano M, Kato
K, Watanabe T, Morita Y, Katsumi A, Itoh T, Takenawa T, et al:
Mutation of ARHGAP9 in patients with coronary spastic angina. J Hum
Genet. 55:42–49. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Katoh M and Katoh M: Identification and
characterization of ARHGAP24 and ARHGAP25 genes in silico. Int J
Mol Med. 14:333–338. 2004.PubMed/NCBI
|
|
24
|
Kim YJ, Choi WI, Jeon BN, Choi KC, Kim K,
Kim TJ, Ham J, Jang HJ, Kang KS and Ko H: Stereospecific effects of
ginsenoside 20-Rg3 inhibits TGF-β1-induced epithelial-mesenchymal
transition and suppresses lung cancer migration, invasion and
anoikis resistance. Toxicology. 322:23–33. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tian L, Shen D, Li X, Shan X, Wang X, Yan
Q and Liu J: Ginsenoside Rg3 inhibits epithelial-mesenchymal
transition (EMT) and invasion of lung cancer by down-regulating
FUT4. Oncotarget. 7:1619–1632. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Junmin S, Hongxiang L, Zhen L, Chao Y and
Chaojie W: Ginsenoside Rg3 inhibits colon cancer cell migration by
suppressing nuclear factor kappa B activity. J Tradit Chin Med.
35:440–444. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu TM, Cui MH, Xin Y, Gu LP, Jiang X, Su
MM, Wang DD and Wang WJ: Inhibitory effect of ginsenoside Rg3 on
ovarian cancer metastasis. Chin Med J (Engl). 121:1394–1397.
2008.PubMed/NCBI
|
|
28
|
Chen QJ, Zhang MZ and Wang LX: Gensenoside
Rg3 inhibits hypoxia-induced VEGF expression in human cancer cells.
Cell Physiol Biochem. 26:849–858. 2011. View Article : Google Scholar
|
|
29
|
Lee YS, Kang YS, Lee SH and Kim JA: Role
of NAD(P)H oxidase in the tamoxifen-induced generation of reactive
oxygen species and apoptosis in HepG2 human hepatoblastoma cells.
Cell Death Differ. 7:925–932. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang QH, Wu CF, Duan L and Yang JY:
Protective effects of ginsenoside Rg(3) against
cyclosphosphamide-induced DNA damage and cell apoptosisin mice.
Arch Toxicol. 82:117–123. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kim JH: Pharmacological and medical
applications of Panax ginseng and ginsenosides: A review for use in
cardiovascular diseases. J Ginseng Res. 42:264–269. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lü JM, Yao Q and Chen C: Ginseng
compounds: an update on their molecular mechanisms and medical
applications. Curr Vasc Pharmacol. 7:293–302. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huang JY, Sun Y, Fan QX and Zhang YQ:
Efficacy of Shenyi Capsule combined with gemcitabine plus cisplatin
in treatment of advanced esophageal cancer: A randomized controlled
trial. Zhong Xi Yi Jie He Xue Bao. 7:1047–1051. 2009.(In Chinese).
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Forner A, Llovet JM and Bruix J:
Hepatocellularcarcinoma. Lancet. 379:1245–1255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sun Y, Lin H, Zhu Y, Feng J, Chen Z, Li G,
Zhang X, Zhang Z, Tang J, Shi M, Hao X and Han H: A randomized,
prospective, multi-centre clinical trial of NP regimen
(vinorelbine+cisplatin) plus Gensing Rg3 in the treatment of
advanced non-small cell lung cancer patients. Zhongguo Fei Ai Za
Zhi. 9:254–258. 2006.(In Chinese). PubMed/NCBI
|
|
36
|
Jiang JW, Chen XM, Chen XH and Zheng SS:
Ginsenoside Rg3 inhibit hepatocellular carcinoma growth via
intrinsic apoptotic pathway. World J Gastroenterol. 17:3605–3613.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shan X, Aziz F, Tian LL, Wang XQ, Yan Q
and Liu JW: Ginsenoside Rg3-induced EGFR/MAPK pathway deactivation
inhibits melanoma cell proliferation by decreasing FUT4/LeY
expression. Int J Oncol. 46:1667–1676. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shan X, Fu YS, Aziz F, Wang XQ, Yan Q and
Liu JW: Ginsenoside Rg3 inhibits melanoma cell proliferation
through down-regulation of histone deacetylase 3 (HDAC3) and
increase of p53 acetylation. PLoS One. 9:e1154012014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kim SM, Lee SY, Cho JS, Son SM, Choi SS,
Yun YP, Yoo HS, Yoon DY, Oh KW, Han SB and Hong JT: Combination of
ginsenoside Rg3 with docetaxel enhances the susceptibility of
prostate cancer cells via inhibition of NF-kappaB. Eur J Pharmacol.
631:1–9. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang X, Chen L, Wang T, Jiang X, Zhang H,
Li P, Lv B and Gao X: Ginsenoside Rg3 antagonizes
adriamycin-induced cardiotoxicity by improving endothelial
dysfunction from oxidative stress via upregulating the Nrf2-ARE
pathway through the activation of akt. Phytomedicine. 22:875–884.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Park D, Bae DK, Jeon JH, Lee J, Oh N, Yang
G, Yang YH, Kim TK, Song J, Lee SH, et al: Immunopotentiation and
antitumor effects of a ginsenoside Rg(3)-fortified red ginseng
preparation in mice bearing H460 lung cancer cells. Environ Toxicol
Pharmacol. 31:397–405. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Amin E, Jaiswal M, Derewenda U, Reis K,
Nouri K, Koessmeier KT, Aspenström P, Somlyo AV, Dvorsky R and
Ahmadian MR: Deciphering the molecular and functional Basis of
RHOGAP family proteins A systematic approach toward selective
inactivation of rho family proteINS. J Biol Chem. 291:20353–20371.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tripathi BK, Qian X, Mertins P, Wang D,
Papageorge AG, Carr SA and Lowy DR: CDK5 is a major regulator of
the tumor suppressor DLC1. J Cell Biol. 207:627–642. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Katoh Y and Katoh M: Identification and
characterization of ARHGAP27 gene in silico. Int J Mol Med.
14:943–947. 2004.PubMed/NCBI
|
|
45
|
Guo F, Liu Y, Huang J, Li Y, Zhou G, Wang
D, Li Y, Wang J, Xie P and Li G: Identification of Rho GTPase
activating protein 6 isoform 1 variant as a new molecular marker in
human colorectal tumors. Pathol Oncol Res. 16:319–326. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li J, Liu Y and Yin Y: Inhibitory effects
of Arhgap6 on cervical carcinoma cells. Tumour Biol. 37:1411–1425.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Luo N, Guo J, Chen L, Yang W, Qu X and
Cheng Z: ARHGAP10, downregulated in ovarian cancer, suppresses
tumorigenicity of ovarian cancer cells. Cell Death Dis.
7:e21572016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Saito K, Ozawa Y, Hibino K and Ohta Y:
FilGAP, a Rho/Rho-associated protein kinase-regulated
GTPase-activating protein for Rac, controls tumor cell migration.
Mol Biol Cell. 23:4739–4750. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Xu G, Lu X, Huang T and Fan J: ARHGAP24
inhibits cell cycle progression, induces apoptosis and suppresses
invasion in renal cell carcinoma. Oncotarget. 7:51829–51839;.
2016.PubMed/NCBI
|
|
50
|
Nishi T, Takahashi H, Hashimura M, Yoshida
T, Ohta Y and Saegusa M: FilGAP, a Rac-specific Rho
GTPase-activating protein, is a novel prognostic factor for
follicular lymphoma. Cancer Med. 4:808–818. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Xu J, Zhou X, Wang J, Li Z, Kong X, Qian
J, Hu Y and Fang JY: RhoGAPs attenuate cell proliferation by direct
interaction with p53 tetramerization domain. Cell Rep. 3:1526–1538.
2013. View Article : Google Scholar : PubMed/NCBI
|